Origination of the Protein Fold Repertoire from Oily Pluripotent Peptides
Abstract
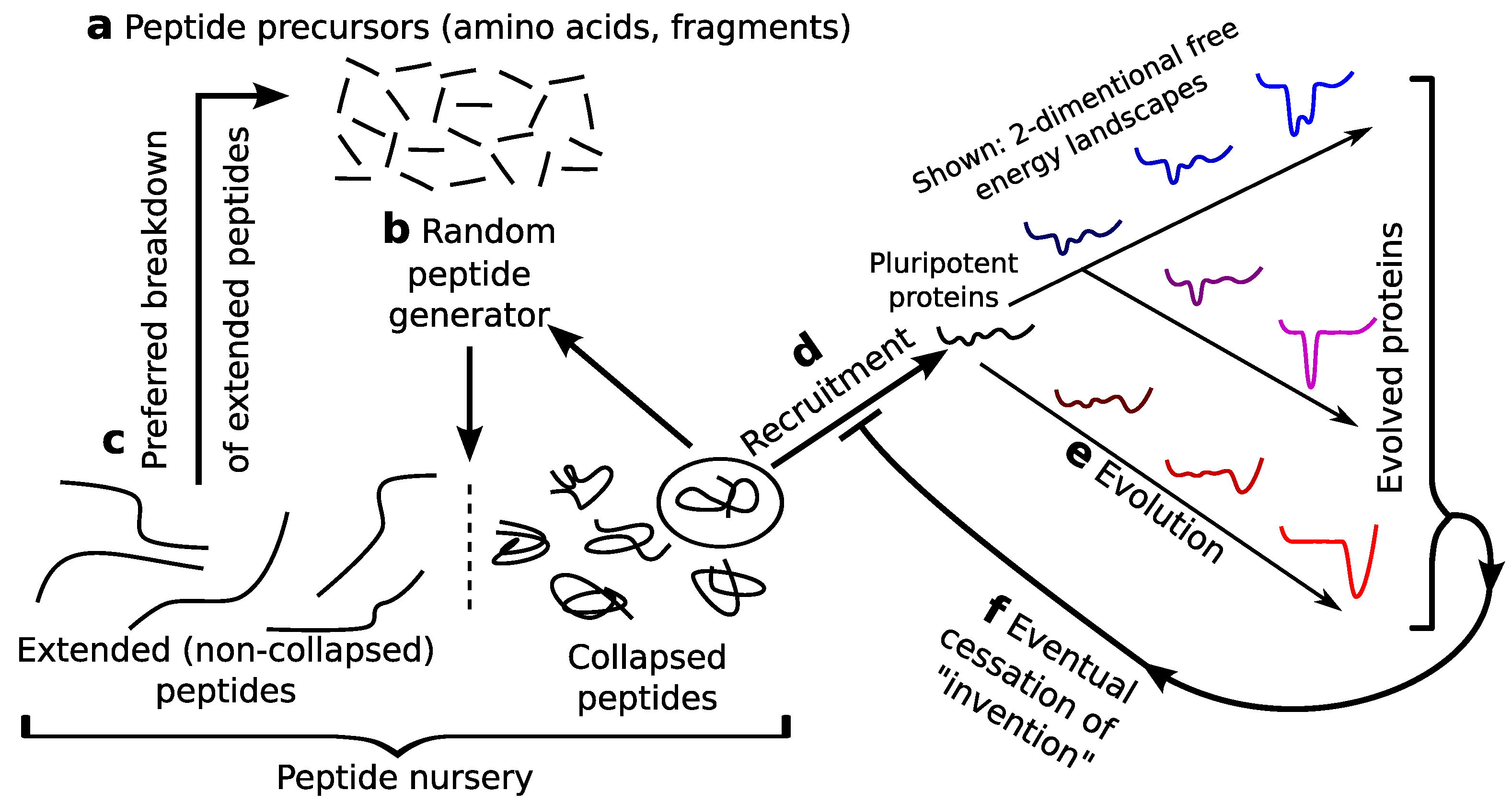
:1. Introduction
“The earliest evolution of the protein repertoire must have involved the ab initio invention of new proteins. At a very low level, this may still take place. But it is clear that [today] the dominant mechanisms for expansion of the protein repertoire, in biology as we now know it, are gene duplication, divergence, and recombination” (Chothia et al., 2003, Science, vol 300 p 1701; italics added).
| BOX 1 |
| Hypothesis: Dynamic, degenerate and collapsed peptides (especially oily peptides) are the best substrates for the origination of protein folds. |
| Evidence can be found in: (i) traces within proteomes [22]; (ii) lattice model simulations [21]; and (iii) diverse reports (reviewed in [23]) on the utility of dynamism in evolvability [24,25,26,27,28,29,30,31,32]. |
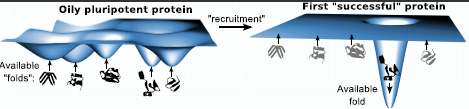
2. Pluripotent Hypothesis
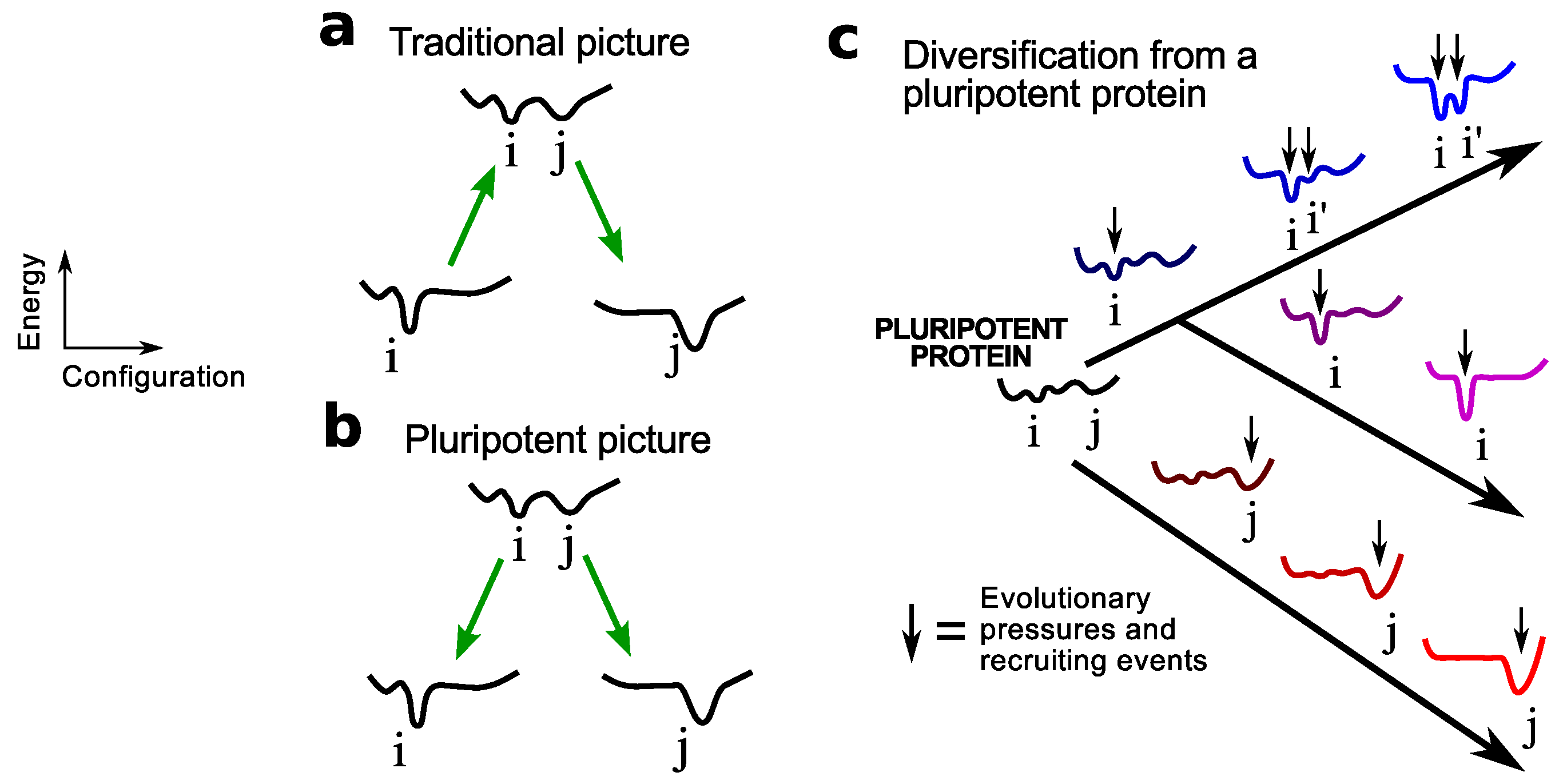
3. Evidence
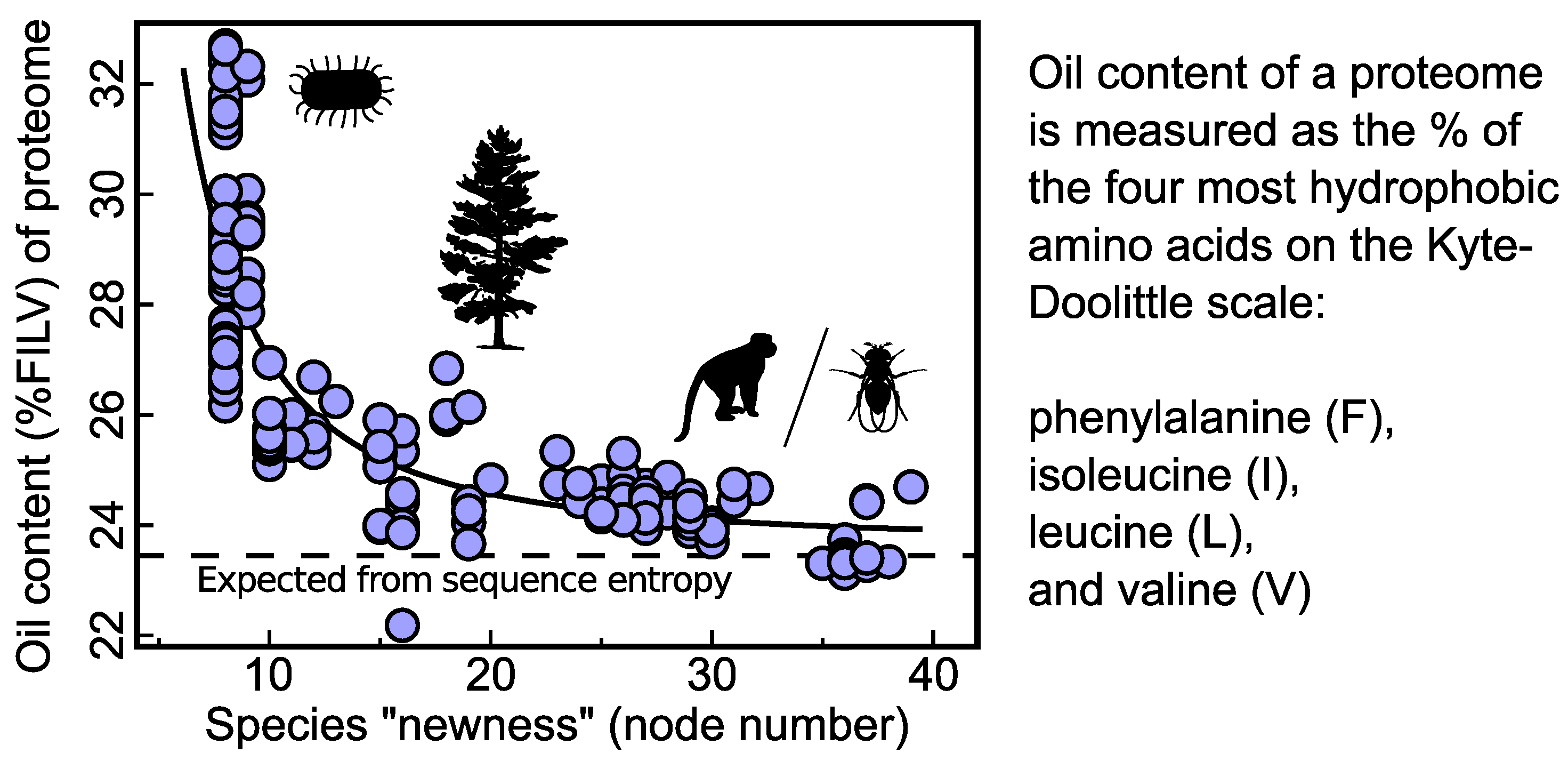
3.1. Evidence of an Oily Ancestor within Today’s Genomes
3.2. Lattice Models Indicate Oily Proteins Are Universally Fold-Evolvable
3.3. Dynamism in Extant Proteins
4. Discussions
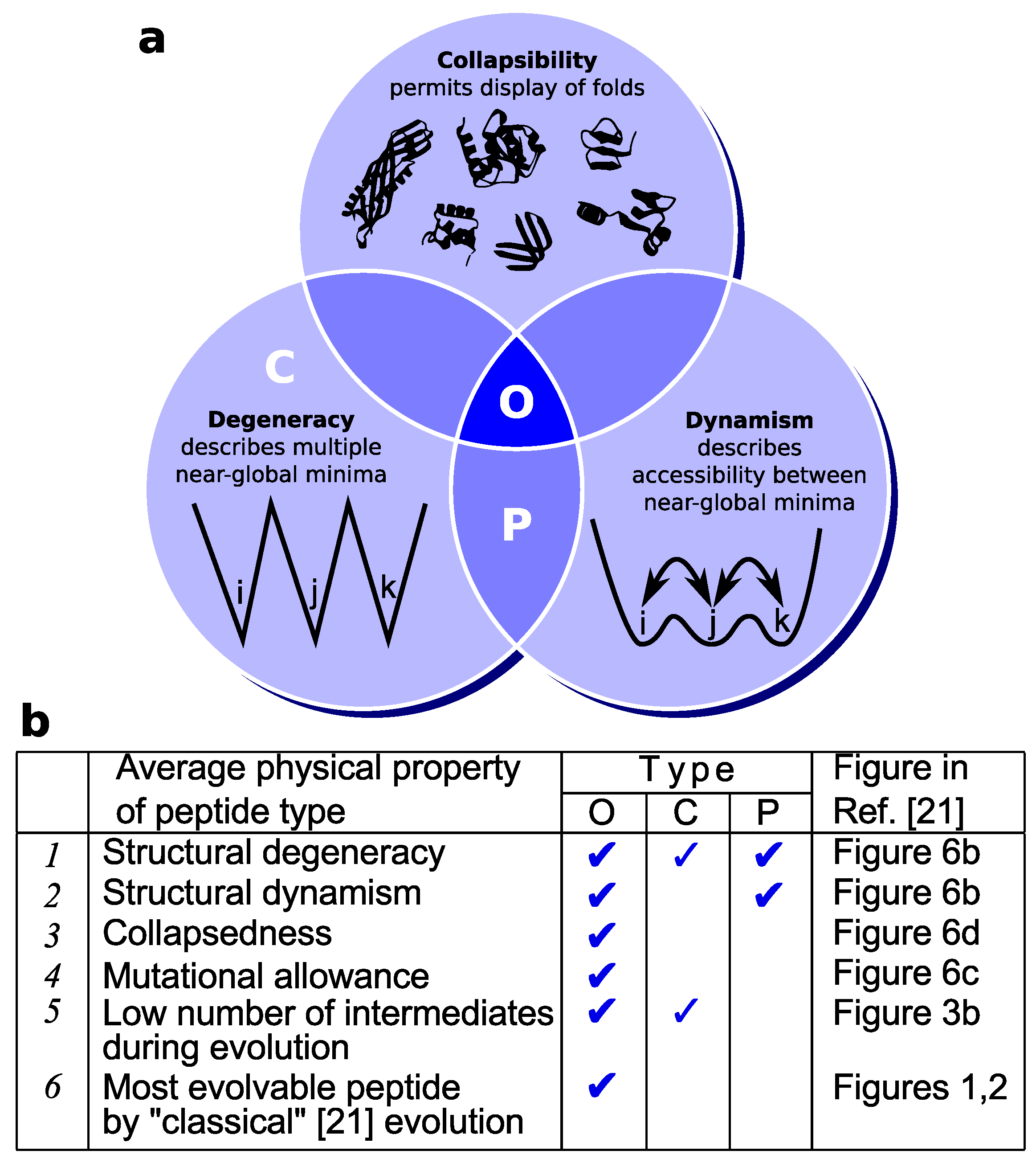
4.1. Different Types of Frustration
4.2. Aggregation
4.3. Expected Extinction of Pluripotent Peptides and Cessation of Mass Invention
4.4. Intrinsically Disordered Proteins Today
4.5. Backbone Hydrogen Bonds Could Discretize Random Collapsed Structures into Proto-Folds
4.6. Detailed Mechanisms
4.7. Importance of Charged and Polar Amino Acids
4.8. Recruiting Events
5. Discussions Regarding Biogenesis
5.1. Pre-Proteome: A Rich Protein Repertoire, Prelife
5.2. Potentially the First Recruiting Event
5.3. An Open Call to Experimentalists
5.4. Ending Remarks
“Life precursors—high molecular compounds, probionts and primitive living entities—repeatedly developed, disintegrated and emerged again in different places and at various times. Therefore, primitive organisms must have co-existed for a long time (probably, for many hundred million years) with simpler representatives of earlier biopoetic stages that had developed in other ‘subvital territories’. This modern concept, assuming multiplicity of the emergence of living beings, rules out entirely the former hypothesis that postulated an accidental emergence of life—the rarest phenomenon which could have happened only once during the whole period of the existence of the Earth”(Oparin, 1976, Origins of Life, vol. 7 p. 3)
Acknowledgments
Conflicts of Interest
References
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, International Edition, 7th ed.; WH Freeman & Co.: New York, NY, USA, 2010. [Google Scholar]
- Cooper, G.M.; Hausman, R.E. The Cell: A Molecular Approach, 6th ed.; Sinauer Associates, Inc.: Sunderland, MA, USA, 2013. [Google Scholar]
- Chothia, C.; Hubbard, T.; Brenner, S.; Barns, H.; Murzin, A. Protein folds in the all-beta and all-alpha classes. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 597–627. [Google Scholar] [CrossRef] [PubMed]
- Anders, E. Pre-biotic organic matter from comets and asteroids. Nature 1989, 342, 255–257. [Google Scholar] [CrossRef] [PubMed]
- Zahnle, K.; Grinspoon, D. Comet dust as a source of amino acids at the Cretaceous/Tertiary boundary. Nature 1990, 348, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Oró, J.; Miller, S.L.; Lazcano, A. The origin and early evolution of life on Earth. Annu. Rev. Earth Planet. Sci. 1990, 18, 317–356. [Google Scholar] [CrossRef] [PubMed]
- Glavin, D.; Dworkin, J.; Sandford, S. Detection of cometary amines in samples returned by Stardust. Meteorit. Planet. Sci. 2008, 43, 399–413. [Google Scholar] [CrossRef]
- Miller, S.L. A production of amino acids under possible primitive earth conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef] [PubMed]
- Matthews, C.N.; Moser, R.E. Peptide synthesis from hydrogen cyanide and water. Nature 1967, 215, 1230–1234. [Google Scholar] [CrossRef] [PubMed]
- Plankensteiner, K.; Reiner, H.; Rode, B.M. Catalytically increased prebiotic peptide formation: Ditryptophan, dilysine, and diserine. Orig. Life Evol. Biosph. 2005, 35, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Wachtershauser, G. On the chemistry and evolution of the pioneer organism. Chem. Biodivers. 2007, 4, 584–602. [Google Scholar] [CrossRef] [PubMed]
- Leman, L.; Orgel, L.; Ghadiri, M.R. Carbonyl sulfide-mediated prebiotic formation of peptides. Science 2004, 306, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Chothia, C.; Gough, J.; Vogel, C.; Teichmann, S. Evolution of the Protein Repertoire. Science 2003, 300, 1701–1703. [Google Scholar] [CrossRef] [PubMed]
- Blanck, A.; Oesterhelt, D.; Ferrando, E.; Schegk, E.S.; Lottspeich, F. Primary structure of sensory rhodopsin I, a prokaryotic photoreceptor. EMBO J. 1989, 8, 3963–3971. [Google Scholar] [PubMed]
- Gough, J. Convergent evolution of domain architectures (is rare). Bioinformatics 2005, 21, 1464–1471. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Aravind, L.; Kondrashov, A.S. The impact of comparative genomics on our understanding of evolution. Cell 2000, 101, 573–576. [Google Scholar] [CrossRef]
- Kurland, C.G.; Canback, B.; Berg, O.G. Horizontal gene transfer: A critical view. Proc. Natl. Acad. Sci. USA 2003, 100, 9658–9662. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F. Evolution and tinkering. Science 1977, 196, 1161. [Google Scholar] [CrossRef] [PubMed]
- Abkevich, V.I.; Gutin, A.M.; Shakhnovich, E.I. How the first biopolymers could have evolved. Proc. Natl. Acad. Sci. USA 1996, 93, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Wang, M.; Caetano-Anollés, D.; Mittenthal, J.E. The origin, evolution and structure of the protein world. Biochem. J. 2009, 417, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Mannige, R.V. Two regimes of protein evolution and their unique dependencies on sequence composition. Phys. Rev. E 2013, 87, e062714. [Google Scholar] [CrossRef]
- Mannige, R.V.; Brooks, C.L., III; Shakhnovich, E.I. A universal trend among proteomes indicates an oily last common ancestor. PLoS Comput. Biol. 2012, 8, e1002839. [Google Scholar] [CrossRef] [PubMed]
- Mannige, R. Dynamic new world: Refining our view of protein structure, function and evolution. Proteomes 2014, 2, 128–153. [Google Scholar] [CrossRef]
- James, L.C.; Roversi, P.; Tawfik, D.S. Antibody multispecificity mediated by conformational diversity. Science 2003, 299, 1362–1367. [Google Scholar] [CrossRef] [PubMed]
- James, L.C.; Tawfik, D.S. Conformational diversity and protein evolution–a 60-year-old hypothesis revisited. Trends Biochem. Sci. 2003, 28, 361–368. [Google Scholar] [CrossRef]
- Tokuriki, N.; Tawfik, D.S. Protein dynamism and evolvability. Science 2009, 324, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Soskine, M.; Tawfik, D.S. Mutational effects and the evolution of new protein functions. Nat. Genet. 2010, 11, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Yadid, I.; Kirshenbaum, N.; Sharon, M.; Dym, O.; Tawfik, D.S. Metamorphic proteins mediate evolutionary transitions of structure. Proc. Natl. Acad. Sci. USA 2010, 107, 7287–7292. [Google Scholar] [CrossRef] [PubMed]
- Pauling, L. A Theory of the Structure and Process of Formation of Antibodies. J. Am. Chem. Soc. 1940, 62, 2643–2657. [Google Scholar] [CrossRef]
- Landsteiner, K. The Specificity of Serological Reactions; Dover Publications (reprinted 1962): Mineola, NY, USA, 1936. [Google Scholar]
- Zimmermann, J.; Oakman, E.L.; Thorpe, I.F.; Shi, X.; Abbyad, P.; Brooks, C.L., III; Boxer, S.G.; Romesberg, F.E. Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc. Natl. Acad. Sci. USA 2006, 103, 13722–13727. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, I.F.; Brooks, C.L., III. Molecular evolution of affinity and flexibility in the immune system. Proc. Natl. Acad. Sci. USA 2007, 104, 8821–8826. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976, 30, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.A.; Gu, W. Evolutionary recruitment of biochemically specialized subdivisions of Family I within the protein superfamily of aminotransferases. J. Bacteriol. 1996, 178, e2161. [Google Scholar]
- Bentley, S.D.; Parkhill, J. Comparative genomic structure of prokaryotes. Annu. Rev. Genet. 2004, 38, 771–792. [Google Scholar] [CrossRef] [PubMed]
- Jordan, I.K.; Kondrashov, F.A.; Adzhubei, I.A.; Wolf, Y.I.; Koonin, E.V.; Kondrashov, A.S.; Sunyaev, S. A universal trend of amino acid gain and loss in protein evolution. Nature 2005, 433, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Zeldovich, K.B.; Berezovsky, I.N.; Shakhnovich, E.I. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput. Biol. 2007, 3, e5. [Google Scholar] [CrossRef] [PubMed]
- Note that affinity maturation is only used as an example to indicate the importance of a transition from a dynamic to rigid state; the actual mode of maturation is far too specialized to be generalized as a relevant mechanism of evolution.
- Bryngelson, J.D.; Wolynes, P.G. Spin glasses and the statistical mechanics of protein folding. Proc. Natl. Acad. Sci. USA 1987, 84, 7524–7528. [Google Scholar] [CrossRef] [PubMed]
- Onuchic, J.N.; Luthey-Schulten, Z.; Wolynes, P.G. Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 1997, 48, 545–600. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J. Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 2001, 26, 597–604. [Google Scholar] [CrossRef]
- Hanczyc, M.M.; Toyota, T.; Ikegami, T.; Packard, N.; Sugawara, T. Fatty acid chemistry at the oil-water interface: Self-propelled oil droplets. J. Am. Chem. Soc. 2007, 129, 9386–9391. [Google Scholar] [CrossRef] [PubMed]
- Toyota, T.; Maru, N.; Hanczyc, M.M.; Ikegami, T.; Sugawara, T. Self-propelled oil droplets consuming “fuel” surfactant. J. Am. Chem. Soc. 2009, 131, 5012–5013. [Google Scholar] [CrossRef] [PubMed]
- Horibe, N.; Hanczyc, M.; Ikegami, T. Mode Switching and Collective Behavior in Chemical Oil Droplets. Entropy 2011, 13, 709–719. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Babu, M.M.; Barbar, E.; Blackledge, M.; Bondos, S.E.; Dosztányi, Z.; Dyson, H.J.; Forman-Kay, J.; Fuxreiter, M.; Gsponer, J.; et al. What’s in a name? Why these proteins are intrinsically disordered? Intrinsically Disord. Proteins 2013, 1, e24157. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Schad, E.; Tompa, P.; Hegyi, H. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 2011, 12, R120. [Google Scholar] [CrossRef] [PubMed]
- Pauling, L.; Corey, R.B.; Branson, H.R. The structure of proteins: Two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Pizzarello, S.; Schrader, D.L.; Monroe, A.A.; Lauretta, D.S. Large enantiomeric excesses in primitive meteorites and the diverse effects of water in cosmochemical evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 11949–11954. [Google Scholar] [CrossRef] [PubMed]
- Longo, L.M.; Lee, J.; Blaber, M. Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein. Proc. Natl. Acad. Sci. USA 2013, 110, 2135–2139. [Google Scholar] [CrossRef] [PubMed]
- Trifonov, E.N. Consensus temporal order of amino acids and evolution of the triplet code. Gene 2000, 261, 139–151. [Google Scholar] [CrossRef]
- Trifonov, E.N. The triplet code from first principles. J. Biomol. Struct. Dyn. 2004, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bar-Nun, A.; Kochavi, E.; Bar-Nun, S. Assemblies of Free Amino Acids as Possible Prebiotic Catalysts. J. Mol. Evol. 1994, 39, 116–122. [Google Scholar]
- Mirny, L.A.; Abkevich, V.I.; Shakhnovich, E.I. How evolution makes proteins fold quickly. Proc. Natl. Acad. Sci. USA 1998, 95, 4976–4981. [Google Scholar] [CrossRef] [PubMed]
- Mirny, L.A.; Shakhnovich, E.I. Universally conserved positions in protein folds: Reading evolutionary signals about stability, folding kinetics and function. J. Mol. Biol. 1999, 291, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Mirny, L.; Shakhnovich, E. Evolutionary conservation of the folding nucleus. J. Mol. Biol. 2001, 308, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.F.; Kong, D.X.; Shen, L.; Chen, L.L.; Ma, B.G.; Zhang, H.Y. Distribution patterns of small-molecule ligands in the protein universe and implications for origin of life and drug discovery. Genome Biol. 2007, 8, R176. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.F.; Chen, L.; Jiang, Y.Y.; Zhang, H.Y. Evolutionary formation of new protein folds is linked to metallic cofactor recruitment. Bioessays 2009, 31, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Oparin, A.I. Proiskhozhdenie zhizny [Origin of life], Izd; Moskovski Rabochii: Moscow, USSR, 1924. Translated in The Origin of Life; Bernal, J.D., Ed.; World: Cleveland, OH, USA, 1967. [Google Scholar]
- Oparin, A.I. Evolution of the concepts of the origin of life, 1924–1974. Orig. Life 1976, 7, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Cairns-Smith, A.G. The origin of life and the nature of the primitive gene. J. Theor. Biol. 1966, 10, 53–88. [Google Scholar] [CrossRef]
- King, G.A. Symbiosis and the origin of life. Orig. Life 1977, 8, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, S.A. Autocatalytic sets of proteins. J. Theor. Biol. 1986, 119, 1–24. [Google Scholar] [CrossRef]
- Eigen, M. Molecular self-organization and the early stages of evolution. Experientia 1971, 27, 149–212. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E. The origin of life on the earth. Sci. Am. 1994, 271, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Lazcano, A.; Miller, S.L. The origin and early evolution of life: Prebiotic chemistry, the pre-RNA world, and time. Cell 1996, 85, 793–798. [Google Scholar] [CrossRef]
- Abkevich, V.I.; Gutin, A.M.; Shakhnovich, E.I. Computer simulations of prebiotic evolution. Pac. Symp. Biocomput. 1997, 97, 27–38. [Google Scholar]
- Marahiel, M.A.; Stachelhaus, T.; Mootz, H.D. Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem. Rev. 1997, 97, 2651–2674. [Google Scholar] [CrossRef] [PubMed]
- Lautru, S.; Challis, G.L. Substrate recognition by nonribosomal peptide synthetase multi-enzymes. Microbiology 2004, 150, 1629–1636. [Google Scholar] [CrossRef] [PubMed]
- Straight, P.D.; Fischbach, M.A.; Walsh, C.T.; Rudner, D.Z.; Kolter, R. A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2007, 104, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Strieker, M.; Tanović, A.; Marahiel, M.A. Nonribosomal peptide synthetases: Structures and dynamics. Curr. Opin. Struct. Biol. 2010, 20, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Szostak, J.W. Functional proteins from a random-sequence library. Nature 2001, 410, 715–718. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mannige, R.V. Origination of the Protein Fold Repertoire from Oily Pluripotent Peptides. Proteomes 2014, 2, 154-168. https://doi.org/10.3390/proteomes2020154
Mannige RV. Origination of the Protein Fold Repertoire from Oily Pluripotent Peptides. Proteomes. 2014; 2(2):154-168. https://doi.org/10.3390/proteomes2020154
Chicago/Turabian StyleMannige, Ranjan V. 2014. "Origination of the Protein Fold Repertoire from Oily Pluripotent Peptides" Proteomes 2, no. 2: 154-168. https://doi.org/10.3390/proteomes2020154