Plant Polygalacturonases Involved in Cell Elongation and Separation—The Same but Different?
Abstract
:1. Introduction
2. Polygalacturonases
3. Polygalacturonases Involved in Fruit Ripening and Cell Separation
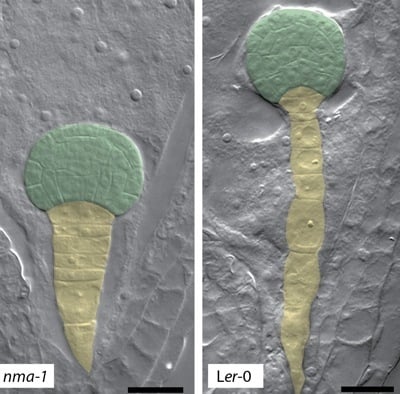
4. Polygalacturonases Involved in Cell Expansion


5. Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Forouzesh, E.; Goel, A.; Mackenzie, S.A.; Turner, J.A. In vivo extraction of Arabidopsis cell turgor pressure using nanoindentation in conjunction with finite element modeling. Plant J. 2013, 73, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.H. Regulation of cell wall biosynthesis. Curr. Opin. Plant Biol. 2007, 10, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Mcneil, M.; Darvill, A.G.; Fry, S.C.; Albersheim, P. Structure and function of the primary-cell walls of plants. Annu. Rev. Biochem. 1984, 53, 625–663. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Langan, P.; Chanzy, H. Crystal structure and hydrogen-bonding system in cellulose Ibeta from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 2002, 124, 9074–9082. [Google Scholar] [CrossRef] [PubMed]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Dick-Perez, M.; Zhang, Y.; Hayes, J.; Salazar, A.; Zabotina, O.A.; Hong, M. Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 2011, 50, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol. 2012, 158, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Braybrook, S.; Hofte, H. Cell wall mechanics and growth control in plants: The role of pectins revisited. Front. Plant Sci. 2012, 3, 121. [Google Scholar] [CrossRef] [PubMed]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D. Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef] [PubMed]
- Atmodjo, M.A.; Hao, Z.; Mohnen, D. Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 2013, 64, 747–779. [Google Scholar] [CrossRef] [PubMed]
- Senechal, F.; Wattier, C.; Rusterucci, C.; Pelloux, J. Homogalacturonan-modifying enzymes: Structure, expression, and roles in plants. J. Exp. Bot. 2014, 65, 5125–5160. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Louvet, R.; Johansen, J.N.; Hofte, H.; Laufs, P.; Pelloux, J.; Mouille, G. Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr. Biol. 2008, 18, 1943–1948. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Braybrook, S.A.; le Guillou, L.; Bron, E.; Kuhlemeier, C.; Hofte, H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 2011, 21, 1720–1726. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Greiner, S. Growth control by cell wall pectins. Protoplasma 2012, 249, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Mouille, G.; Pelloux, J. Homogalacturonan methyl-esterification and plant development. Mol. Plant 2009, 2, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Willats, W.G.; McCartney, L.; Mackie, W.; Knox, J.P. Pectin: Cell biology and prospects for functional analysis. Plant Mol. Biol. 2001, 47, 9–27. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, K.A.; Bennett, A.B. Polygalacturonases: Many genes in search of a function. Plant Physiol. 1998, 117, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Torki, M.; Mandaron, P.; Mache, R.; Falconet, D. Characterization of a ubiquitous expressed gene family encoding polygalacturonase in Arabidopsis thaliana. Gene 2000, 242, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Park, K.C.; Kwon, S.J.; Kim, P.H.; Bureau, T.; Kim, N.S. Gene structure dynamics and divergence of the polygalacturonase gene family of plants and fungus. Genome 2008, 51, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Shiu, S.H.; Thoma, S.; Li, W.H.; Patterson, S.E. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006, 7, R87. [Google Scholar] [CrossRef] [PubMed]
- Park, K.C.; Kwon, S.J.; Kim, N.S. Intron loss mediated structural dynamics and functional differentiation of the polygalacturonase gene family in land plants. Genes Genomics 2010, 32, 570–577. [Google Scholar] [CrossRef]
- Markovic, O.; Janecek, S. Pectin degrading glycoside hydrolases of family 28: Sequence-structural features, specificities and evolution. Protein Eng. 2001, 14, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Protsenko, M.A.; Buza, N.L.; Krinitsyna, A.A.; Bulantseva, E.A.; Korableva, N.P. Polygalacturonase-inhibiting protein is a structural component of plant cell wall. Biochemistry (Mosc.) 2008, 73, 1053–1062. [Google Scholar] [CrossRef]
- Clausen, M.H.; Willats, W.G.T.; Knox, J.P. Synthetic methyl hexagalacturonate hapten inhibitors of antihomogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 2003, 338, 1797–1800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Pierce, M.L.; Mort, A.J. Changes in homogalacturonans and enzymes degrading them during cotton cotyledon expansion. Phytochemistry 2007, 68, 1094–1103. [Google Scholar] [CrossRef] [PubMed]
- Abbott, D.W.; Boraston, A.B. The structural basis for exopolygalacturonase activity in a family 28 glycoside hydrolase. J. Mol. Biol. 2007, 368, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Pressey, R.; Hinton, D.M.; Avants, J.K. Development of Polygalacturonase Activity and Solubilization of Pectin in Peaches during Ripening. J. Food Sci. 1971, 36, 1070–1073. [Google Scholar] [CrossRef]
- Smith, C.J.; Watson, C.F.; Morris, P.C.; Bird, C.R.; Seymour, G.B.; Gray, J.E.; Arnold, C.; Tucker, G.A.; Schuch, W.; Harding, S.; et al. Inheritance and effect on ripening of antisense polygalacturonase genes in transgenic tomatoes. Plant Mol. Biol. 1990, 14, 369–379. [Google Scholar] [CrossRef]
- Smith, C.J.S.; Watson, C.F.; Ray, J.; Bird, C.R.; Morris, P.C.; Schuch, W.; Grierson, D. Antisense RNA inhibition of polygalacturonase gene-expression in transgenic tomatoes. Nature 1988, 334, 724–726. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Sutherland, P.W.; Johnston, S.L.; Gunaseelan, K.; Hallett, I.C.; Mitra, D.; Brummell, D.A.; Schroder, R.; Johnston, J.W.; Schaffer, R.J. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol. 2012, 12, 129. [Google Scholar] [CrossRef] [PubMed]
- Pose, S.; Paniagua, C.; Cifuentes, M.; Blanco-Portales, R.; Quesada, M.A.; Mercado, J.A. Insights into the effects of polygalacturonase FaPG1 gene silencing on pectin matrix disassembly, enhanced tissue integrity, and firmness in ripe strawberry fruits. J. Exp. Bot. 2013, 64, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- Quesada, M.A.; Blanco-Portales, R.; Pose, S.; Garcia-Gago, J.A.; Jimenez-Bermudez, S.; Munoz-Serrano, A.; Caballero, J.L.; Pliego-Alfaro, F.; Mercado, J.A.; Munoz-Blanco, J. Antisense down-regulation of the FaPG1 gene reveals an unexpected central role for polygalacturonase in strawberry fruit softening. Plant Physiol. 2009, 150, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Tucker, G.A.; Schindler, C.B.; Roberts, J.A. Flower abscission in mutant tomato plants. Planta 1984, 160, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Carranza, Z.H.; Elliott, K.A.; Roberts, J.A. Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 3719–3730. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Sander, L.; Child, R.; van Onckelen, H.; Ulvskov, P.; Borkhardt, B. Isolation and characterisation of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol. Biol. 1996, 31, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, R.G.; Schroder, R.; Hallett, I.C.; Cohen, D.; MacRae, E.A. Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 2002, 129, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ma, Y.; Chen, N.; Guo, S.; Liu, H.; Guo, X.; Chong, K.; Xu, Y. Overexpression of stress-inducible OsBURP16, the beta subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environ. 2014, 37, 1144–1158. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Osborne, E.; Poindexter, P.D.; Somerville, C.R. Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 2003, 133, 1170–1180. [Google Scholar] [PubMed]
- Ogawa, M.; Kay, P.; Wilson, S.; Swain, S.M. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 2009, 21, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Somerville, C.; Anderson, C.T. POLYGALACTURONASE INVOLVED IN EXPANSION1 functions in cell elongation and flower development in Arabidopsis. Plant Cell 2014, 26, 1018–1035. [Google Scholar] [CrossRef] [PubMed]
- Sitrit, Y.; Hadfield, K.A.; Bennett, A.B.; Bradford, K.J.; Downie, A.B. Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol. 1999, 121, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Riov, J. A polygalacturonase from citrus leaf explants: role in abscission. Plant Physiol. 1974, 53, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Cao, J. The pectin lyases in Arabidopsis thaliana: Evolution, selection and expression profiles. PLoS One 2012, 7, e46944. [Google Scholar] [CrossRef] [PubMed]
- Babu, Y.; Musielak, T.; Henschen, A.; Bayer, M. Suspensor length determines developmental progression of the embryo in Arabidopsis. Plant Physiology 2013, 162, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Irshad, M.; Canut, H.; Borderies, G.; Pont-Lezica, R.; Jamet, E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 2008, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Le, B.H.; Cheng, C.; Bui, A.Q.; Wagmaister, J.A.; Henry, K.F.; Pelletier, J.; Kwong, L.; Belmonte, M.; Kirkbride, R.; Horvath, S.; et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 8063–8070. [Google Scholar] [CrossRef] [PubMed]
- Kalaitzis, P.; Solomos, T.; Tucker, M.L. Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol. 1997, 113, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a systems approach to understanding plant cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- Burgert, I.; Keplinger, T. Plant micro- and nanomechanics: Experimental techniques for plant cell-wall analysis. J. Exp. Bot. 2013, 64, 4635–4649. [Google Scholar] [CrossRef] [PubMed]
- Knox, J.P.; Linstead, P.J.; King, J.; Cooper, C.; Roberts, K. Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 1990, 181, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babu, Y.; Bayer, M. Plant Polygalacturonases Involved in Cell Elongation and Separation—The Same but Different? Plants 2014, 3, 613-623. https://doi.org/10.3390/plants3040613
Babu Y, Bayer M. Plant Polygalacturonases Involved in Cell Elongation and Separation—The Same but Different? Plants. 2014; 3(4):613-623. https://doi.org/10.3390/plants3040613
Chicago/Turabian StyleBabu, Yashodar, and Martin Bayer. 2014. "Plant Polygalacturonases Involved in Cell Elongation and Separation—The Same but Different?" Plants 3, no. 4: 613-623. https://doi.org/10.3390/plants3040613