Redox Modulation Matters: Emerging Functions for Glutaredoxins in Plant Development and Stress Responses
Abstract
:1. Introduction

| Species | CGFS-Type | CPYC-Type | CC-Type |
|---|---|---|---|
| Chlamydomonas reinhardtii (green alga) | 4 | 2 | 0 |
| Synechocystis sp. PCC6803 (cyanobacteria) | 1 | 2 | 0 |
| Physcomitrella patens (moss) | 6 | 4 | 2 |
| Pinus taeda (loblolly pine) | 8 | 4 | 5 |
| Oryza sativa (rice) | 5 | 5 | 17 |
| Populus trichocarpa (black cottonwood) | 6 | 5 | 22 |
| Arabidopsis thaliana (Arabidopsis) | 4 | 6 | 21 |
2. GRXs with a CGYC or CGFS Active Site Act in the Assembly of Iron-Sulfur Clusters
3. Land Plant-Specific CC-Type GRXs in Flower Development and Defense Responses
3.1. CC-Type GRXs Are Required for Floral Organ Development and Microspore Formation
3.2. CC-Type GRXs Participate in Disease Resistance
3.3. Functional Redundancy of a Subset of CC-Type GRXs with a Conserved C-Terminal Motif
4. GRXs Participate in Abiotic Stress Responses
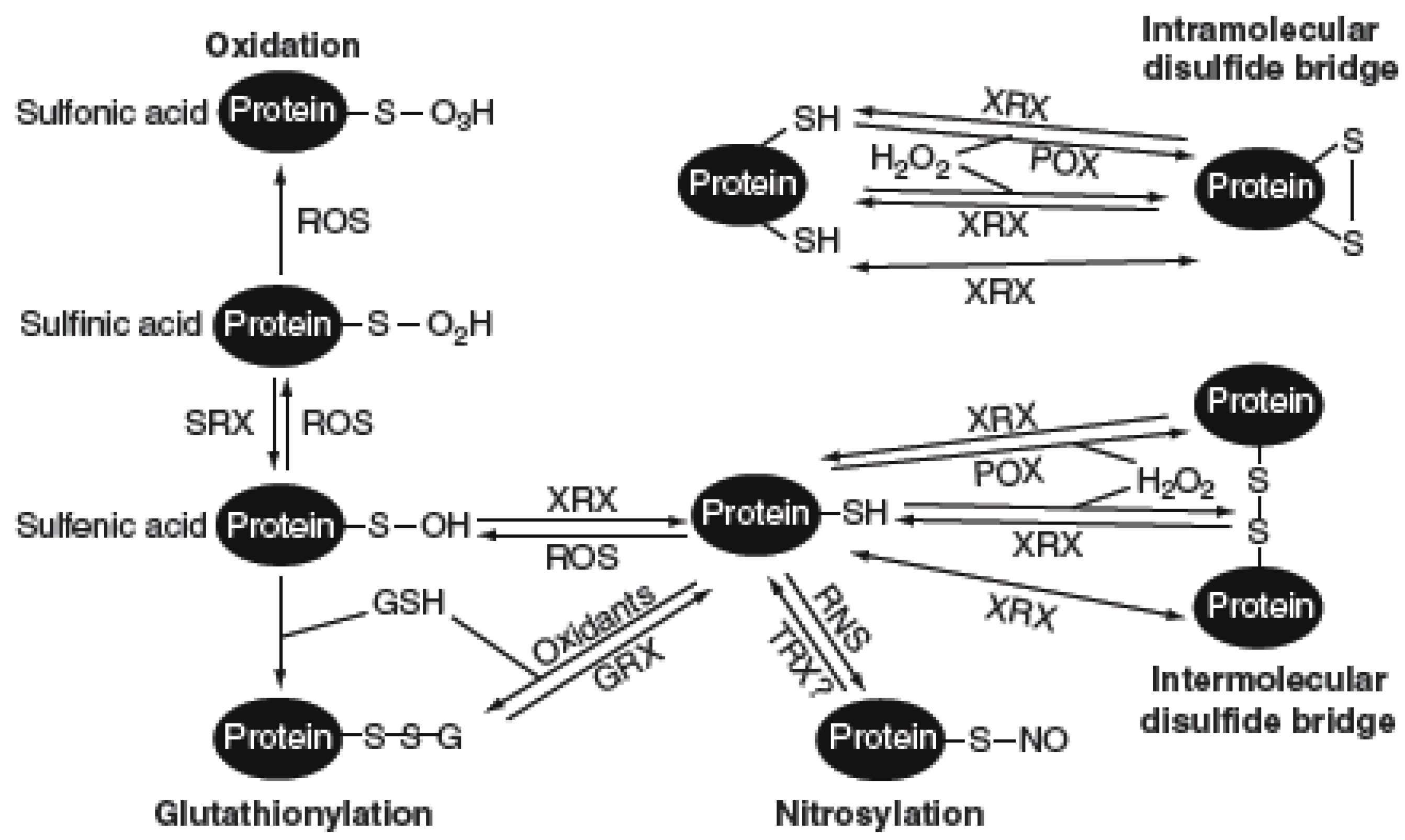
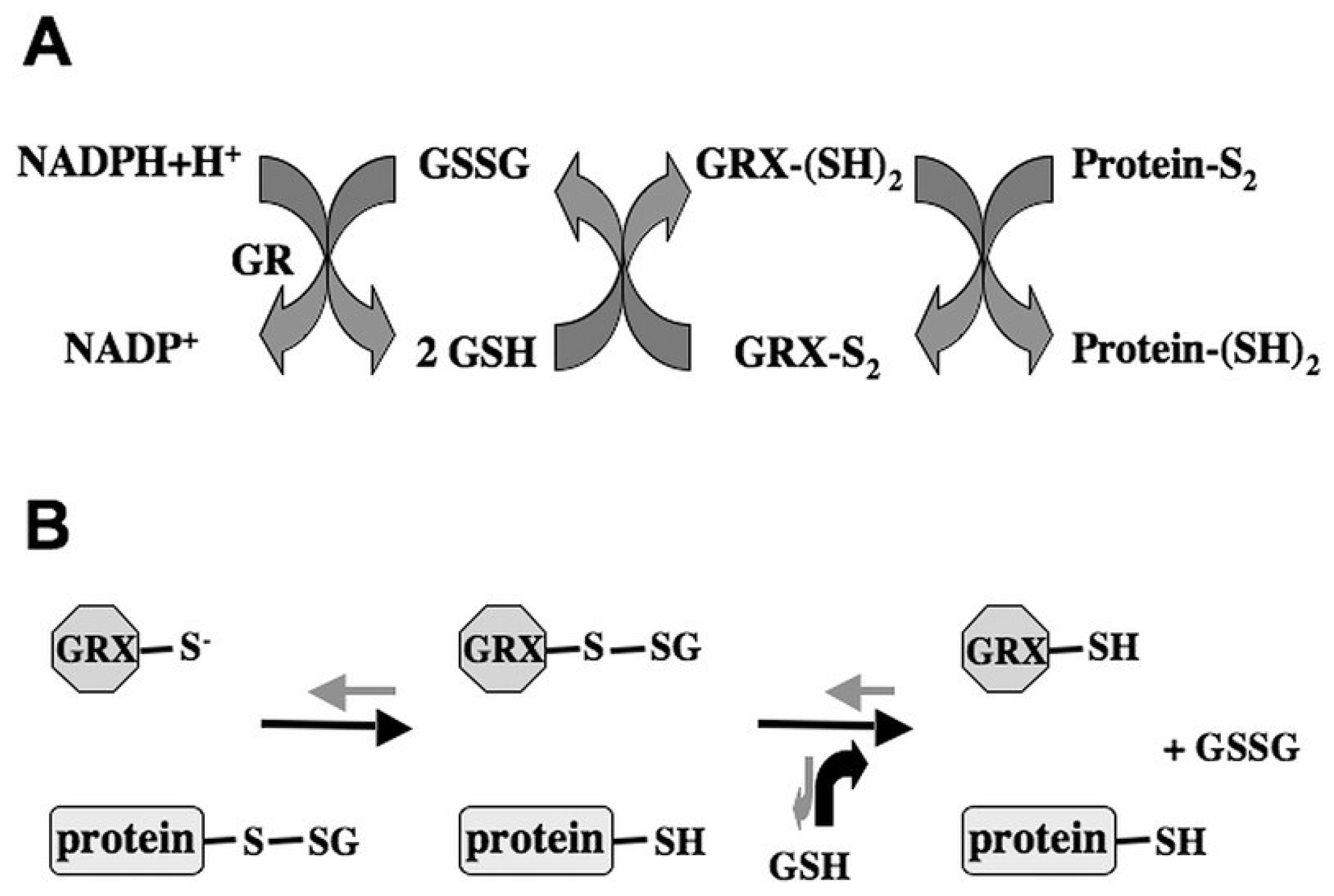
5. GRXs Cross-Talk with TRXs in Plant Development and Stress Responses
6. Conclusions and Perspectives
| GRX | Type | Species | Function * |
|---|---|---|---|
| GRXC7/ROXY1 a | CC | Arabidopsis | petal initiation and organogenesis, anther development and microspore formation [7,11,46] |
| GRXC8/ROXY2 a | CC | Arabidopsis | anther development and microspore formation [7,46] |
| OsMIL1 a | CC | rice | anther development and microspore formation [16] |
| MSCA1 a | CC | maize | anther development and microspore formation [15] |
| GRXC11/ROXY4 b | CC | Arabidopsis | GA-signaling and floral organ development [64] |
| GRXS13/ROXY18 a | CC | Arabidopsis | disease susceptibility, photooxidative stresses [55,71,100] |
| GRXC9/ROXY19 b | CC | Arabidopsis | Crosstalk between the SA and JA/ET defense pathways, disease susceptibility [12,60,100] |
| PvGRX5 c | CGFS | fern | arsenic resistance, oxidative abiotic stresses [20,76,77] |
| SlGRX1 a | CGFS | tomato | abiotic oxidative stresses [73] |
| GRXS17 a | CGFS | Arabidopsis | temperature-dependent postembryonic growth and development, thermotolerance [70,72] |
| GRXS14 a | CGFS | Arabidopsis | protection against protein oxidation [18] |
| GRXS14, 16 d | CGFS | poplar | assembly and transfer of Fe-S clusters [17] |
| GRXS15 d | CGFS | Arabidopsis | abiotic oxidative stresses [69] |
| GRXC1 a,d | CPYC | Arabidopsis | assembly of Fe-S clusters, early steps after pollination [34] |
| GRXC2 a | CPYC | Arabidopsis | early steps after pollination [34] |
| GRXC1 d | CPYC | poplar | assembly of Fe-S clusters [8,19] |
Acknowledgments
Conflicts of Interest
References
- Fernandes, A.P.; Holmgren, A. Glutaredoxins: Glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 2004, 6, 63–74. [Google Scholar]
- Buchanan, B.B.; Balmer, Y. Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 2005, 56, 187–220. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.L. Thioredoxin—A fold for all reasons. Structure 1995, 3, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, S.D. The glutaredoxin family in oxygenic photosynthetic organisms. Photosyn. Res. 2004, 79, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Heine, G.F.; Hernandez, J.M.; Grotewold, E. Two cysteines in plant R2R3 MYB domains participate in redox-sensitive DNA binding. J. Biol. Chem. 2004, 279, 37878–37885. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Gelhaye, E.; Jacquot, J.P. Plant glutaredoxins: Still mysterious reducing systems. Cell. Mol. Life Sci. 2004, 61, 1266–1277. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Zachgo, S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 2008, 53, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhong, N.; Rouhier, N.; Hase, T.; Kusunoki, M.; Jacquot, J.P.; Jin, C.; Xia, B. Structural insight into poplar glutaredoxin C1 with a bridging iron-sulfur cluster at the active site. Biochemistry 2006, 45, 7998–8008. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Couturier, J.; Jacquot, J.P. Genome-wide analysis of plant glutaredoxin systems. J. Exp. Bot. 2006, 57, 1685–1696. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Lauri, A.; Zachgo, S. Redox regulation and flower development: A novel function for glutaredoxins. Plant Biol. 2006, 8, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Rosso, M.G.; Zachgo, S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 2005, 132, 1555–1565. [Google Scholar] [CrossRef] [PubMed]
- Ndamukong, I.; Abdallat, A.A.; Thurow, C.; Fode, B.; Zander, M.; Weigel, R.; Gatz, C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007, 50, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lauri, A.; Ziemann, M.; Busch, A.; Bhave, M.; Zachgo, S. Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 2009, 21, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xing, S.; Birkenbihl, R.P.; Zachgo, S. Conserved functions of Arabidopsis and rice CC-type glutaredoxins in flower development and pathogen response. Mol. Plant 2009, 2, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Kelliher, T.; Walbot, V. Hypoxia triggers meiotic fate acquisition in maize. Science 2012, 337, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Tang, D.; Zhu, K.; Wang, K.; Li, M.; Cheng, Z. Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell 2012, 24, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Gama, F.; Molina-Navarro, M.M.; Gualberto, J.M.; Claxton, R.; Natik, S.G.; Huynh, B.H.; Herrero, E.; Jacquot, J.P.; et al. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008, 27, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.H.; Liu, J.Z.; Brock, A.; Nelson, R.S.; Hirschi, K.D. AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J. Biol. Chem. 2006, 281, 26280–26288. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Unno, H.; Bandyopadhyay, S.; Masip, L.; Kim, S.K.; Hirasawa, M.; Gualberto, J.M.; Lattard, V.; Knaff, D.B.; Georgiou, G.; Hase, T.; Johnson, M.K.; et al. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe–2S] cluster in poplar glutaredoxin C1. Proc. Natl. Acad. Sci. USA 2007, 104, 7379–7384. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, S.; Rathinasabapathi, B.; Ma, L.Q.; Rosen, B.P. An arsenate-activated glutaredoxin from the arsenic hyperaccumulator fern Pteris vittata L. regulates intracellular arsenite. J. Biol. Chem. 2008, 283, 6095–6101. [Google Scholar] [CrossRef] [PubMed]
- Cobbett, C.S.; May, M.J.; Howden, R.; Rolls, B. The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in gamma-glutamylcysteine synthetase. Plant J. 1998, 16, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Vernoux, T.; Wilson, R.C.; Seeley, K.A.; Reichheld, J.P.; Muroy, S.; Brown, S.; Maughan, S.C.; Cobbett, C.S.; Montagu, M.V.; Inze, D.; et al. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 2000, 12, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Tasaka, Y.; Mino, M.; Tanaka, Y.; Iwabuchi, M. Association of glutathione with flowering in Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Hatano-Iwasaki, A.; Yanagida, M.; Iwabuchi, M. Level of glutathione is regulated by ATP-dependent ligation of glutamateand cysteine through photosynthesis in Arabidopsis thaliana: Mechanism of strong interaction of light intensity with flowering. Plant Cell Physiol. 2004, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Reichheld, J.P.; Khafif, M.; Riondet, C.; Droux, M.; Bonnard, G.; Meyer, Y. Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 2007, 19, 1851–1865. [Google Scholar] [CrossRef] [PubMed]
- Bashandy, T.; Guilleminot, J.; Vernoux, T.; Caparros-Ruiz, D.; Ljung, K.; Meyer, Y.; Reichheld, J.P. Interplay between the NADP-Linked thioredoxin and glutathione systems in Arabidopsis auxin signalling. Plant Cell 2010, 22, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Beinert, H.; Kiley, P.J. Fe-S proteins in sensing and regulatory functions. Curr. Opin. Chem. Bio. 1999, 3, 152–157. [Google Scholar] [CrossRef]
- Beinert, H.; Holm, R.H.; Münck, E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Lill, R.; Kispal, G. Maturation of cellular Fe-S proteins: An essential function of mitochondria. Trends Biochem. Sci. 2000, 25, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.K.; Smith, A.D. Iron-sulfur proteins. In Encyclopedia of Inorganic Chemistry; King, R.B., Ed.; John Wiley & Sons: Chichester, UK, 2005; pp. 2589–2619. [Google Scholar]
- Balk, J.; Lobreaux, S. Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 2005, 10, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Abdel-Ghany, S.E.; Anderson, T.D.; Pilon-Smits, E.A.; Pilon, M. CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J. Biol. Chem. 2006, 281, 8958–8969. [Google Scholar] [CrossRef] [PubMed]
- Mühlenhoff, U.; Gerber, J.; Richhardt, N.; Lill, R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003, 22, 4815–4825. [Google Scholar] [PubMed]
- Riondet, C.; Desouris, J.P.; Montoya, J.G.; Chartier, Y.; Meyer, Y.; Reichheld, J. A dicotyledon-specific glutaredoxin GRXC1 family with dimer-dependent redox regulation is functionally redundant with GRXC2. Plant Cell Environ. 2012, 35, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Picciocchi, A.; Saguez, C.; Boussac, A.; Cassier-Chauvat, C.; Chauvat, F. CGFS type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry 2007, 46, 15018–15026. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Manzaneque, M.T.; Tamarit, J.; Belli, G.; Ros, J.; Herrero, E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell. 2002, 13, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Didierjean, C.; Jacqot, J.P.; Rouhier, N. Engineered mutated glutaredoxins mimicking peculiar plant class III glutaredoxins bind iron-sulfur centers and possess reductase activity. Biochem. Biophys. Res. Commun. 2010, 403, 435–441. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 1, 106–111. [Google Scholar] [CrossRef]
- Després, C.; Chubak, C.; Rochon, A.; Clark, R.; Bethune, T.; Desveaux, D.; Fobert, P.R. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 2003, 15, 2181–2191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Tessaro, M.J.; Lassner, M.; Li, X. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 2003, 15, 2647–2653. [Google Scholar] [CrossRef] [PubMed]
- Kesarwani, M.; Yoo, J.; Dong, X. Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 2007, 144, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Gatz, C. From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant Microbe Interact. 2013, 26, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Running, M.P.; Meyerowitz, E.M. Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 1996, 122, 1261–1269. [Google Scholar] [PubMed]
- Chuang, C.F.; Running, M.P.; Williams, R.W.; Meyerowitz, E.M. The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 1999, 13, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Murmu, J.; Bush, M.J.; DeLong, C.; Li, S.; Xu, M.; Khan, M.; Malcolmson, C.; Fobert, P.R.; Zachgo, S.; Hepworth, S.R. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 2010, 154, 1492–1504. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gutsche, N.; Zachgo, S. The ROXY1 C-terminal L ** LL motif is essential for the interaction with TGA transcription factors. Plant Physiol. 2011, 157, 2056–2068. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zachgo, S. Glutaredoxins in development and stress responses of plants. Adv. Bot. Res. 2009, 52, 333–361. [Google Scholar]
- Schenk, P.M.; Kazan, K.; Wilson, I.; Anderson, J.P.; Richmond, T.; Somerville, S.C.; Manners, J.M. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 2000, 97, 11655–11660. [Google Scholar] [CrossRef] [PubMed]
- De Vos, M.; van Oosten, V.R.; van Poecke, R.M.P.; van Pelt, J.A.; Pozo, M.J.; Mueller, M.J.; Buchala, A.J.; Metraux, J.P.; van Loon, L.C.; et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant-Microbe Interact. 2005, 18, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Johnson, J.S.; Dond, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef] [PubMed]
- Sticher, L.; Mauch-Mani, B.; Metraux, J.P. Systemic acquired resistance. Ann. Rev. Phytopathol. 1997, 35, 235–270. [Google Scholar] [CrossRef]
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef] [PubMed]
- La Camera, S.; L’haridon, F.; Astier, J.; Zander, M.; Abou-Mansour, E.; Page, G.; Thurow, C.; Wendehenne, D.; Gatz, C.; Métraux, J.P.; et al. The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants. Plant J. 2011, 68, 507–519. [Google Scholar] [CrossRef]
- Blanco, F.; Salinas, P.; Cecchini, N.M.; Jordana, X.; Van Hummelen, P.; Alvarez, M.E.; Holuigue, L. Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 2009, 70, 79–102. [Google Scholar] [CrossRef] [PubMed]
- Zarei, A.; Körbes, A.P.; Younessi, P.; Montiel, G.; Champion, A.; Memelink, J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 2011, 75, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Pre, M.; Atallah, M.; Champion, A.; de Vos, M.; Pieterse, C.M.J.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Leon-Reyes, A.; Du, Y.; Koornneef, A.; Proietti, S.; Körbes, A.P.; Memelink, J.; Pieterse, C.M.J.; Ritsema, T. Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol. Plant Microbe Interact. 2010, 23, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Zander, M.; Chen, S.; Imkampe, J.; Thurow, C.; Gatz, C. Repression of the Arabidopsis thaliana jasmonic acid/ethylene induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Mol. Plant 2012, 5, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Xiao, J.; Xie, W.; Liu, H.; Li, X.; Xiong, L.; Wang, S. Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol. Plant 2008, 1, 538–551. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Xiao, J.; Ding, B.; Xiong, M.; Cai, M.; Cao, Y.; Li, X.; Xu, C.; Wang, S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmoate-dependent signalling. Mol. Plant Microbe Interact. 2007, 20, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Zander, M.; La Camera, S.; Lamotte, O.; Métraux, J.P.; Gatz, C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 2010, 61, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.L.; Hu, W.W.; Shen, L.S.; Lee, L.Y.C.; Tao, Z.; Han, J.H.; Yu, H. Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiol. 2008, 147, 1126–1142. [Google Scholar] [CrossRef] [PubMed]
- Ziemann, M.; Bhave, M.; Zachgo, S. Origin and diversification of land plant CC-type glutaredoxins. Genome Biol. Evol. 2009, 1, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Michelet, L.; Zaffagnini, M.; Massot, V.; Keryer, E.; Vanacker, H.; Miginiac-Maslow, M.; Issakidis-Bourguet, E.; Lemaire, S.D. Thioredoxins, glutaredoxins, and glutathionylation: New crosstalks to explore. Photosynth. Res. 2006, 89, 225–245. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F.V. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 10, 1360–1385. [Google Scholar]
- Cheng, N.H. AtGRX4, an Arabidopsis chloroplastic monothiol glutaredoxin, is able to suppress yeast grx5 mutant phenotypes and respond to oxidative stress. FEBS Lett. 2008, 582, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.H.; Liu, J.Z.; Liu, X.; Wu, Q.; Thompson, S.M.; Lin, J.; Chang, J.; Whitham, S.A.; Park, S.; Cohen, J.D.; et al. Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response. J. Biol. Chem. 2011, 286, 20398–20406. [Google Scholar]
- Laporte, D.; Olate, E.; Salinas, P.; Salazar, M.; Jordana, X.; Holuigue, L. Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J. Exp. Bot. 2012, 63, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Lin, J.; Liu, J.Z.; Wang, X.; Lim, W.; Oh, M.; Park, J.; Rajashekar, C.B.; Whitham, S.A.; Cheng, N.H.; et al. Ecotopic expression of Arabidopsis glutaredoxin AtGRXS17 enhances thermotolerance in tomato. Plant Biotechnol. J. 2012, 10, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, C.; Xie, Y.; Song, F.; Zhou, Y. A tomato glutaredoxin gene SlGRX1 regulates plant responses to oxidative, drought and salt stresses. Planta 2010, 232, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.Q.; Komar, K.M.; Tu, C.; Zhang, W.; Cai, Y.; Kennelley, E.D. A fern that hyperaccumulates arsenic. Nature 2001, 409. [Google Scholar] [CrossRef]
- Liu, Z.; Shen, J.; Carbrey, J.M.; Mukhopadhyay, R.; Agre, P.; Rosen, B.P. Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP9. Proc. Natl. Acad. Sci. USA 2002, 99, 6053–6058. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, S.; Wu, S.; Ma, L.Q.; Rathinasabapathi, B. Expression of a Pteris vittata glutaredoxin PvGRX5 in transgenic Arabidopsis thaliana increases plant arsenic tolerance and decreases arsenic accumulation in the leaves. Plant Cell Environ. 2009, 32, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, S.; Rathinasabapathi, B. Transgenic expression of fern Pteris vittata glutaredoxin PvGrx5 in Arabidopsis thaliana increases plant tolerance to high temperature stress and reduces oxidative damage to proteins. Planta 2010, 231, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Villarejo, A.; Srivastava, M.; Gelhaye, E.; Keech, O.; Droux, M.; Finkemeier, I.; Samuelsson, G.; Dietz, K.J.; Jacquot, J.P.; et al. Identification of plant glutaredoxin targets. Antioxid. Redox Signal. 2005, 7, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Meyer, Y.; Belin, C.; Delorme-Hinoux, V.; Reichheld, J.P.; Riondet, C. Thioredoxin and glutaredoxin systems in plants: Molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 2012, 17, 1124–1160. [Google Scholar] [CrossRef] [PubMed]
- Arsova, B.; Hoja, U.; Wimmelbacher, M.; Greiner, E.; Ustun, S.; Melzer, M.; Petersen, K.; Lein, W.; Bornke, F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: Evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 2010, 22, 1498–1515. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Schwendtmayer, C.; Schurmann, P.; Ramaswamy, S.; Eklund, H. Redox signalling in chloroplasts: Cleavage of disulfides by an iron-sulfur cluster. Science 2000, 287, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Marty, L.; Siala, W.; Schwarzlaender, M.; Fricker, M.D.; Wirtz, M.; Sweetlove, L.J.; Meyer, Y.; Meyer, A.J.; Reichheld, J.P.; Hell, R. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 9109–9114. [Google Scholar] [CrossRef] [PubMed]
- Cairns, N.G.; Pasternak, M.; Wachter, A.; Cobbett, C.S.; Meyer, A.J. Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 2006, 141, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Howden, R.; Andersen, C.R.; Goldsbrough, P.B.; Cobbett, C.S. A cadmium-sensitive, glutathione-deficient mutant of Arabidopsis thaliana. Plant Physiol. 1995, 107, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Fernandez, R.; Fricker, M.; Corben, L.B.; White, N.S.; Sheard, N.; Leaver, C.J.; Montagu, M.V.; Inze, D.; May, M.J. Cell proliferation an hair tip growth in the Arabidopsis root are under mechanistically different forms of redox control. Proc. Natl. Acad. Sci. USA 1997, 94, 2745–2750. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Cobbert, C.S. Phytochelatin biosynthesis and function in heavy-metal detoxification. Curr. Opin. Plant Biol. 2000, 3, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.C.; Seeley, K.; Sung, Z.R. RML1 and RML2 Arabidopsis genes required for cell proliferation at the root tip. Plant Physiol. 1995, 107, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Parisy, V.; Poinssot, B.; Owsianowski, L.; Buchala, A.; Glazebrook, J.; Mauch, F. Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 2007, 49, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Reichheld, J.P.; Meyer, E.; Khafif, M.; Bonnard, G.; Meyer, Y. AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett. 2005, 579, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Gelhaye, E.; Rounier, N.; Jacquot, J.P. Evidence for a subgroup of thioredoxin h that requires GSH/GRX for its reduction. FEBS Lett. 2003, 555, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.S.M.; Alvarez, J.; Bossinger, G.; Smyth, D.R. Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 1995, 8, 505–520. [Google Scholar] [CrossRef]
- Christensen, S.K.; Dagenais, N.; Chory, J.; Weigel, D. Regulation of auxin response by the protein kinase PINOID. Cell 2000, 100, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Gälweiler, L.; Guan, C.; Müller, A.; Wisman, E.; Mendgen, K.; Yephremov, A.; Palme, K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 1998, 282, 2226–2230. [Google Scholar] [CrossRef] [PubMed]
- Hardtke, C.S.; Ckurshumova, W.; Vidaurre, D.P.; Singh, S.A.; Stamatiou, G.; Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J.; Berleth, T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 2004, 131, 1089–1100. [Google Scholar] [CrossRef]
- Cheng, Y.; Qin, G.; Dai, X.; Zhao, Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 18825–18829. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zachgo, S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013, 76, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014, 9, e27522. [Google Scholar] [CrossRef] [PubMed]
- Backstrom, S.; Elfving, N.; Nilsson, R.; Wingsle, G.; Bjorklund, S. Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 2007, 26, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Schluttenhofer, C.M.; Bhide, K.; Shreve, J.; Thimmapuram, J.; Lee, S.Y.; Yun, J.D.; Mengiste, T. MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 2014, 5, 3064. [Google Scholar] [CrossRef]
- Lorang, J.M.; Carkaci-Salli, N.; Wolpert, T.J. Identification and characterization of victorin sensitivity in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2004, 17, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Schwarzer, C.; Lally, E.; Zhang, S.; Ruzin, S.; Machen, T.; Remington, S.J.; Feldman, L. Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol. 2006, 141, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.J.; Brach, T.; Marty, L.; Kreye, S.; Rouhier, N.; Jacquot, J.P.; Hell, R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007, 52, 973–986. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.J.; Dick, T.P. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 2010, 13, 621–650. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Jhanwar, S.; Tyagi, A.K.; Jain, M. Genome-wide survey and expression analysis suggests diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 2010, 17, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Belin, C.; Bashandy, T.; Cela, J.; Delorme-Hinoux, V.; Riondet, C.; Reichheld, J.P. A comprehensive study of thiol reduction gene expression under stress conditions in Arabidopsis thaliana. Plant Cell Environ. 2014. [Google Scholar] [CrossRef]
- Meyer, Y.; Siala, W.; Bashandy, T.; Riondet, C.; Vignols, F.; Reichheld, J.P. Glutaredoxins and thioredoxins in plants. Biochim. Biophys. Acta 2008, 1783, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Iwabuchi, M.; Ogawa, K. The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: Detection using biotinylated glutathione. Plant Cell Physiol. 2003, 44, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.M.; Jones, D.P. The redox proteome. J. Biochem. Chem. 2013, 288, 26512–26520. [Google Scholar]
- Lee, K.; Lee, J.; Kim, Y.; Bae, D.; Kang, K.Y.; Yoon, S.C.; Lim, D.B. Defining the plant disulfide proteome. Electrophoresis 2004, 25, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Skipsey, M.; Grundy, N.M.; Edwards, R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 2005, 138, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.; Zhu, N.; Zhu, M.; Chen, S. Profiling thiol redox proteome using isotope tagging mass spectrometry. J. Visual. Exp. 2012, 61, e376661. [Google Scholar]
- Zaffagnini, M.; Michelet, L.; Massot, V.; Trost, P.; Lemaire, S.D. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J. Biol. Chem. 2008, 283, 8868–8876. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N. Plant glutaredoxins: Pivotal players in redox biology and iron-sulphur centre assembly. New Phytol. 2010, 186, 362–372. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S. Redox Modulation Matters: Emerging Functions for Glutaredoxins in Plant Development and Stress Responses. Plants 2014, 3, 559-582. https://doi.org/10.3390/plants3040559
Li S. Redox Modulation Matters: Emerging Functions for Glutaredoxins in Plant Development and Stress Responses. Plants. 2014; 3(4):559-582. https://doi.org/10.3390/plants3040559
Chicago/Turabian StyleLi, Shutian. 2014. "Redox Modulation Matters: Emerging Functions for Glutaredoxins in Plant Development and Stress Responses" Plants 3, no. 4: 559-582. https://doi.org/10.3390/plants3040559
APA StyleLi, S. (2014). Redox Modulation Matters: Emerging Functions for Glutaredoxins in Plant Development and Stress Responses. Plants, 3(4), 559-582. https://doi.org/10.3390/plants3040559




