Responses of African Grasses in the Genus Sporobolus to Defoliation and Sodium Stress: Tradeoffs, Cross-Tolerance, or Independent Responses?
Abstract
:1. Introduction
2. Results
2.1. Growth Rate & Photosynthetic Responses
| Response | Na+ treatment (β's) | Defoliation (β's) | Na+ x Defoliation (β's) | ||||
|---|---|---|---|---|---|---|---|
| 100 mM | 200 mM | 400 mM | Clipped | Clipped | Clipped | Clipped | |
| 100 mM | 200 mM | 400 mM | |||||
| RGR | 0.001 | −0.004 | −0.008 | −0.01 | |||
| Day 2 | |||||||
| A0 | 1.09 | −3.96 | −9.11 | ||||
| Gs | −0.021 | −0.01 | −0.029 | ||||
| E | −1.23 | −0.67 | −1.8 | ||||
| Day 14 | |||||||
| A0 | −4.84 | −7.44 | 0.52 | ||||
| Gs | −0.022 | −0.026 | −0.002 | ||||
| E | −1.59 | −1.8 | −0.16 | ||||
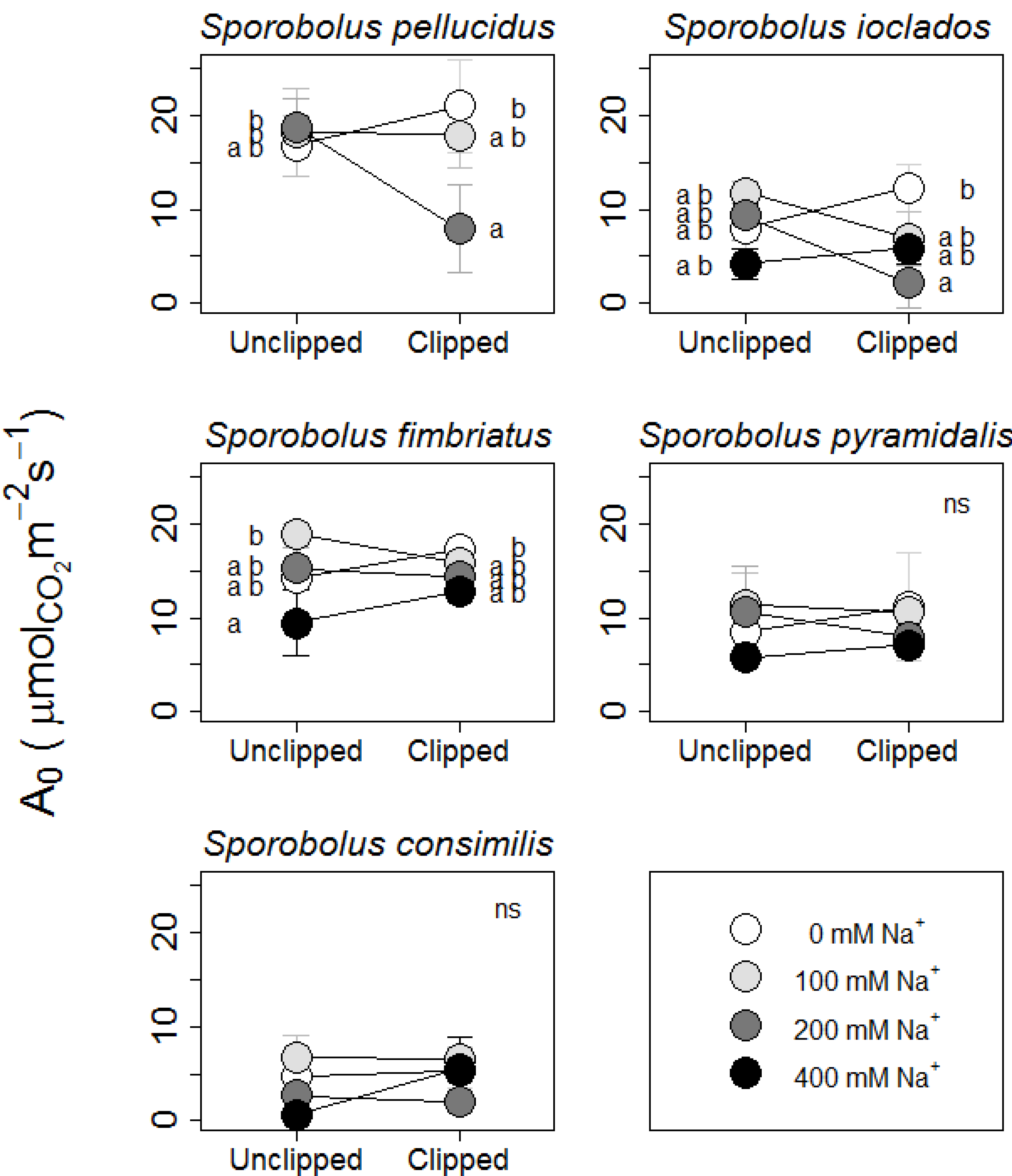
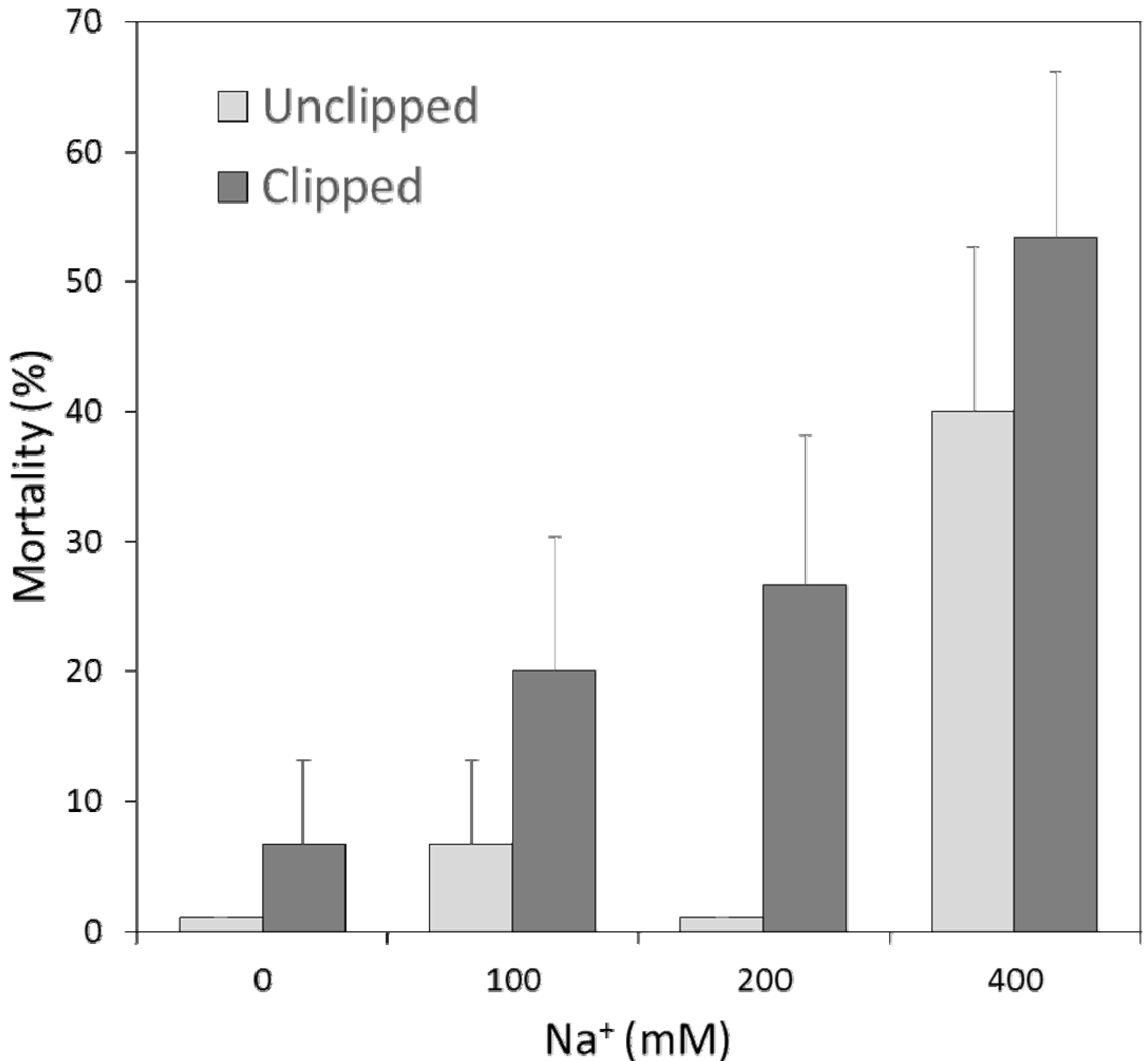
2.2. Association between Survivorship & Traits

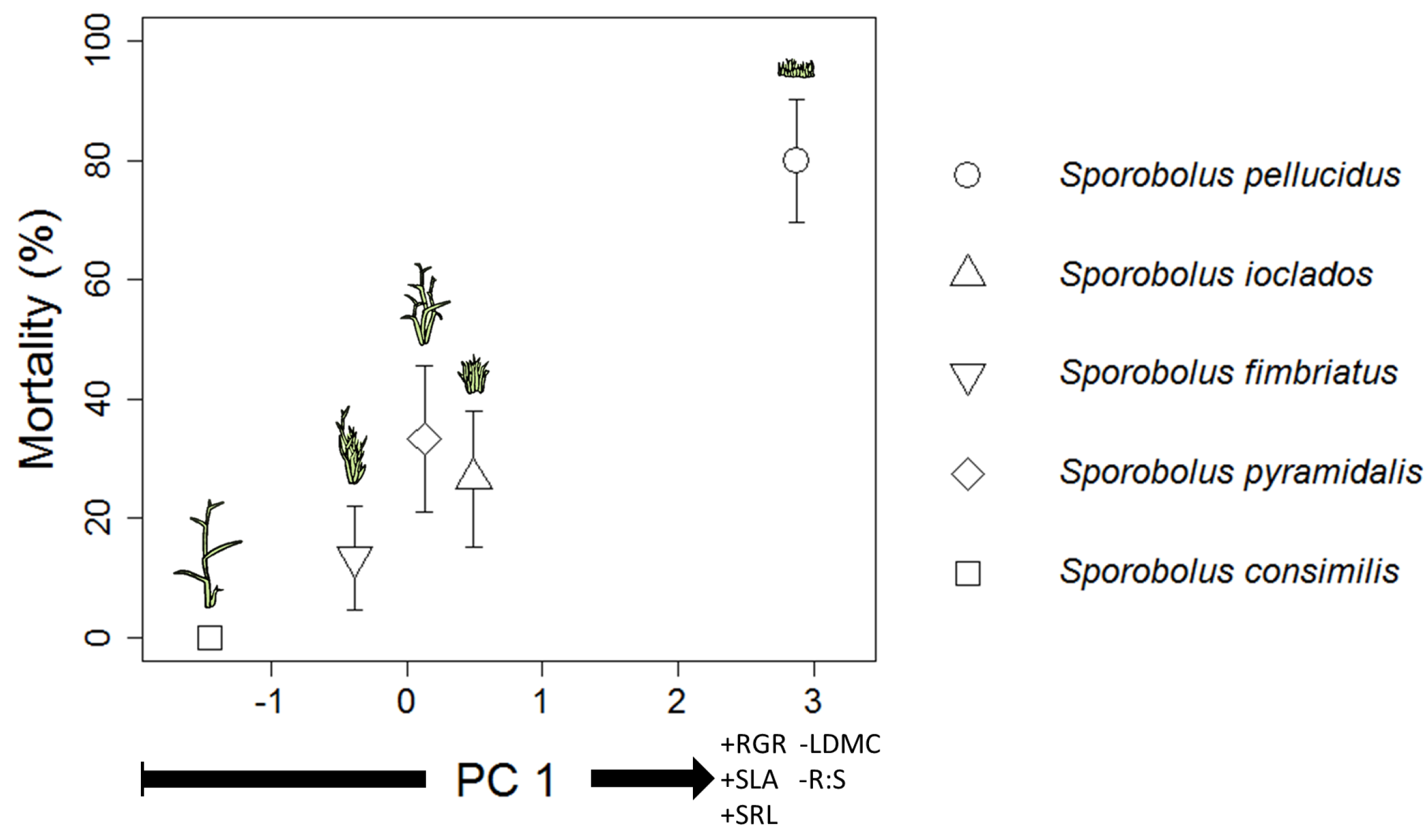
3. Discussion
4. Experimental Section
4.1. Plant Material and Greenhouse
4.2. Treatments
4.3. Measurements
4.4. Statistical Analysis
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Maestre, F.T.; Callaway, R.M.; Valladares, F.; Lortie, C.J. Refining the stress-gradient hypothesis for competition and facilitation in plant communities. J. Ecol. 2009, 97, 199–205. [Google Scholar] [CrossRef]
- Cipollini, D. Stretching the limits of plasticity: Can a plant defend against both competitors and herbivores? Ecology 2004, 85, 28–37. [Google Scholar] [CrossRef]
- Agrawal, A.A.; Conner, J.K.; Rasmann, S. Tradeoffs and Negative Correlations in Evolutionary Ecology. In Evolution since Darwin: The First 150 Years; Sinauer Associates: Sunderland, MA, USA, 2010; pp. 243–268. [Google Scholar]
- Fine, P.V.A. Herbivores promote habitat specialization by trees in amazonian forests. Science 2004, 305, 663–665. [Google Scholar] [CrossRef]
- Fine, P.V.; Miller, Z.J.; Mesones, I.; Irazuzta, S.; Appel, H.M.; Stevens, M.H.; Sääksjärvi, I.; Schultz, J.C.; Coley, P.D. The growth-defense trade-off and habitat specialization by plants in Amazonian forests. Ecology 2006, 87, 150–162. [Google Scholar] [CrossRef]
- Tilman, D. Plant Strategies and the Dynamics and Structure of Plant Communities; Princeton University Press: Princeton, NJ, USA, 1988. [Google Scholar]
- Tilman, D. The Maintenance of Species Richness in Plant Communities. In Species Diversity in Ecological Communities; University of Chicago Press: Chicago, IL, USA, 1993; pp. 13–25. [Google Scholar]
- Kneitel, J.M.; Chase, J.M. Trade-offs in community ecology: Linking spatial scales and species coexistence. Ecol. Lett. 2004, 7, 69–80. [Google Scholar] [CrossRef]
- Capiati, D.A. Wounding increases salt tolerance in tomato plants: Evidence on the participation of calmodulin-like activities in cross-tolerance signalling. J. Exp. Bot. 2006, 57, 2391–2400. [Google Scholar] [CrossRef]
- Armbruster, W.S.; Lee, J.; Baldwin, B.G. Macroevolutionary patterns of defense and pollination in Dalechampia vines: Adaptation, exaptation, and evolutionary novelty. Proc. Natl. Acad. Sci.USA 2009, 106, 18085–18090. [Google Scholar] [CrossRef]
- Losos, J.B. Convergence, adaptation, and constraint. Evolution 2011, 65, 1827–1840. [Google Scholar] [CrossRef]
- Armbruster, W.S. Exaptations link evolution of plant-herbivore and plant-pollinator interactions: A phylogenetic inquiry. Ecology 1997, 78, 1661–1672. [Google Scholar]
- Armbruster, W.S. Evolution of floral form: Electrostatic forces, pollination, and adaptive compromise. New Phytol. 2001, 152, 181–183. [Google Scholar] [CrossRef]
- Ackerly, D.D.; Schwilk, D.W.; Webb, C.O. Niche evolution and adaptive radiation: Testing the order of trait divergence. Ecology 2006, 87, 50–61. [Google Scholar] [CrossRef]
- Coughenour, M. Graminoid Responses to grazing by large herbivores: Adaptations, exaptations, and interacting processes. Ann. Mo. Bot. Gard. 1985, 72, 852–863. [Google Scholar] [CrossRef]
- Zhang, S.-B.; Cao, K.-F.; Fan, Z.-X.; Zhang, J.-L. Potential hydraulic efficiency in angiosperm trees increases with growth-site temperature but has no trade-off with mechanical strength. Glob. Ecol. Biogeogr. 2013, 22, 971–981. [Google Scholar] [CrossRef]
- Coley, P.D.; Bryant, J.P.; Chapin, F.S. Resource availability and plant antiherbivore defense. Science 1985, 230, 895–899. [Google Scholar]
- Maschinski, J.; Whitham, T.G. The continuum of plant responses to herbivory: The influence of plant association, nutrient availability, and timing. Am. Nat. 1989, 134, 1–19. [Google Scholar]
- McNaughton, S.J. Promotion of the cycling of diet-enhancing nutrients by African Grazers. Science 1997, 278, 1798–1800. [Google Scholar] [CrossRef]
- Anderson, T.M.; Hopcraft, J.G.; Eby, S.; Ritchie, M.; Grace, J.B.; Olff, H. Landscape-scale analyses suggest both nutrient and antipredator advantages to Serengeti herbivore hotspots. Ecology 2010, 91, 1519–1529. [Google Scholar] [CrossRef]
- Belsky, A.J. Regional influences on small-scale vegetational heterogeneity within grasslands in the Serengeti National Park, Tanzania. Plant Ecol. 1988, 74, 3–10. [Google Scholar] [CrossRef]
- Belsky, J. Population and community processes in a mosaic grassland in the Serengeti, Tanzania. J. Ecol. 1986, 74, 841–856. [Google Scholar] [CrossRef]
- Dawson, J.B. The geology of oldoinyo lengai. Bull. Volcanol. 1962, 24, 349–387. [Google Scholar] [CrossRef]
- Hamilton, E.W., III. Na Stress and Grazing: Molecular, Physiological and Growth Responses in four Serengeti C4 Grasses. PhD Thesis, Syracuse University, Syracuse, NY, USA, 1999. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Ruess, R.W. The interaction of defoliation and nutrient uptake in Sporobolus kentrophyllus, a short-grass species from the Serengeti Plains. Oecologia 1988, 77, 550–556. [Google Scholar] [CrossRef]
- McNaughton, S.J. Mineral nutrition and spatial concentrations of African ungulates. Nature 1988, 334, 343–345. [Google Scholar] [CrossRef]
- Oesterheld, M.; McNaughton, S.J. Intraspecific variation in the response of Themeda triandra to defoliation: The effect of time of recovery and growth rates on compensatory growth. Oecologia 1988, 77, 181–186. [Google Scholar] [CrossRef]
- Anderson, T.M.; Shaw, J.; Olff, H. Ecology’s cruel dilemma, phylogenetic trait evolution and the assembly of Serengeti plant communities. J. Ecol. 2011, 99, 797–806. [Google Scholar] [CrossRef]
- Belsky, A.J. Effects of grazing, competition, disturbance and fire on species composition and diversity in grassland communities. J. Veg. Sci. 1992, 3, 187–200. [Google Scholar] [CrossRef]
- Ghazanfar, S.A.; Beentje, H.J. Sabkha Regions of Tropical East Africa. In Sabkha Ecosystems; Öztürk, M., Böer, B., Barth, H.J., Clüsener-Godt, M., Khan, M.A., Breckle, S.W., Eds.; Springer: Dordrecht, The Netherlands, 2010; Volume 46, pp. 1–7. [Google Scholar]
- Belsky, A.J. Long-term vegetation monitoring in the Serengeti National Park, Tanzania. J. Appl. Ecol. 1985, 22, 449–460. [Google Scholar] [CrossRef]
- Hartvigsen, A.G.; McNaughton, S.J. Tradeoff between height and relative growth rate in a dominant grass from the Serengeti ecosystem. Ecology 1995, 102, 273–276. [Google Scholar]
- Hamilton, E.W., III; McNaughton, S.J.; Coleman, J.S. Molecular, physiological, and growth responses to sodium stress in C4 grasses from a soil salinity gradient in the Serengeti ecosystem. Am. J. Bot. 2001, 88, 1258–1265. [Google Scholar] [CrossRef]
- McNaughton, S.J. Ecology of a grazing ecosystem: The Serengeti. Ecol. Monogr. 1985, 55, 260–294. [Google Scholar] [CrossRef]
- Belsky, A.J. Small-scale pattern in grassland communities in the Serengeti National Park, Tanzania. Plant Ecol. 1984, 55, 141–151. [Google Scholar] [CrossRef]
- Belsky, A.J. Revegetation of Artificial disturbances in grasslands of the Serengeti-National-Park, Tanzania: I. Colonization of grazed and ungrazed plots. J. Ecol. 1986, 74, 419–437. [Google Scholar] [CrossRef]
- Wood, J.N.; Gaff, D.F. Salinity studies with drought-resistant species of Sporobolus. Oecologia 1989, 78, 559–564. [Google Scholar] [CrossRef]
- Anderson, T.M.; Kumordzi, B.B.; Fokkema, W.; Fox, H.V.; Olff, H. Distinct physiological responses underlie defoliation tolerance in African lawn and bunch grasses. Int. J. Plant Sci. 2013, 174, 769–778. [Google Scholar] [CrossRef]
- McNaughton, S.J. Grazing as an optimization process: Grass-ungulate relationships in the Serengeti. Am. Nat. 1979, 113, 691–703. [Google Scholar]
- Anderson, T.M.; Schütz, M.; Risch, A.C. Endozoochorous seed dispersal and germination strategies of Serengeti plants. J. Veg. Sci. 2013. [Google Scholar] [CrossRef]
- Vaieretti, M.V.; Diaz, S.; Vile, D.; Garnier, E. Two measurement methods of leaf dry matter content produce similar results in a broad range of species. Ann. Bot. 2007, 99, 955–958. [Google Scholar] [CrossRef]
- Cornelissen, J.H.C.; Lavorel, S.; Garnier, E.; Díaz, S.; Buchmann, N.; Gurvich, D.E.; Reich, P.B.; ter Steege, H.; Morgan, H.D.; Heijden, M.G.A.; et al. A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust. J. Bot. 2003, 51, 335–380. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Coskun, D.; Schulze, L.M.; Wong, J.R.; Britto, D.T. Sodium as nutrient and toxicant. Plant Soil 2013, 369, 1–23. [Google Scholar] [CrossRef]
- Siemens, D.H.; Duvall-Jisha, J.; Jacobs, J.; Manthey, J.; Haugen, R.; Matzner, S. Water deficiency induces evolutionary tradeoff between stress tolerance and chemical defense allocation that may help explain range limits in plants. Oikos 2012, 121, 790–800. [Google Scholar] [CrossRef]
- Anderson, T.M.; Ritchie, M.E.; Mayemba, E.; Eby, S.; Grace, J.B.; McNaughton, S.J. Forage nutritive quality in the Serengeti ecosystem: The roles of fire and herbivory. Am. Nat. 2007, 170, 343–357. [Google Scholar] [CrossRef]
- McNaughton, S.J. Mineral nutrition and seasonal movements of African migratory ungulates. Nature 1990, 345, 613–615. [Google Scholar] [CrossRef]
- Ostonen, I.; Püttsepp, Ü.; Biel, C.; Alberton, O.; Bakker, M.R.; Lõhmus, K.; Majdi, H.; Metcalfe, D.; Olsthoorn, A.F.M.; Pronk, A.; et al. Specific root length as an indicator of environmental change. Plant Biosyst. 2007, 141, 426–442. [Google Scholar] [CrossRef]
- The R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012. [Google Scholar]
- Pinheiro, J.C.; Bates, D.M. Mixed-Effects Models in S and S-PLUS; Springer: New York, NY, USA, 2000. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; Team, R.D.C. NLME: Linear and Nonlinear Mixed Effects Models; Springer: New York, NY, USA, 2012. [Google Scholar]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom. J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Therneau, T.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- Meyer, D.; Zeileis, A.; Hornik, K. VCD: Visualizing Categorical Data. Available online: http://www.cran.r-project.org/web/packages/vcd (accessed on 30 October 2013).
- Verhoeven, K.J.F.; Simonsen, K.L.; McIntyre, L.M. Implementing false discovery rate control: Increasing your power. Oikos 2005, 108, 643–647. [Google Scholar] [CrossRef]
- Pike, N. Using false discovery rates for multiple comparisons in ecology and evolution: False discovery rates for multiple comparisons. Methods Ecol. Evol. 2011, 2, 278–282. [Google Scholar] [CrossRef]
- Tester, M. Na+ Tolerance and Na+ Transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef]
Appendix
| Response | Na+ treatment | Defoliation | Na+ x Defoliation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| F | df | p | F | df | p | F | df | p | |
| RGR | 8.71 | 3,78 | <0.0001 | 84.86 | 1,78 | <0.0001 | |||
| Day 2 | |||||||||
| A0 | 31.25 | 3,102 | <0.0001 | ||||||
| Gs | 3.09 | 3,98 | 0.0306 | ||||||
| E | 2.85 | 3,98 | 0.0414 | ||||||
| Day 14 | |||||||||
| A0 | 6.77 | 3,88 | 0.0004 | ||||||
| Gs | 5.77 | 3,88 | 0.0012 | ||||||
| E | 5.54 | 3,88 | 0.0016 | ||||||
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Griffith, D.M.; Anderson, T.M. Responses of African Grasses in the Genus Sporobolus to Defoliation and Sodium Stress: Tradeoffs, Cross-Tolerance, or Independent Responses? Plants 2013, 2, 712-725. https://doi.org/10.3390/plants2040712
Griffith DM, Anderson TM. Responses of African Grasses in the Genus Sporobolus to Defoliation and Sodium Stress: Tradeoffs, Cross-Tolerance, or Independent Responses? Plants. 2013; 2(4):712-725. https://doi.org/10.3390/plants2040712
Chicago/Turabian StyleGriffith, Daniel M., and T. Michael Anderson. 2013. "Responses of African Grasses in the Genus Sporobolus to Defoliation and Sodium Stress: Tradeoffs, Cross-Tolerance, or Independent Responses?" Plants 2, no. 4: 712-725. https://doi.org/10.3390/plants2040712
APA StyleGriffith, D. M., & Anderson, T. M. (2013). Responses of African Grasses in the Genus Sporobolus to Defoliation and Sodium Stress: Tradeoffs, Cross-Tolerance, or Independent Responses? Plants, 2(4), 712-725. https://doi.org/10.3390/plants2040712




