The Epicardium in the Embryonic and Adult Zebrafish
Abstract
:1. Introduction
1.1. The Zebrafish Epicardium: Embryonic Development and Role during Injury Responses in the Adult
1.2. Transgenic Lines Used for the Study of Epicardium Formation in the Zebrafish
2. Experimental Section
2.1. Animal Handling
2.2. Immunostaining
2.3. Immunohistochemistry on Sections Was Performed As Described in [16]
Image Acquisition
3. Results and Discussion
3.1. Characterization of Wt1 Transgenic Reporter Lines during Epicardium Formation
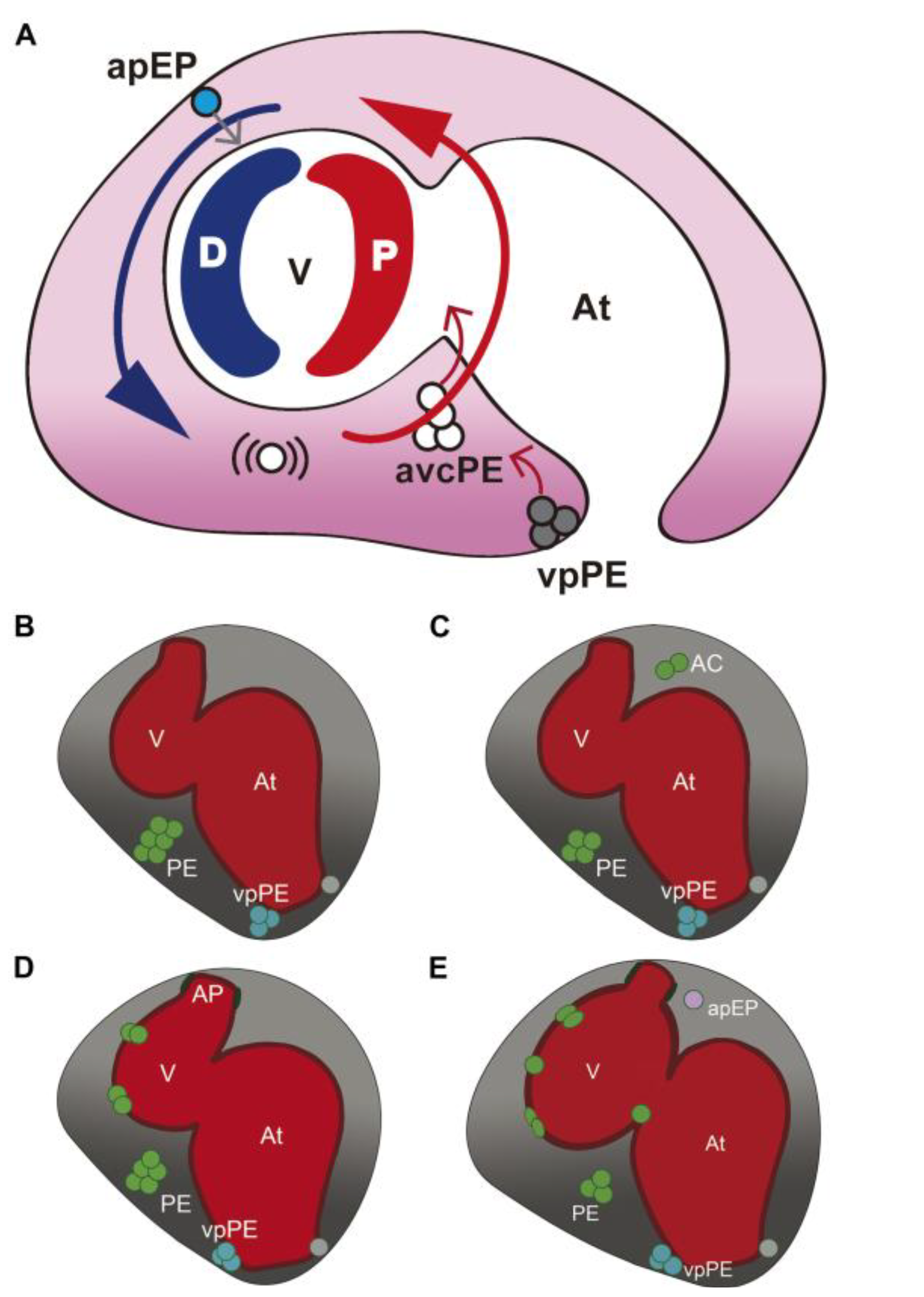
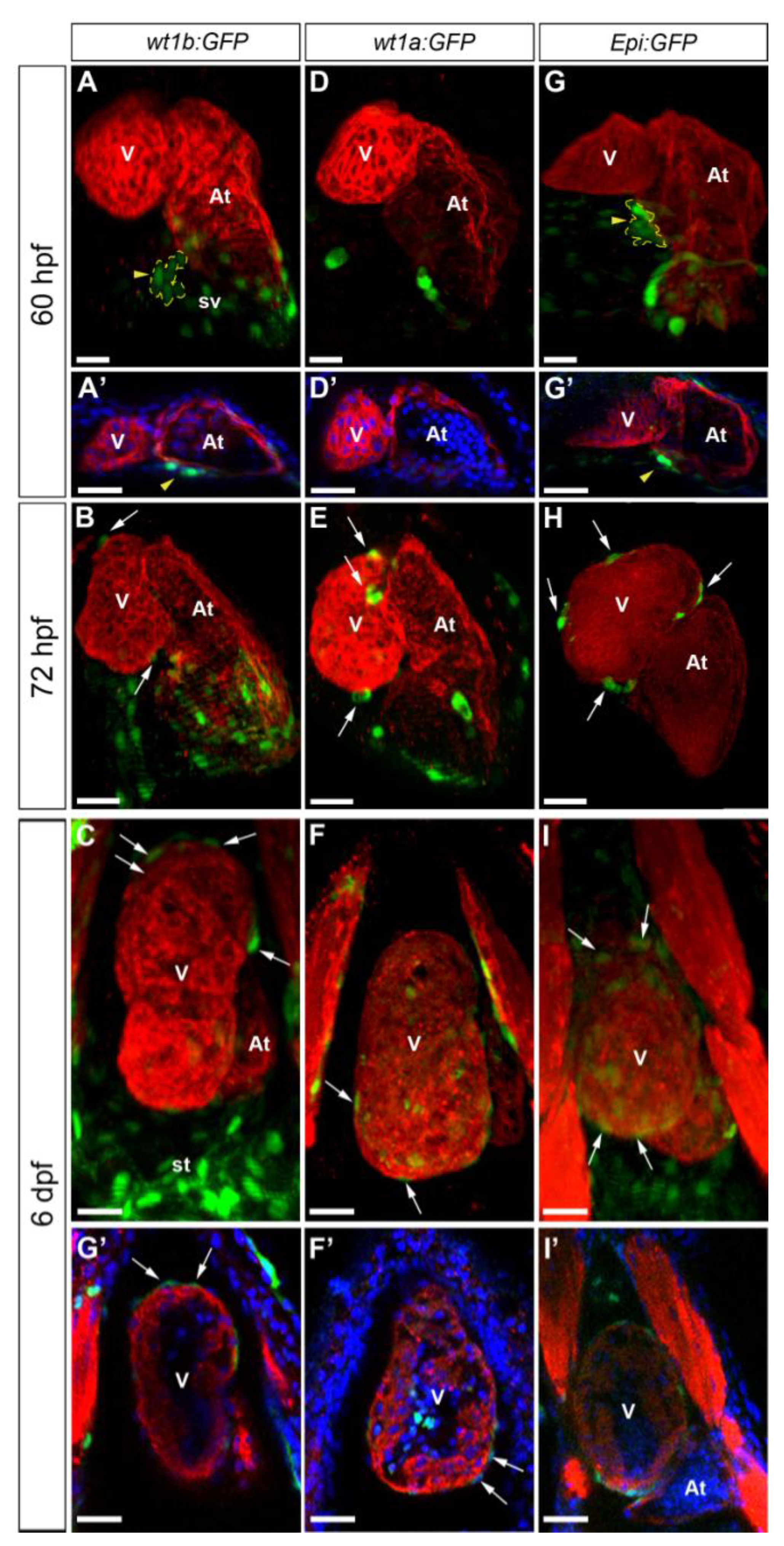
3.2. Characterization of Wt1 Transgenic Reporter Lines during Cardiac Regeneration

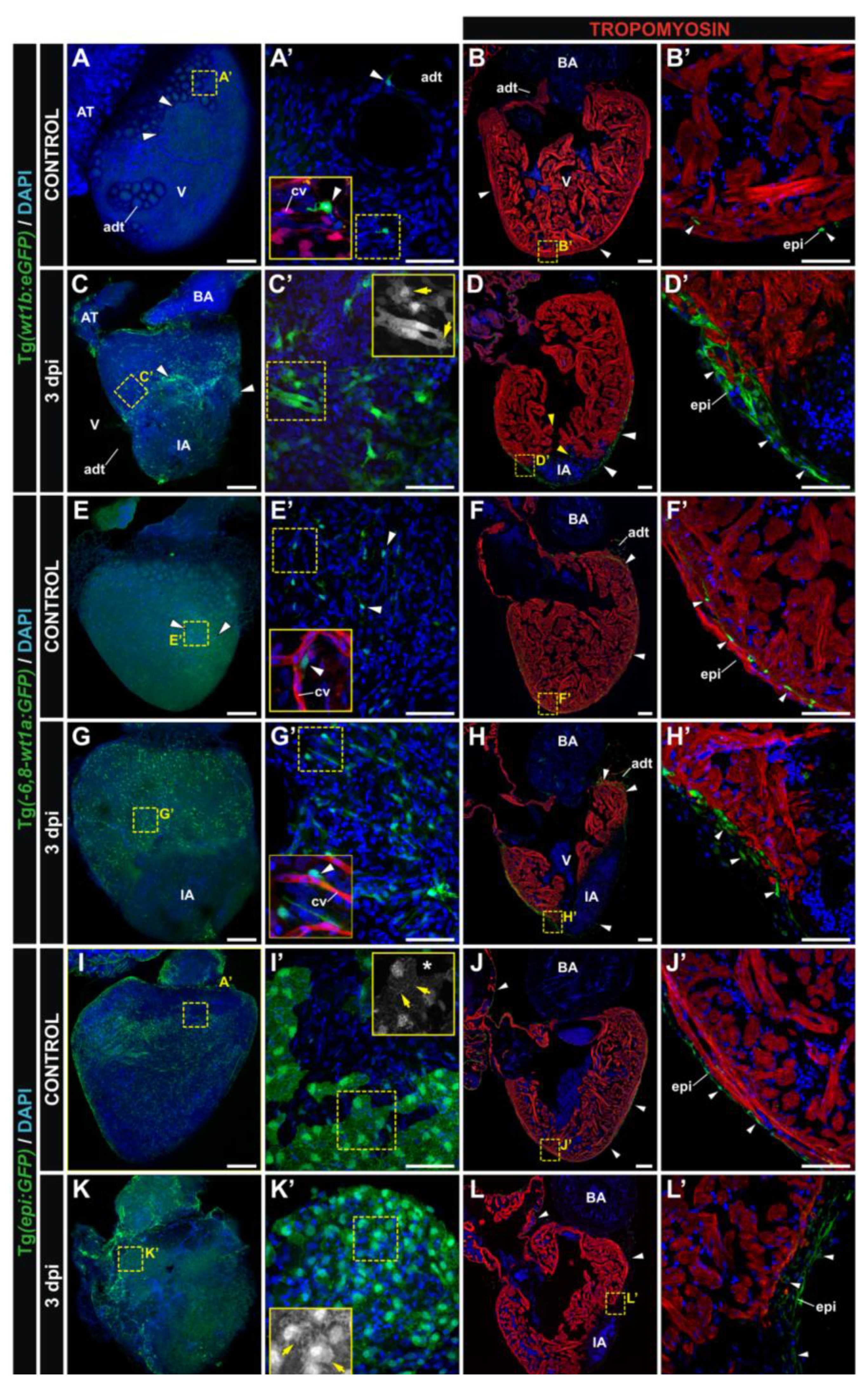
4. Conclusions
| Lines | Expression | Development | Adult Homeostasis | Adult cryoinjury |
|---|---|---|---|---|
| proepicardium | ++ | |||
| Tg(wt1b:GFP) | epicardium | ++ | – | +++ |
| pericardium | +++ | – | ++ | |
| sinus venosus | +++ | – | – | |
| proepicardium | + | |||
| Tg( -6.8kbwt1a:GFP) | epicardium | ++ | – | – |
| pericardium | ++ | – | – | |
| sinus venosus | + | – | – | |
| proepicardium | +++ | |||
| Et(-26.5Hsa.WT1-1gata2:EGFP)cn1 | epicardium | +++ | +++ | ++ |
| (Epi:GFP) | pericardium | +++ | ++ | ++ |
| sinus venosus | ++ | ++ | ++ |
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Bakkers, J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 2011, 91, 279–288. [Google Scholar] [CrossRef]
- Liu, J.; Stainier, D.Y. Zebrafish in the study of early cardiac development. Circ. Res. 2012, 110, 870–874. [Google Scholar] [CrossRef]
- Choi, W.Y.; Poss, K.D. Cardiac regeneration. Curr. Top. Dev. Biol. 2012, 100, 319–344. [Google Scholar] [CrossRef]
- Gemberling, M.; Bailey, T.J.; Hyde, D.R.; Poss, K.D. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013, 29, 611–620. [Google Scholar] [CrossRef]
- Grimes, A.C.; Stadt, H.A.; Shepherd, I.T.; Kirby, M.L. Solving an enigma: arterial pole development in the zebrafish heart. Dev. Biol. 2006, 290, 265–276. [Google Scholar] [CrossRef]
- de Pater, E.; Clijsters, L.; Marques, S.R.; Lin, Y.F.; Garavito-Aguilar, Z.V.; Yelon, D.; Bakkers, J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 2009, 136, 1633–1641. [Google Scholar] [CrossRef]
- Hami, D.; Grimes, A.C.; Tsai, H.J.; Kirby, M.L. Zebrafish cardiac development requires a conserved secondary heart field. Development 2011, 138, 2389–2398. [Google Scholar] [CrossRef]
- Zhou, Y.; Cashman, T.J.; Nevis, K.R.; Obregon, P.; Carney, S.A.; Liu, Y.; Gu, A.; Mosimann, C.; Sondalle, S.; Peterson, R.E.; et al. Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature 2011, 474, 645–648. [Google Scholar] [CrossRef]
- Serluca, F.C. Development of the proepicardial organ in the zebrafish. Dev. Biol. 2008, 315, 18–27. [Google Scholar] [CrossRef]
- Schlueter, J.; Brand, T. Epicardial Progenitor Cells in Cardiac Development and Regeneration. J. Cardiovasc. Transl. Res. 2012. [Google Scholar]
- Ruiz-Villalba, A.; Perez-Pomares, J.M. The expanding role of the epicardium and epicardial-derived cells in cardiac development and disease. Curr. Opin. Pediatr. 2012, 5, 641–653. [Google Scholar]
- Hu, N.; Sedmera, D.; Yost, H.J.; Clark, E.B. Structure and function of the developing zebrafish heart. Anatomical Rec. 2000, 260, 148–157. [Google Scholar]
- Wills, A.A.; Holdway, J.E.; Major, R.J.; Poss, K.D. Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development 2008, 135, 183–192. [Google Scholar]
- Kikuchi, K.; Gupta, V.; Wang, J.; Holdway, J.E.; Wills, A.A.; Fang, Y.; Poss, K.D. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 2011, 138, 2895–2902. [Google Scholar] [CrossRef]
- Lepilina, A.; Coon, A.N.; Kikuchi, K.; Holdway, J.E.; Roberts, R.W.; Burns, C.G.; Poss, K.D. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 2006, 127, 607–619. [Google Scholar] [CrossRef]
- Gonzalez-Rosa, J.M.; Martin, V.; Peralta, M.; Torres, M.; Mercader, N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 2011, 138, 1663–1674. [Google Scholar] [CrossRef]
- Schnabel, K.; Wu, C.C.; Kurth, T.; Weidinger, G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PloS one 2011, 6, e18503. [Google Scholar] [CrossRef]
- Kim, J.; Wu, Q.; Zhang, Y.; Wiens, K.M.; Huang, Y.; Rubin, N.; Shimada, H.; Handin, R.I.; Chao, M.Y.; Tuan, T.L.; et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. PNAS 2010, 107, 17206–17210. [Google Scholar] [CrossRef]
- Kikuchi, K.; Holdway, J.E.; Werdich, A.A.; Anderson, R.M.; Fang, Y.; Egnaczyk, G.F.; Evans, T.; Macrae, C.A.; Stainier, D.Y.; Poss, K.D. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010, 464, 601–605. [Google Scholar] [CrossRef]
- Jopling, C.; Sleep, E.; Raya, M.; Marti, M.; Raya, A.; Belmonte, J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar]
- Gonzalez-Rosa, J.M.; Peralta, M.; Mercader, N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev. Biol. 2012, 370, 173–186. [Google Scholar] [CrossRef]
- Kikuchi, K.; Holdway, J.E.; Major, R.J.; Blum, N.; Dahn, R.D.; Begemann, G.; Poss, K.D. Retinoic Acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 2011, 20, 397–404. [Google Scholar] [CrossRef]
- Itou, J.; Oishi, I.; Kawakami, H.; Glass, T.J.; Richter, J.; Johnson, A.; Lund, T.C.; Kawakami, Y. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 2012, 139, 4133–4142. [Google Scholar] [CrossRef]
- Nesbitt, T.; Lemley, A.; Davis, J.; Yost, M.J.; Goodwin, R.L.; Potts, J.D. Epicardial development in the rat: A new perspective. Microsc. Microanal. 2006, 12, 390–398. [Google Scholar]
- Komiyama, M.; Ito, K.; Shimada, Y. Origin and development of the epicardium in the mouse embryo. Anat. Embryol. (Berl) 1987, 176, 183–189. [Google Scholar] [CrossRef]
- Viragh, S.; Challice, C.E. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat. Rec. 1981, 201, 157–168. [Google Scholar] [CrossRef]
- Manner, J. The development of pericardial villi in the chick embryo. Anat. Embryol. (Berl) 1992, 186, 379–385. [Google Scholar] [CrossRef]
- Jahr, M.; Schlueter, J.; Brand, T.; Manner, J. Development of the proepicardium in Xenopus laevis. Dev. Dyn. 2008, 237, 3088–3096. [Google Scholar] [CrossRef]
- Hirakow, R. Epicardial formation in staged human embryos. Kaibogaku Zasshi 1992, 67, 616–622. [Google Scholar]
- Zhou, B.; von Gise, A.; Ma, Q.; Rivera-Feliciano, J.; Pu, W.T. Nkx2–5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem. Biophys. Res. Commun. 2008, 375, 450–453. [Google Scholar] [CrossRef]
- Liu, J.; Stainier, D.Y. Tbx5 and Bmp signaling are essential for proepicardium specification in zebrafish. Circ. Res. 2010, 106, 1818–1828. [Google Scholar] [CrossRef]
- van Wijk, B.; van den Berg, G.; Abu-Issa, R.; Barnett, P.; van der Velden, S.; Schmidt, M.; Ruijter, J.M.; Kirby, M.L.; Moorman, A.F.; van den Hoff, M.J. Epicardium and Myocardium Separate From a Common Precursor Pool by Crosstalk Between Bone Morphogenetic Protein- and Fibroblast Growth Factor-Signaling Pathways. Circ. Res. 2009, 105, 431–441. [Google Scholar] [CrossRef]
- Nahirney, P.C.; Mikawa, T.; Fischman, D.A. Evidence for an extracellular matrix bridge guiding proepicardial cell migration to the myocardium of chick embryos. Dev. Dyn. 2003, 227, 511–523. [Google Scholar] [CrossRef]
- Munoz-Chapuli, R.; Macias, D.; Ramos, C.; de Andres, V.; Gallego, A.; Navarro, P. Cardiac development in the dogfish (Scyliorhinus canicula): a model for the study of vertebrate cardiogenesis. Cardioscience 1994, 5, 245–253. [Google Scholar]
- Peralta, M.; Steed, E.; Harlepp, S.; Gonzalez-Rosa, J.M.; Monduc, F.; Ariza-Cosano, A.; Cortes, A.; Rayon, T.; Gomez-Skarmeta, J.L.; Zapata, A.; et al. Heartbeat-driven pericardiac fluid forces contribute to epicardium morphogenesis. Curr. Biol. 2013, 23, 1726–1735. [Google Scholar] [CrossRef]
- Perez-Pomares, J.M.; Phelps, A.; Sedmerova, M.; Wessels, A. Epicardial-like cells on the distal arterial end of the cardiac outflow tract do not derive from the proepicardium but are derivatives of the cephalic pericardium. Dev. Dyn. 2003, 227, 56–68. [Google Scholar] [CrossRef]
- Poon, K.L.; Liebling, M.; Kondrychyn, I.; Garcia-Lecea, M.; Korzh, V. Zebrafish cardiac enhancer trap lines: New tools for in vivo studies of cardiovascular development and disease. Dev. Dyn. 2010, 239, 914–926. [Google Scholar] [CrossRef]
- Bollig, F.; Perner, B.; Besenbeck, B.; Kothe, S.; Ebert, C.; Taudien, S.; Englert, C. A highly conserved retinoic acid responsive element controls wt1a expression in the zebrafish pronephros. Development 2009, 136, 2883–2892. [Google Scholar] [CrossRef]
- Perner, B.; Englert, C.; Bollig, F. The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev. Biol. 2007, 309, 87–96. [Google Scholar] [CrossRef]
- Gonzalez-Rosa, J.M.; Mercader, N. Cryoinjury as a myocardial infarction model for the study of cardiac regeneration in the zebrafish. Nat. Protoc. 2012, 7, 782–788. [Google Scholar] [CrossRef]
- Bessa, J.; Tena, J.J.; de la Calle-Mustienes, E.; Fernandez-Minan, A.; Naranjo, S.; Fernandez, A.; Montoliu, L.; Akalin, A.; Lenhard, B.; Casares, F.; et al. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev. Dyn. 2009, 238, 2409–2417. [Google Scholar] [CrossRef]
- Arrenberg, A.B.; Stainier, D.Y.; Baier, H.; Huisken, J. Optogenetic control of cardiac function. Science 2010, 330, 971–974. [Google Scholar] [CrossRef]
- Tessadori, F.; van Weerd, J.H.; Burkhard, S.B.; Verkerk, A.O.; de Pater, E.; Boukens, B.J.; Vink, A.; Christoffels, V.M.; Bakkers, J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PloS one 2012, 7, e47644. [Google Scholar] [CrossRef]
- Gittenberger-de Groot, A.C.; Vrancken Peeters, M.P.; Bergwerff, M.; Mentink, M.M.; Poelmann, R.E. Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circ. Res. 2000, 87, 969–971. [Google Scholar] [CrossRef]
- Manner, J. Experimental study on the formation of the epicardium in chick embryos. Anat. Embryol. (Berl) 1993, 187, 281–289. [Google Scholar] [CrossRef]
- Icardo, J.M.; Guerrero, A.; Duran, A.C.; Colvee, E.; Domezain, A.; Sans-Coma, V. The development of the epicardium in the sturgeon Acipenser naccarii. Anat. Rec. (Hoboken) 2009, 292, 1593–1601. [Google Scholar]
- Kim, J.; Rubin, N.; Huang, Y.; Tuan, T.L.; Lien, C.L. In vitro culture of epicardial cells from adult zebrafish heart on a fibrin matrix. Nat. Protoc. 2012, 7, 247–255. [Google Scholar] [CrossRef]
- Rudat, C.; Kispert, A. Wt1 and epicardial fate mapping. Circ. Res. 2012, 111, 165–169. [Google Scholar] [CrossRef]
- Wang, J.; Panakova, D.; Kikuchi, K.; Holdway, J.E.; Gemberling, M.; Burris, J.S.; Singh, S.P.; Dickson, A.L.; Lin, Y.F.; Sabeh, M.K.; et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 2011, 138, 3421–3430. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Peralta, M.; González-Rosa, J.M.; Marques, I.J.; Mercader, N. The Epicardium in the Embryonic and Adult Zebrafish. J. Dev. Biol. 2014, 2, 101-116. https://doi.org/10.3390/jdb2020101
Peralta M, González-Rosa JM, Marques IJ, Mercader N. The Epicardium in the Embryonic and Adult Zebrafish. Journal of Developmental Biology. 2014; 2(2):101-116. https://doi.org/10.3390/jdb2020101
Chicago/Turabian StylePeralta, Marina, Juan Manuel González-Rosa, Inês Joao Marques, and Nadia Mercader. 2014. "The Epicardium in the Embryonic and Adult Zebrafish" Journal of Developmental Biology 2, no. 2: 101-116. https://doi.org/10.3390/jdb2020101




