Direct Identification of Functional Amyloid Proteins by Label-Free Quantitative Mass Spectrometry
Abstract
:1. Introduction
2. Results
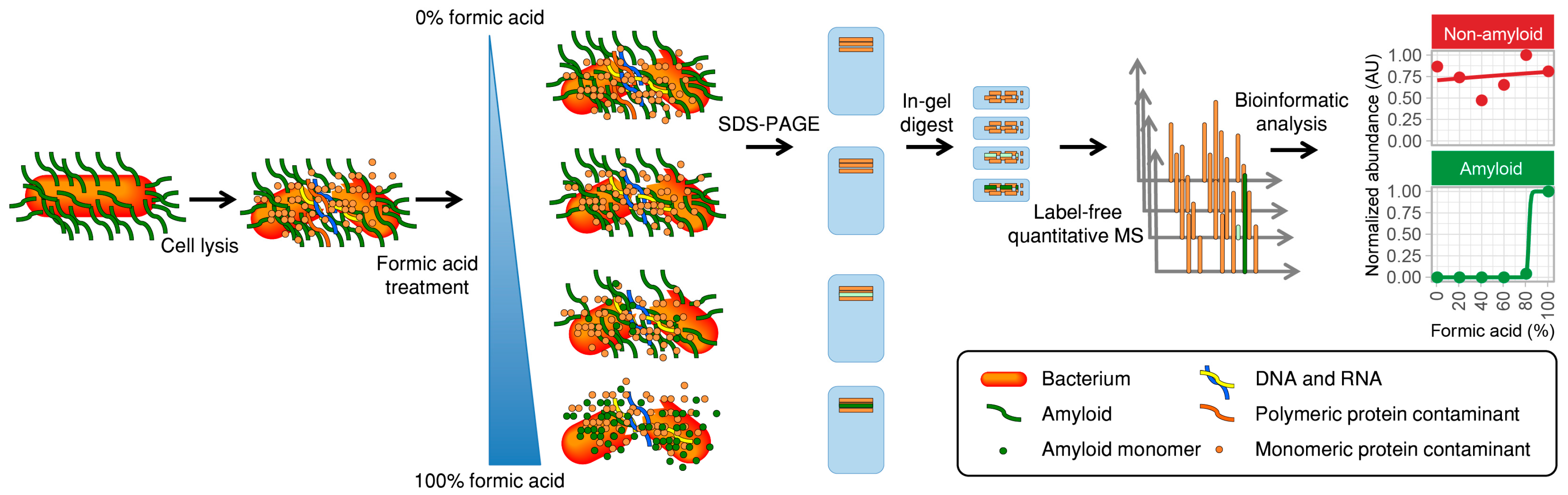
2.1. Description of the Method
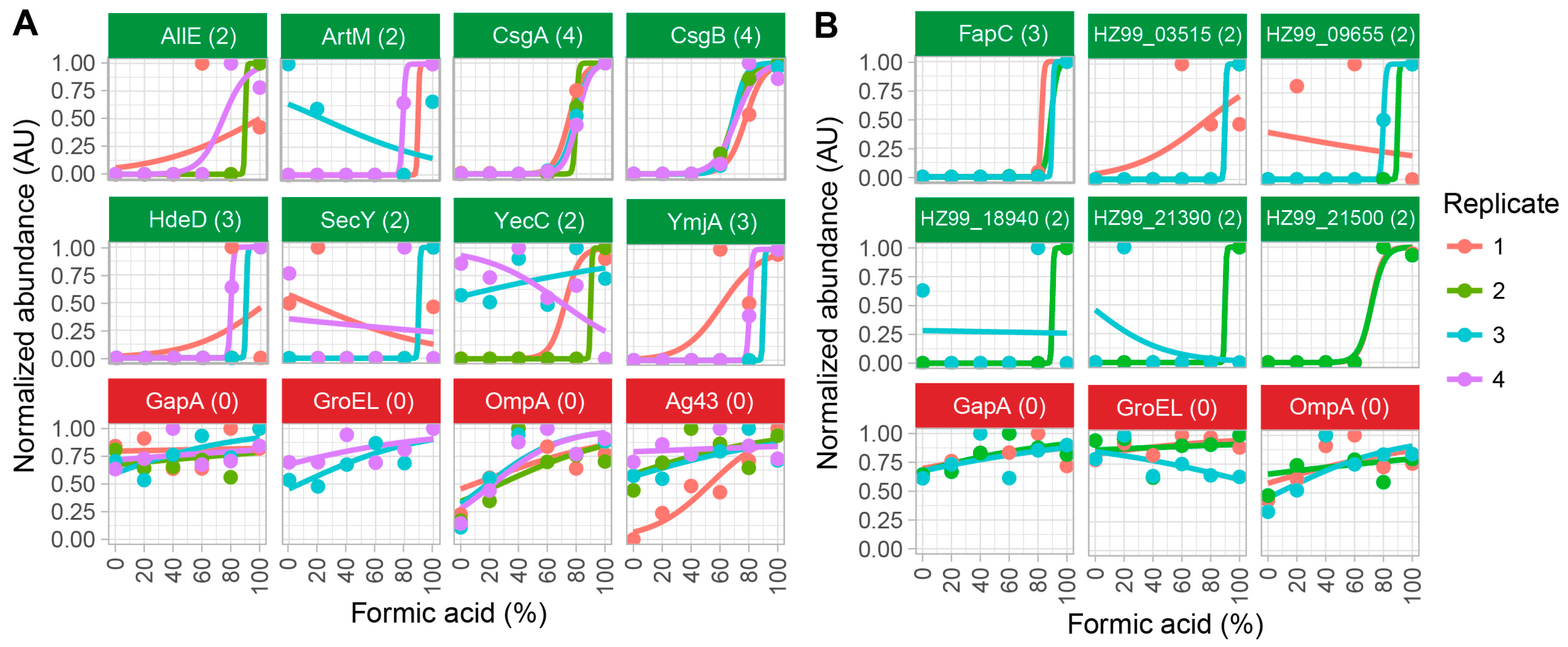
2.2. Method Validation with Cell Lysates of E. coli SM2258
2.3. Method Validation with Cell Lysates of Pseudomonas sp. UK4
3. Discussion
4. Materials and Methods
4.1. Bacterial Cultures and Activated Sludge
4.2. Sample Preparation for Mass Spectrometry
4.3. Mass Spectrometry
4.4. Data Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fändrich, M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell. Mol. Life Sci. 2007, 64, 2066–2078. [Google Scholar] [CrossRef] [PubMed]
- Makin, O.S.; Atkins, E.; Sikorski, P.; Johansson, J.; Serpell, L.C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. USA 2005, 102, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Blanco, L.P.; Evans, M.L.; Smith, D.R.; Badtke, M.P.; Chapman, M.R. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012, 20, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Nielsen, P.H. Amyloids—A neglected child of the slime. In The Perfect Slime: Microbial Extracellular Polymeric Substances (EPS); Flemming, H.-C., Neu, T.R., Wingender, J., Eds.; IWA Publishing: London, UK, 2016; pp. 113–133. ISBN 9781780407425. [Google Scholar]
- Nguyen, P.Q.; Botyanszki, Z.; Tay, P.K.R.; Joshi, N.S. Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 2014, 5, 4945. [Google Scholar] [CrossRef] [PubMed]
- Wösten, H.A.B.; Scholtmeijer, K. Applications of hydrophobins: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.E.L.; Henneberger, R.; Huber, H.; Moissl-Eichinger, C. Microbial syntrophy: Interaction for the common good. FEMS Microbiol. Rev. 2013, 37, 384–406. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Malone, M.; Bjarnsholt, T.; McBain, A.J.; James, G.A.; Stoodley, P.; Leaper, D.; Tachi, M.; Schultz, G.; Swanson, T.; Wolcott, R.D. The prevalence of biofilms in chronic wounds: A systematic review and meta-analysis of published data. J. Wound Care 2017, 26, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.; Nielsen, J.L.; Dueholm, M.S.; Wetzel, R.; Otzen, D.; Nielsen, P.H. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 2007, 9, 3077–3090. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.; Nielsen, J.L.; Otzen, D.; Nielsen, P.H. Amyloid-like adhesins produced by floc-forming and filamentous bacteria in activated sludge. Appl. Environ. Microbiol. 2008, 74, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Taglialegna, A.; Lasa, I.; Valle, J. Amyloid structures as biofilm matrix scaffolds. J. Bacteriol. 2016, 198, 2579–2588. [Google Scholar] [CrossRef] [PubMed]
- Wiehlmann, L.; Munder, A.; Adams, T.; Juhas, M.; Kolmar, H.; Salunkhe, P.; Tummler, B. Functional genomics of Pseudomonas aeruginosa to identify habitat-specific determinants of pathogenicity. Int. J. Med. Microbiol. 2007, 297, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Collinson, S.K.; Emody, L.; Muller, K.H.; Trust, T.J.; Kay, W.W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 1991, 173, 4773–4781. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Petersen, S.V.; Sønderkaer, M.; Larsen, P.; Christiansen, G.; Hein, K.L.; Enghild, J.J.; Nielsen, J.L.; Nielsen, K.L.; Nielsen, P.H.; et al. Functional amyloid in Pseudomonas. Mol. Microbiol. 2010, 77, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Claessen, D.; Rink, R.; de Jong, W.; Siebring, J.; de Vreugd, P.; Boersma, F.G.; Dijkhuizen, L.; Wosten, H.A. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003, 17, 1714–1726. [Google Scholar] [CrossRef] [PubMed]
- Jordal, P.B.; Dueholm, M.S.; Larsen, P.; Petersen, S.V.; Enghild, J.J.; Christiansen, G.; Hojrup, P.; Nielsen, P.H.; Otzen, D.E. Widespread abundance of functional bacterial amyloid in mycolata and other Gram-positive bacteria. Appl. Environ. Microbiol. 2009, 75, 4101–4110. [Google Scholar] [CrossRef] [PubMed]
- Cegelski, L.; Pinkner, J.S.; Hammer, N.D.; Cusumano, C.K.; Hung, C.S.; Chorell, E.; Åberg, V.; Walker, J.N.; Seed, P.C.; Almqvist, F.; et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 2009, 5, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Søndergaard, M.T.; Nilsson, M.; Christiansen, G.; Stensballe, A.; Overgaard, M.T.; Givskov, M.; Tolker-Nielsen, T.; Otzen, D.E.; Nielsen, P.H. Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen 2013, 2, 365–382. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Gerstel, U.; Romling, U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 2003, 154, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Albertsen, M.; Otzen, D.; Nielsen, P.H. Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS ONE 2012, 7, e51274. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Otzen, D.; Nielsen, P.H. Evolutionary insight into the functional amyloids of the pseudomonads. PLoS ONE 2013, 8, e76630. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.L.; Chorell, E.; Taylor, J.D.; Åden, J.; Götheson, A.; Li, F.; Koch, M.; Sefer, L.; Matthews, S.J.; Wittung-Stafshede, P.; et al. The bacterial curli system possesses a potent and selective inhibitor of amyloid formation. Mol. Cell. 2015, 57, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Stenvang, M.; Dueholm, M.S.; Vad, B.S.; Seviour, T.; Zeng, G.; Geifman-Shochat, S.; Søndergaard, M.T.; Christiansen, G.; Meyer, R.L.; Kjelleberg, S.; et al. Epigallocatechin gallate remodels overexpressed functional amyloids in Pseudomonas aeruginosa and increases biofilm susceptibility to antibiotic treatment. J. Biol. Chem. 2016, 291, 26540–26553. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Aguilar, C.; Losick, R.; Kolter, R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 2230–2234. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Danielsen, H.N.; Nielsen, P.H. Complete genome sequence of Pseudomonas sp. UK4, a model organism for studies of functional amyloids in Pseudomonas. Genome Announc. 2014, 2, e00898-14. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Vizcaino, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017, 45, D12–D17. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danielsen, H.N.; Hansen, S.H.; Herbst, F.-A.; Kjeldal, H.; Stensballe, A.; Nielsen, P.H.; Dueholm, M.S. Direct Identification of Functional Amyloid Proteins by Label-Free Quantitative Mass Spectrometry. Biomolecules 2017, 7, 58. https://doi.org/10.3390/biom7030058
Danielsen HN, Hansen SH, Herbst F-A, Kjeldal H, Stensballe A, Nielsen PH, Dueholm MS. Direct Identification of Functional Amyloid Proteins by Label-Free Quantitative Mass Spectrometry. Biomolecules. 2017; 7(3):58. https://doi.org/10.3390/biom7030058
Chicago/Turabian StyleDanielsen, Heidi N., Susan H. Hansen, Florian-Alexander Herbst, Henrik Kjeldal, Allan Stensballe, Per H. Nielsen, and Morten S. Dueholm. 2017. "Direct Identification of Functional Amyloid Proteins by Label-Free Quantitative Mass Spectrometry" Biomolecules 7, no. 3: 58. https://doi.org/10.3390/biom7030058






