Inhibition of Bacterial RNase P RNA by Phenothiazine Derivatives
Abstract
:1. Introduction
2. Results
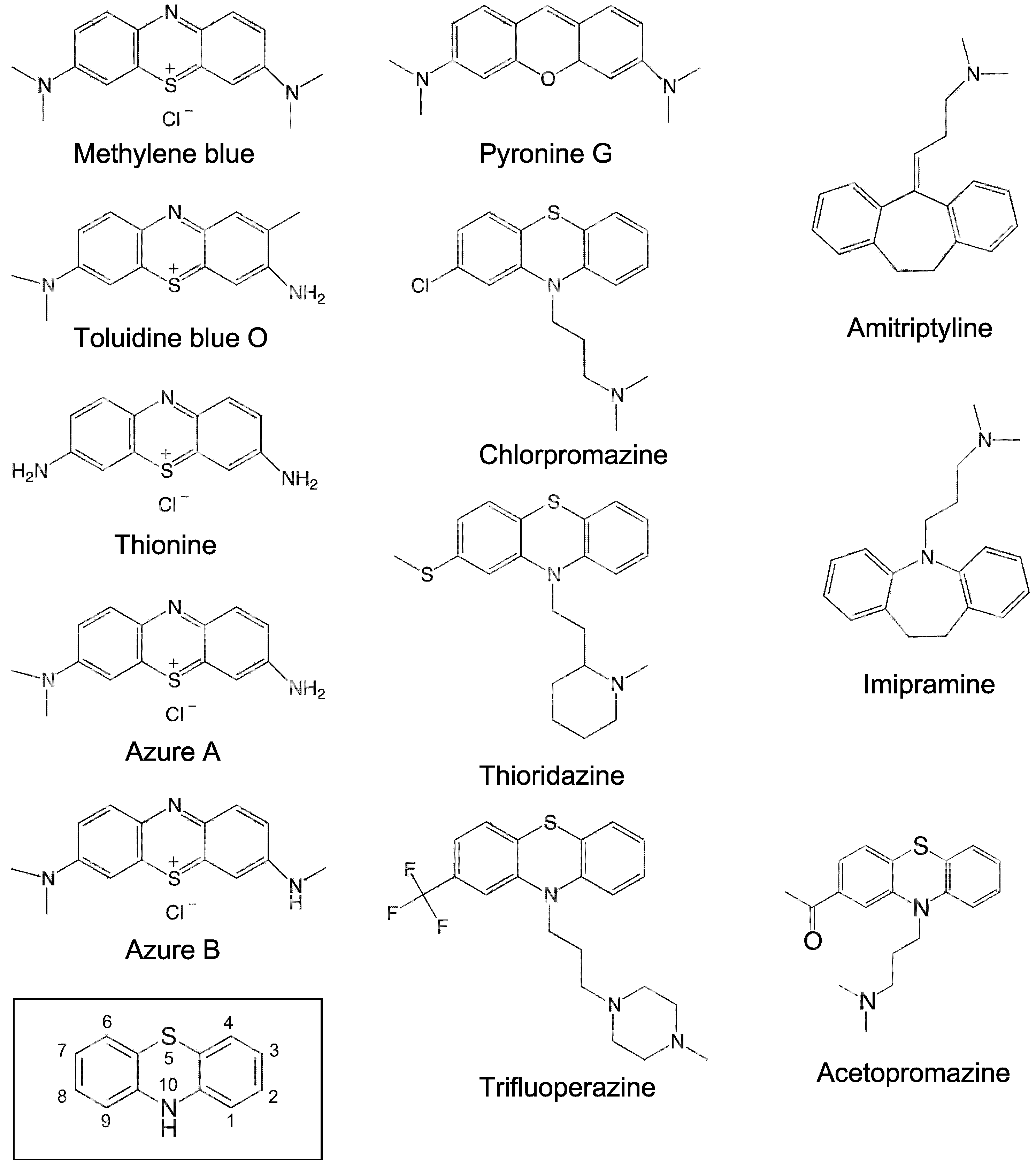
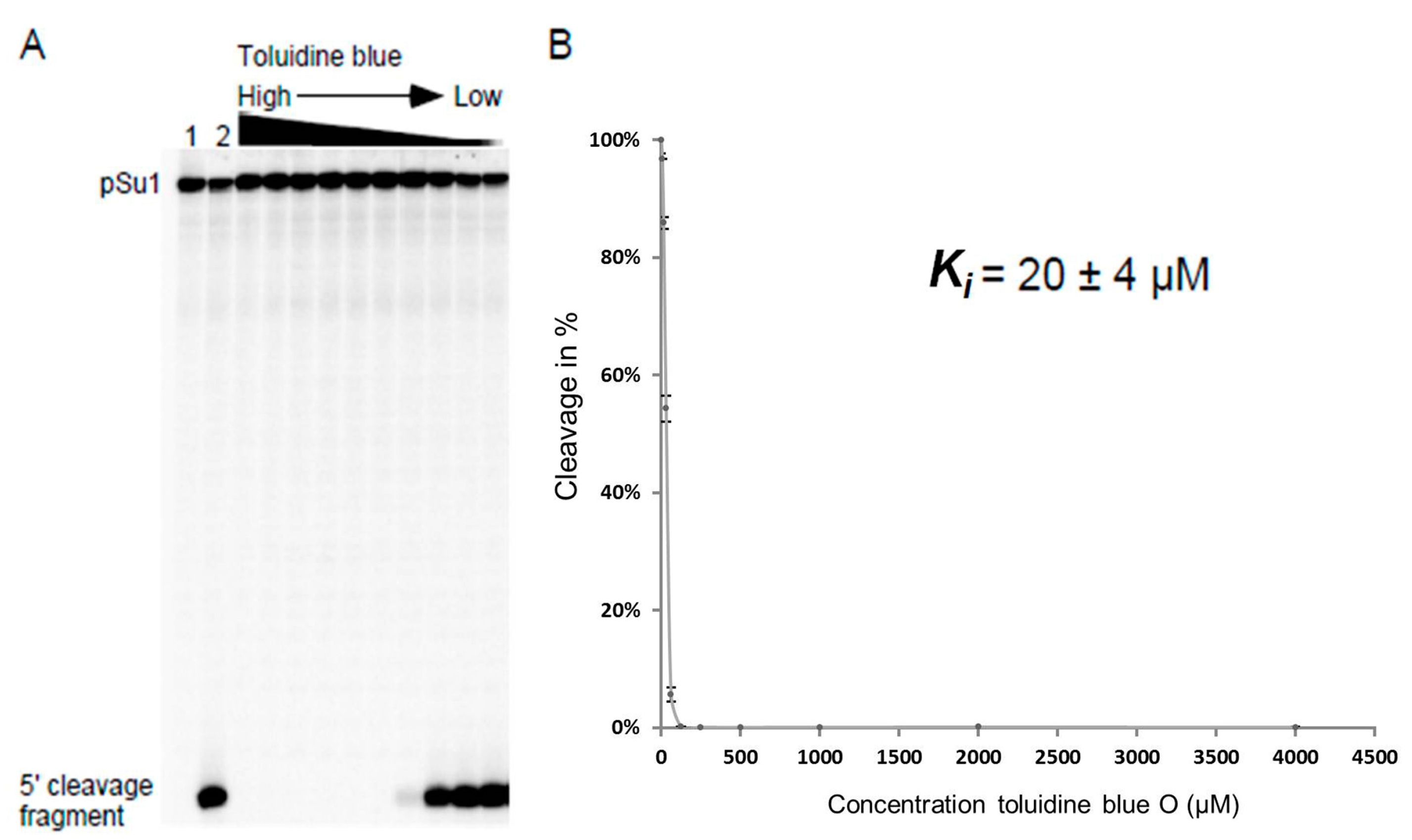
2.1. Inhibition of RNase P RNA Cleavage by Phenothiazine Derivatives
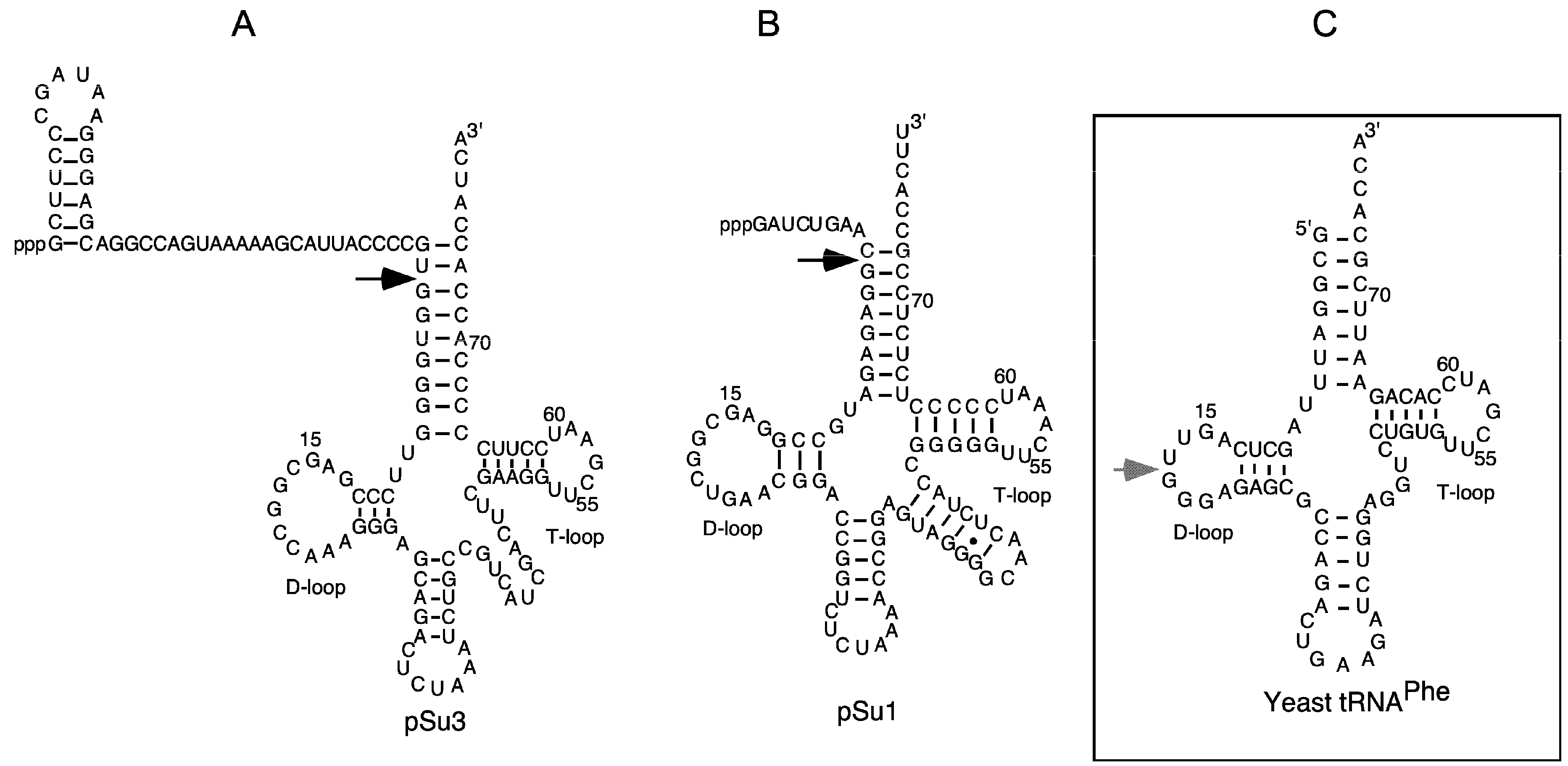
2.2. Lead(II)-Induced Cleavage of Yeast tRNAPhe, P15 RNA, and Eco RPR
2.3. Inhibition of Bacterial RPR Activities by Antipsychotic Phenothiazine Variants
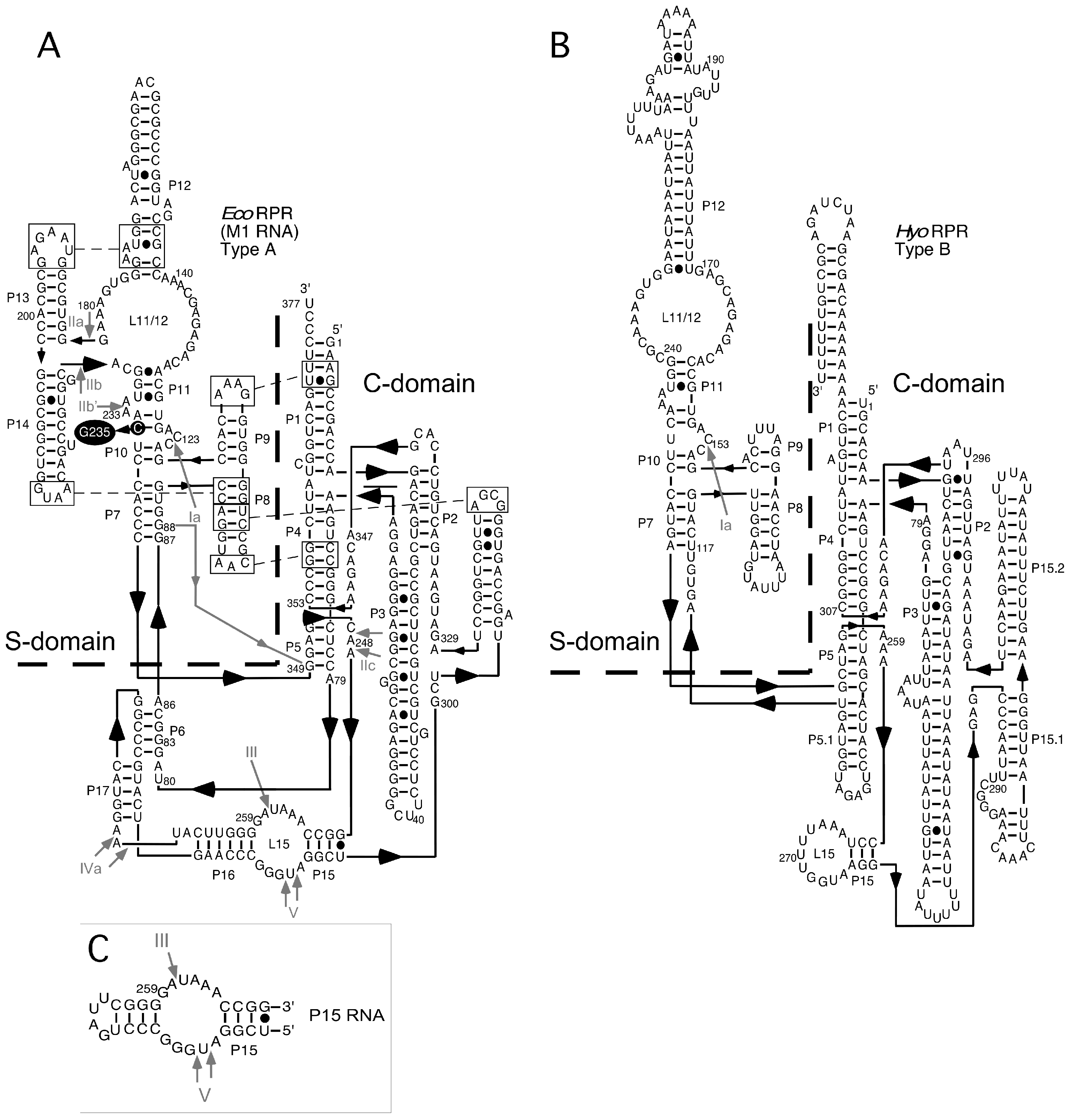
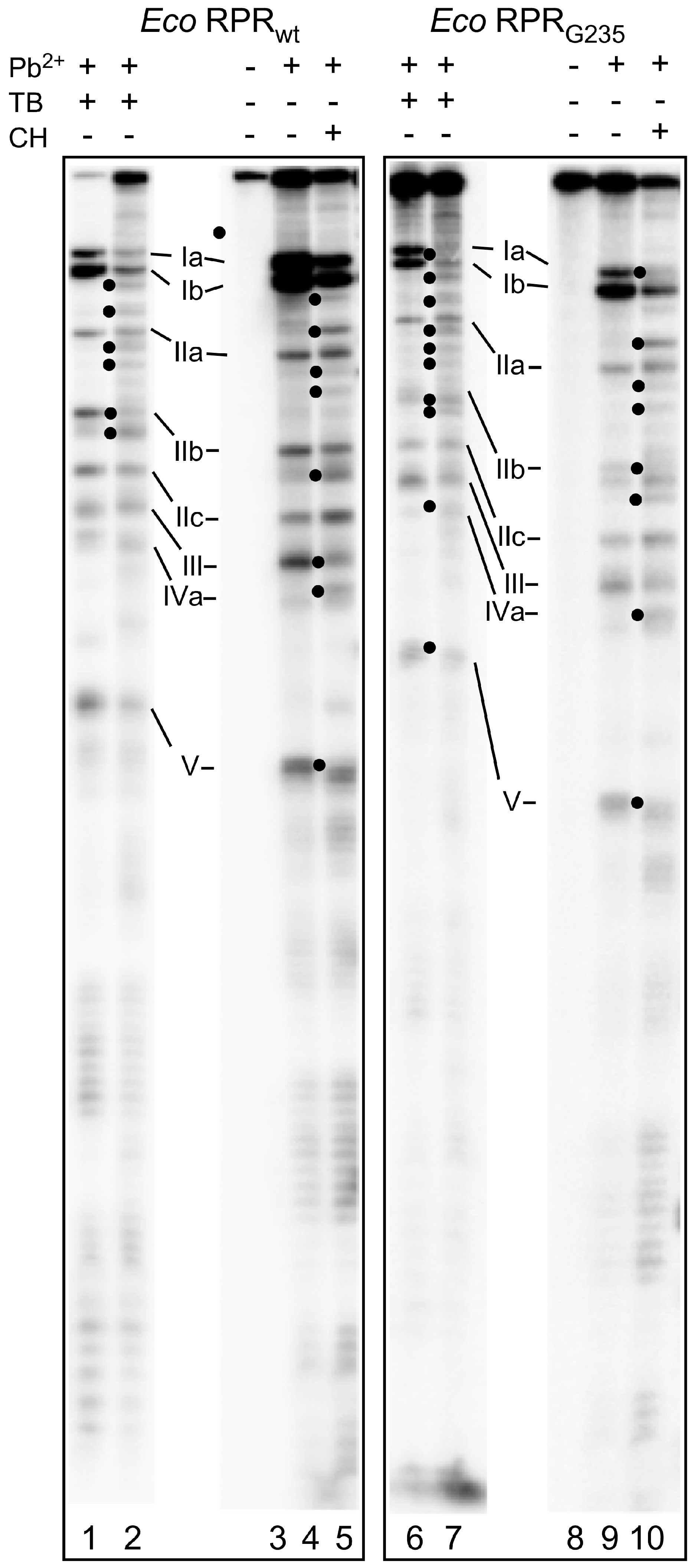
2.4. Binding of Phenothiazine Affects the Structure of Eco RPR
2.5. Phenothiazine Inhibition of Structurally Altered Eco RPR
3. Discussion
3.1. Comparison with Aminoglycoside Interaction with RNA
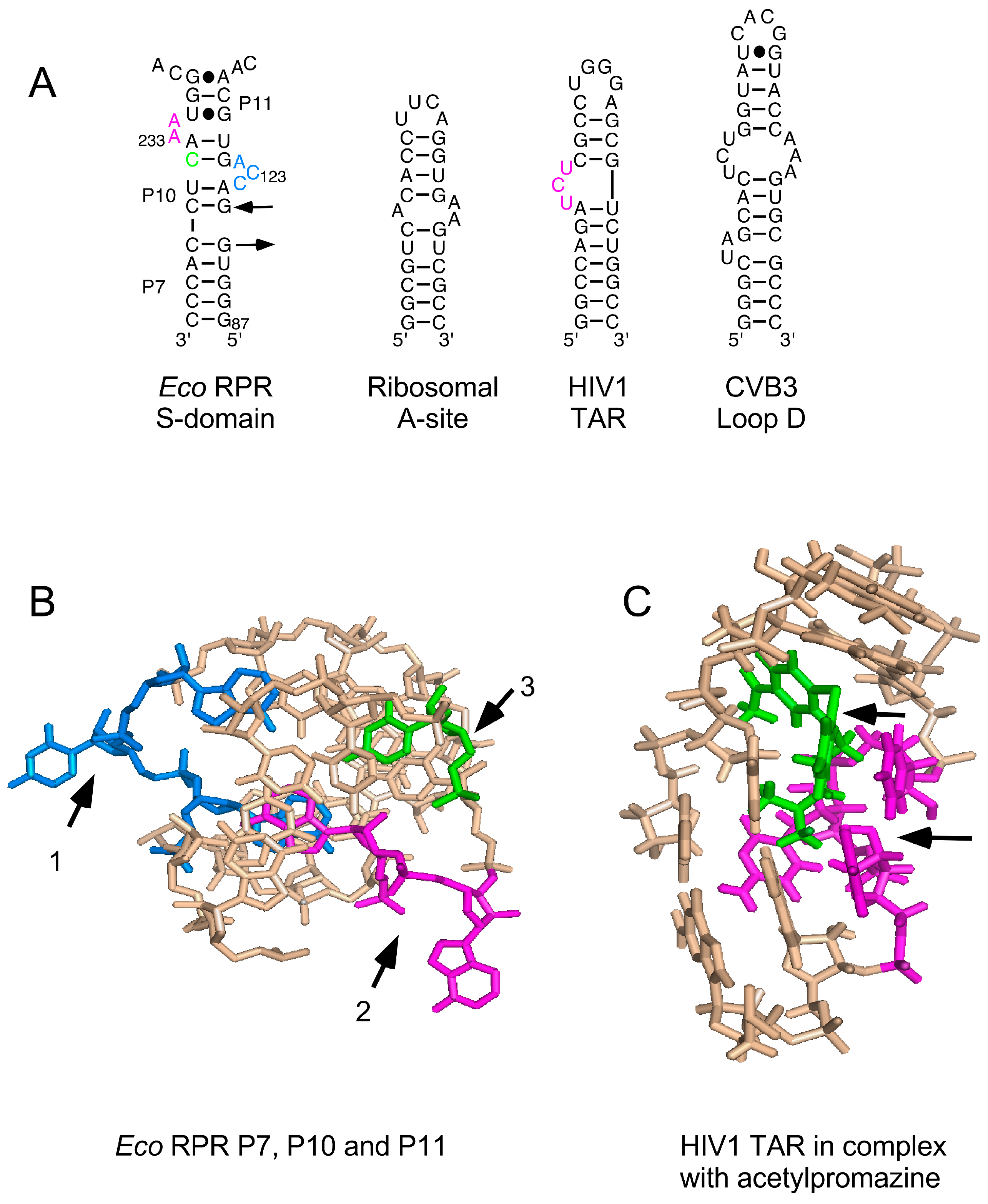
3.2. Phenothiazines Interact Preferentially with Internal RNA Bulges
3.3. Phenothiazine Interaction with the S Domain and the Pre-tRNA D/T Loop
3.4. Phenothiazine Interaction with the C Domain and the Pre-tRNA D/T Loop
3.5. Phenothiazine, a Scaffold with Potential
4. Materials and Methods
4.1. Preparation of RNA
4.2. Preparation of Compounds
4.3. RNase P Assay Conditions
4.4. Lead(II)-Ion Induced Cleavage of Yeast tRNAPhe, Eco RPR and P15 RNA
4.5. Determination of the Inhibition Constant Ki
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kirsebom, L.A.; Trobro, S. RNase P RNA-mediated cleavage. IUBMB Life 2009, 61, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Altman, S. Ribonuclease P (Protein Reviews); Springer: New York, NY, USA; Dordrecht, The Netherlands; Heidelberg, Germany; London, UK, 2010; Volume 10. [Google Scholar]
- Lai, L.B.; Vioque, A.; Kirsebom, L.A.; Gopalan, V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: Challenges and prospects. FEBS Lett. 2010, 584, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Bartkiewicz, M.; Gold, H.; Altman, S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989, 3, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N.; Gopalan, V. Archaeal/eukaryal RNase P: Subunits, functions and RNA diversification. Nucl. Acids Res. 2010, 38, 7885–7894. [Google Scholar] [CrossRef] [PubMed]
- Kirsebom, L.A. RNase P and its substrate. In The Many Faces of RNA; Eggelston, D.S., Prescott, C.D., Pearson, N.D., Eds.; Academic Press: San Diego, CA, USA; London, UK, 1998; pp. 127–144. [Google Scholar]
- Kirsebom, L.A. The structure and function of the ribozyme RNase P RNA is dictated by magnesium(II) ions. In RNA Biochemistry and Biotechnology; Barciszewski, J., Clark, B.F.C., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1999; pp. 89–109. [Google Scholar]
- Kirsebom, L.A.; Svärd, S.G. RNase P processing of tRNA precursors. In Ribozyme Biochemistry and Biotechnology; Krupp, G., Gaur, R.K., Eds.; Eton Publishing: Natick, MA, USA, 2000; pp. 111–131. [Google Scholar]
- Kirsebom, L.A.; Virtanen, A. Inhibition of RNase P processing. In RNA-Binding Antibiotics; Schroeder, R., Wallis, M.G., Eds.; Landes Bioscience: Georgetown, TX, USA, 2001; pp. 56–72. [Google Scholar]
- Eder, P.S.; Hatfield, C.; Vioque, A.; Gopalan, V. Bacterial RNase P as a potential target for novel anti-infectives. Curr. Opin. Investig. Drugs 2003, 4, 937–943. [Google Scholar] [PubMed]
- Kirsebom, L.A.; Virtanen, A.; Mikkelsen, N.E. Aminoglycoside interactions with RNAs and nucleases. Handb. Exp. Pharmacol. 2006, 173, 73–96. [Google Scholar] [PubMed]
- Willkomm, D.K.; Pfeffer, P.; Reuter, K.; Klebe, G.; Hartmann, R.K. RNase P as a drug target. In Ribonuclease P; Liu, F., Altman, S., Eds.; Springer: New York, NY, USA; Dordrecht, The Netherlands; Heidelberg, Germany; London, UK, 2010; Volume 10, pp. 235–256. [Google Scholar]
- Vioque, A. Protein synthesis inhibitors and catalytic RNA. Effect of puromycin on tRNA precursor processing by the RNA component of Escherichia coli RNase P. FEBS Lett. 1989, 246, 137–139. [Google Scholar] [CrossRef]
- Mikkelsen, N.-E.; Brännvall, M.; Virtanen, A.; Kirsebom, L.A. Inhibition of RNase P RNA cleavage by aminoglycosides. Proc. Natl. Acad. Sci. USA 1999, 96, 6155–6160. [Google Scholar] [CrossRef] [PubMed]
- Eubank, T.D.; Biswas, R.; Jovanovic, M.; Litovchick, A.; Lapidot, A.; Gopalan, V. Inhibition of bacterial RNase P by aminoglycosides-arginine conjugates. FEBS Lett. 2002, 511, 107–112. [Google Scholar] [CrossRef]
- Kawamoto, S.A.; Sudhahar, C.G.; Hatfield, C.L.; Sun, J.; Behrman, E.J.; Gopalan, V. Studies on the mechanism of inhibition of bacterial ribonuclease P by aminoglycoside derivatives. Nucleic Acids Res. 2008, 36, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, N.-E.; Johansson, K.; Virtanen, A.; Kirsebom, L.A. Aminoglycoside binding displaces a divalent metal ion in a tRNA-neomycin B complex. Nat. Struct. Biol. 2001, 8, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Papadimou, E.; Georgiou, S.; Tsambaos, D.; Drainas, D. Inhibition of ribonuclease P activity by retinoids. J. Biol. Chem. 1998, 273, 24375–24378. [Google Scholar] [CrossRef] [PubMed]
- Tekos, A.; Tsagla, A.; Stathopoulos, C.; Drainas, D. Inhibition of eukaryotic ribonuclease P activity by aminoglycosides: Kinetic studies. FEBS Lett. 2000, 485, 71–75. [Google Scholar] [CrossRef]
- Schirmer, R.H.; Adler, H.; Pickhardt, M.; Mandelkow, E. “Lest we forget you—Methylene blue…”. Neurobiol. Aging 2011, 32, e7–e16. [Google Scholar]
- Spicer, S.S. Differentiation of nucleic acids by staining at controlled pH and by a Schiff-methylene blue sequence. Strain Technol. 1961, 36, 337–340. [Google Scholar] [CrossRef]
- Wilkinson, M.; Doskow, J.; Lindsey, S. RNA blots: Staining procedures and optimization of conditions. Nucleic Acids Res. 1991, 19, 679. [Google Scholar] [CrossRef] [PubMed]
- Haas, E.S.; Brown, J.W. Evolutionary variation in bacterial RNase P RNAs. Nucleic Acids Res. 1998, 26, 4093–4099. [Google Scholar] [CrossRef] [PubMed]
- Brännvall, M.; Kikovska, E.; Wu, S.; Kirsebom, L.A. Evidence for induced fit in bacterial RNase P RNA-mediated cleavage. J. Mol. Biol. 2007, 372, 1149–1164. [Google Scholar] [CrossRef] [PubMed]
- Svärd, S.G.; Mattsson, J.G.; Johansson, K.-E.; Kirsebom, L.A. Cloning and characterization of the RNase P RNA genes from two porcine mycoplasmas. Mol. Microbiol. 1994, 11, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Svärd, S.G.; Kagardt, U.; Kirsebom, L.A. Phylogenetic comparative mutational analysis of the base-pairing between RNase P RNA and its substrate. RNA 1996, 2, 463–472. [Google Scholar] [PubMed]
- Dooley, K.E.; Phillips, P.P.; Nahid, P.; Hoelscher, M. Challenges in the clinical assessment of novel tuberculosis drugs. Adv. Drug Deliv. Rev. 2016, 102, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Massire, C.; Jaeger, L.; Westhof, E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998, 279, 773–793. [Google Scholar] [CrossRef] [PubMed]
- Kikovska, E.; Wu, S.; Mao, G.; Kirsebom, L.A. Cleavage mediated by the P15 domain of bacterial RNase P RNA. Nucleic Acids Res. 2012, 40, 2224–2233. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Kikovska, E.; Lindell, M.; Kirsebom, L.A. Cleavage mediated by the catalytic domain of bacterial RNase P RNA. J. Mol. Biol. 2012, 422, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Kufel, J.; Kirsebom, L.A. The P15-loop of Escherichia coli RNase P RNA is an autonomous divalent metal ion binding domain. RNA 1998, 4, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Glemarec, C.; Kufel, J.; Földesi, A.; Maltseva, T.; Sandström, A.; Kirsebom, L.A.; Chattopadhyaya, J. The NMR structure of 31mer RNA domain of Escherichia coli RNase P RNA using its non-uniformity deuterium labelled counterpart [the ‘NMR-window’ concept]. Nucleic Acids Res. 1996, 24, 2022–2035. [Google Scholar] [CrossRef] [PubMed]
- Reiter, N.J.; Osterman, A.; Torres-Larios, A.; Swinger, K.K.; Pan, T.; Mondragón, A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 2010, 468, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Lind, K.E.; Du, Z.; Fujinaga, K.; Peterlin, B.M.; James, T.L. Structure-based computational database screening in vitro assay, and NMR assessment of compounds that target TAR RNA. Chem. Biol. 2002, 9, 185–193. [Google Scholar] [CrossRef]
- Du, A.; Lind, K.E.; James, T.L. Structure of TAR RNA complexed with a Tat-TAR interaction nanomolar inhibitor that was identified by computational screening. Chem. Biol. 2002, 9, 707–712. [Google Scholar] [CrossRef]
- Mayer, M.; Lang, T.; Gerber, S.; Madrid, P.B.; Gómez Pinto, I.; Guy, R.K.; James, T.L. Synthesis and testing of a focused phenothiazine library for binding to HIV-1 TAR RNA. Chem. Biol. 2006, 13, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; James, T.L. NMR-based characterization of phenothiazines as a RNA binding scaffold. J. Am. Chem. Soc. 2004, 126, 4453–4460. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, J.E.; Dastidar, S.G.; Palchoudhuri, S.; Sinha Roy, D.; Das, S.; Hendricks, O.; Christensen, J.B. Phenothiazines as a solution for multidrug resistant tuberculosis: From the origin to present. Int. Microbiol. 2015, 18, 1–12. [Google Scholar] [PubMed]
- Hermann, T.; Westhof, E. Aminoglycoside binding to the hammerhead ribozyme: A general model for the interaction of cationic antibiotics with RNA. J. Mol. Biol. 1998, 276, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.R.; Hergenrother, P.J. Targeting RNA with small molecules. Chem. Rev. 2008, 108, 1172–1224. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.; Barbieri, C.M.; Pilch, D.S. Defining the basis for the specificity of aminoglycoside-rRNA recognition: A comparative study of drug binding to the A sites of Escherichia coli and Human rRNA. J. Mol. Biol. 2005, 346, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Pitt, S.W.; Zhang, Q.; Patel, D.J.; Al-Hashimi, H.M. Evidence that electrostatic interactions dictate the ligand-induced arrest of RNA global flexibility. Angew. Chem. Int. Ed. 2005, 44, 3412–3415. [Google Scholar] [CrossRef] [PubMed]
- Krasilnikov, A.S.; Xiao, Y.; Pan, T.; Mondragón, A. Basis for structural diversity in homologous RNAs. Science 2004, 306, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Lindell, M.; Brännvall, M.; Wagner, E.G.H.; Kirsebom, L.A. Lead(II) cleavage analysis of RNase P RNA in vivo. RNA 2005, 11, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Tallsjö, A.; Kufel, J.; Kirsebom, L.A. A novel tertiary interaction in M1 RNA, the catalytic subunit of Escherichia coli RNase P. Nucleic Acids Res. 1993, 21, 3927–3933. [Google Scholar] [CrossRef] [PubMed]
- Zito, K.; Hüttenhofer, A.; Pace, N.R. Lead-catalyzed cleavage of ribonuclease P RNA as a probe for integrity of tertiary interaction. Nucleic Acids Res. 1993, 21, 5916–5920. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.S.; Dewan, J.C.; Klug, A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry 1985, 24, 4785–4801. [Google Scholar] [CrossRef] [PubMed]
- Behlen, L.S.; Sampson, J.R.; DiRenzo, A.B.; Uhlenbeck, O.C. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry 1990, 29, 2515–2523. [Google Scholar] [CrossRef] [PubMed]
- Loria, A.; Pan, T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry 1997, 36, 6317–6325. [Google Scholar] [CrossRef] [PubMed]
- Ciesiolka, J.; Hardt, W.D.; Schlegl, J.; Erdmann, V.A.; Hartmann, R.K. Lead-ion-induced cleavage of RNase P RNA. Eur. J. Biochem. 1994, 219, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Brännvall, M.; Mikkelsen, N.-E.; Kirsebom, L.A. Monitoring the structure of Escherichia coli RNase P RNA in the presence of various divalent metal ions. Nucleic Acids Res. 2001, 29, 1426–1432. [Google Scholar] [CrossRef] [PubMed]
- Childs, J.L.; Poole, A.W.; Turner, D.H. Inhibition of Escherichia coli RNase P by oligonucleotide directed misfolding of RNA. RNA 2003, 9, 1437–1445. [Google Scholar] [CrossRef] [PubMed]
- Gruegelsiepe, H.; Willkommen, D.K.; Goudinakis, O.; Hartmann, R.K. Antisense inhibition of Escherichia coli RNase P RNA: Mechanistic aspects. ChemBioChem 2003, 4, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Gruegelsiepe, H.; Brandt, O.; Hartmann, R.K. Antisense inhibition of RNase P: Mechanistic aspects and application to live bacteria. J. Biol. Chem. 2006, 281, 30613–3060. [Google Scholar] [CrossRef] [PubMed]
- Ratnakar, P.; Murthy, P.S. Trifluoperazine inhibits the incorporation of labelled precursors into lipids, proteins and DNA of Mycobacterium tuberculosis H37RV. FEMS Microbiol. Lett. 1993, 110, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.O.; Austin, S.E.; Chan, C.; Yin, H.; Marks, D.; Vaghjiani, S.N.; Kendrick, H.; Yardley, V.; Croft, S.L.; Douglas, K.T. Use of an additional hydrophobic binding site, the Z site, in the rational drug design of a new class of stronger trypanothione reductase inhibitor, quarternary alkylammonium phenothiazines. J. Med. Chem. 2000, 43, 3148–3156. [Google Scholar] [CrossRef] [PubMed]
- Carfagna, M.A.; Muhoberac, B.B. Interaction of tricyclic drug analogs with synaptic plasma membranes: Structure-mechanism relationships in inhibition of neuronal Na+/K(+)-ATPase activity. Mol. Pharmacol. 1993, 44, 129–144. [Google Scholar] [PubMed]
- Chowdhury, A.K.; Rogers, H.; Skinner, A.; Spector, R.G.; Watts, D.C. The influence of psychotropic drugs on aldolase, mitochondrial malic dehydrogenase and Mg++Na+K+ adenosine triphosphate. Br. J. Pharmacol. 1969, 37, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Amaral, L.; Martins, A.; Molnar, J.; Kristiansen, J.E.; Martins, M.; Viveiros, M.; Rodrigues, L.; Spenglier, G.; Couto, I.; Ramos, J.; et al. Phenothiazines, bacterial efflux pumps and targeting the macrophage for enhanced killing of intracellular XDRTB. In Vivo 2010, 24, 409–424. [Google Scholar] [PubMed]
- Van Soolingen, D.; Hernandez-Pando, R.; Orozco, H.; Aguilar, D.; Magis-Escurra, C.; Amaral, L.; van Ingen, J.; Boeree, M.J. The antipsychotic thioridazine shows promising therapeutic activity in a mouse model of multidrug-resistant tuberculosis. PLoS ONE 2010, 5, e12640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbate, E.; Vescovo, M.; Natiello, M.; Cufré, M.; Garcia, A.; Gonzalez Montaner, P.; Ambroggi, M.; Ritacco, V.; van Soolingen, D. Successful alternative treatment of extensively drug-resistant tuberculosis in Argentina with a combination of linezolid, moxifloxacin and thioridazine. J. Antimicrob. Chemother. 2012, 67, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Varani, L.; Spillantini, M.G.; Goedert, M.; Varani, G. Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics. Nucleic Acids Res. 2000, 28, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Milligan, J.F.; Groebe, D.R.; Whiterell, G.W.; Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987, 15, 8783–8798. [Google Scholar] [CrossRef] [PubMed]
- Kirsebom, L.A.; Svärd, S.G. Identification of a region within M1 RNA of Escherichia coli RNase P important for the location of the site of cleavage on a wild-type tRNA precursor. J. Mol. Biol. 1993, 231, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Brännvall, M.; Kirsebom, L.A. Manganese ions induce miscleavage in the Escherichia coli RNase P RNA-catalyzed reaction. J. Mol. Biol. 1999, 292, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Vioque, A.; Arnez, J.; Altman, S. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J. Mol. Biol. 1988, 202, 835–848. [Google Scholar] [CrossRef]
- Kirsebom, L.A.; Ciesiolka, J. Pb2+-Induced Cleavage of RNA. In Handbook of RNA Biochemistry, 2nd ed.; Hartmann, R.K., Bindereif, A., Schön, A., Westhof, E., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2014; Volume 1, pp. 269–283. [Google Scholar]
| RPR-Mediated Cleavage | Lead(II)-Induced Cleavage | ||||
|---|---|---|---|---|---|
| Eco | Myc | Hyo | P15 RNA | Yeast tRNAPhe | |
| Methylene blue | 14 ± 2.6 | 28 ± 13 | 25 ± 1.2 | 62 ± 6.3 | 98 ± 20 |
| Toluidine blue O | 8.6 ± 1.7 | 12 ± 3.8 | 10 ± 4.9 | 14 ± 8 | 70 ± 20 |
| Thionine acid | 17 ± 0.33 | 8.3 ± 4.3 | 9.3 ± 3.3 | 41 ± 1.3 | 46 ± 51 |
| Azure A | 29 ± 9 | 21 ± 2.3 | 19 ± 0.67 | 28 ± 8.3 | 52 ± 8.3 |
| Azure B | 17 ± 9 | 12 ± 2 | 12 ± 2.7 | 37 ± 10 | 58 ± 28 |
| Pyronin G | 15 ± 1.7 | 44 ± 7.0 | 35 ± 8.6 | 20 ± 3 | 166 ± 42 |
| Thioridazine | 177 ± 23 | 67 ± 6.2 | 125 ± 31 | 80 ± 1.7 | ND |
| Trifluoperazine | 131 ± 22 | 106 ± 3.3 | 151 ± 26 | 132 ± 24 | ND |
| Chlorpromazine | 437 ± 33 | 161 ± 16 | 170 ± 9.2 | 133 ± 19 | ND |
| Eco RPR Variant | Toluidine Blue O (μM) | Chlorpromazine (μM) |
|---|---|---|
| Eco RPRwt | 20 ± 4 | 450 ± 40 |
| Eco RPRG235 | 17 ± 3 | 200 ± 30 |
| Eco CP RPR | 30 ± 5 | ND |
| Inhibitor | Conditions | Order of Addition | Ki Values (μM) |
|---|---|---|---|
| Toluidine blue O | Buffer I, pH 7.2 | RPR + I then S | 16 ± 2 |
| Toluidine blue O | Buffer II, pH 7.2 | RPR + I then S | 8.6 ± 0.9 |
| Toluidine blue O | Buffer II, pH 9.0 | RPR + I then S | 11 ± 2.6 |
| Toluidine blue O | Buffer II, pH 6.0 | RPR + I then S | 9.3 ± 0.67 |
| Toluidine blue O | Buffer II, pH 7.2 | RPR, I and S | 10 ± 1.8 |
| Toluidine blue O | Buffer II, pH 7.2 | S + I then RPR | 2.2 ± 0.4 |
| Thionine acid | Buffer I, pH 7.2 | RPR + I then S | 63 ± 6.8 |
| Thionine acid | Buffer II, pH 7.2 | RPR + I then S | 17 ± 0.3 |
| Thionine acid | Buffer II, pH 7.2 | RPR, I and S | 7.5 ± 3.5 |
| Thionine acid | Buffer II, pH 7.2 | S + I then RPR | 2.2 ± 0.4 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Mao, G.; Kirsebom, L.A. Inhibition of Bacterial RNase P RNA by Phenothiazine Derivatives. Biomolecules 2016, 6, 38. https://doi.org/10.3390/biom6030038
Wu S, Mao G, Kirsebom LA. Inhibition of Bacterial RNase P RNA by Phenothiazine Derivatives. Biomolecules. 2016; 6(3):38. https://doi.org/10.3390/biom6030038
Chicago/Turabian StyleWu, Shiying, Guanzhong Mao, and Leif A. Kirsebom. 2016. "Inhibition of Bacterial RNase P RNA by Phenothiazine Derivatives" Biomolecules 6, no. 3: 38. https://doi.org/10.3390/biom6030038





