RNA-Mediated Regulation of HMGA1 Function
Abstract
:1. Introduction
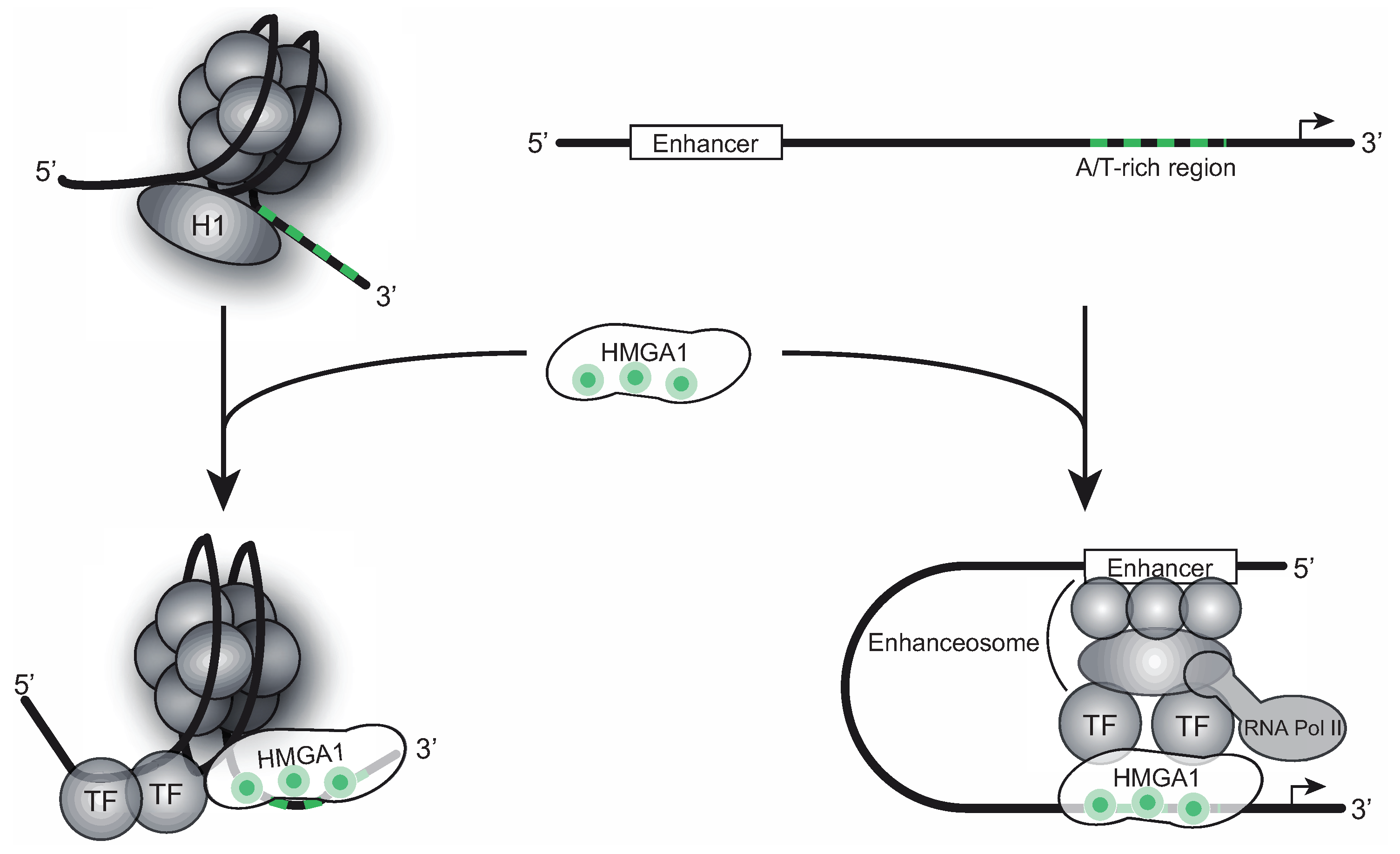
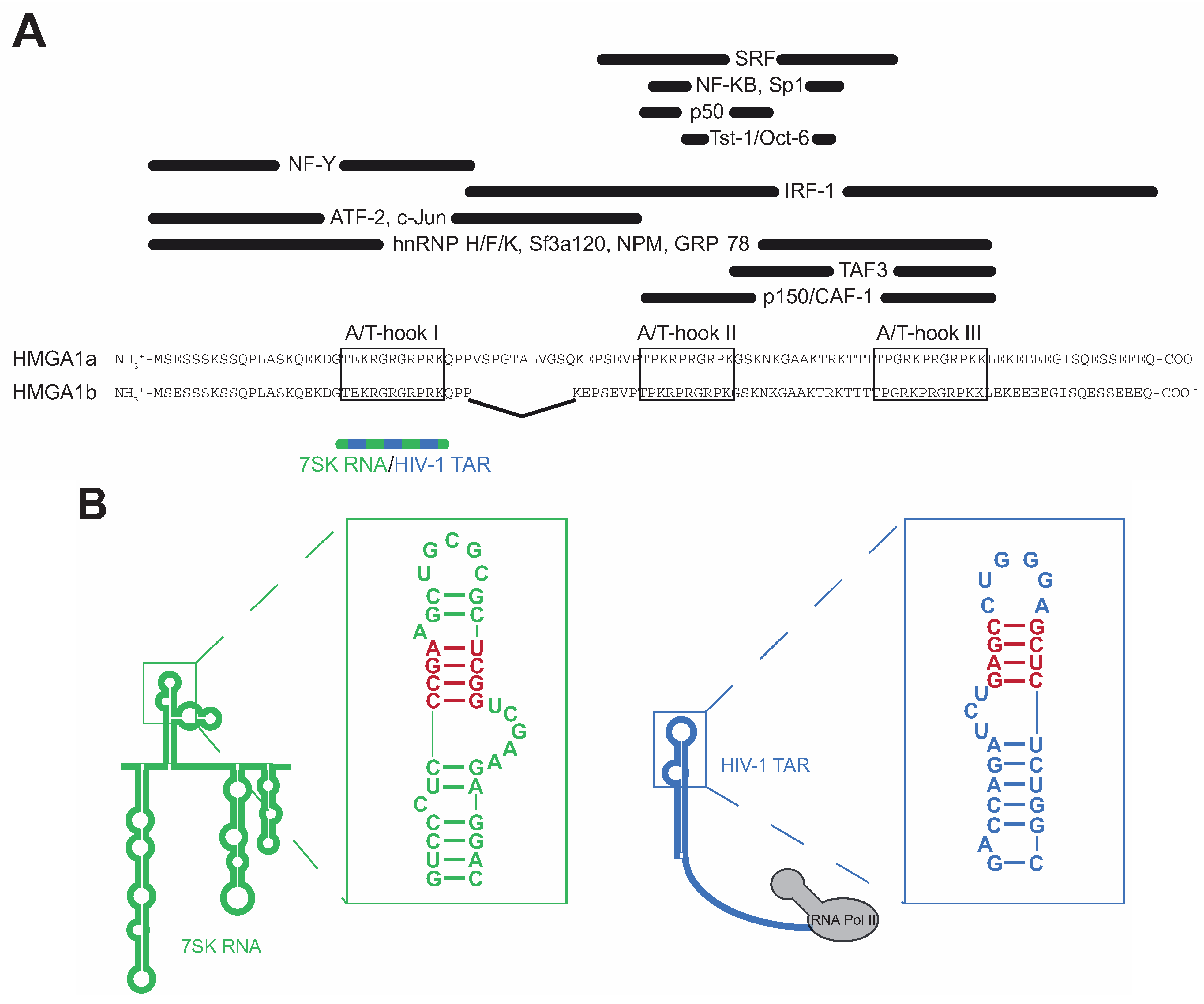
2. HMGA1-RNA Interactions from the Structural Point of View
3. HMGA1-Regulation by 7SK RNA
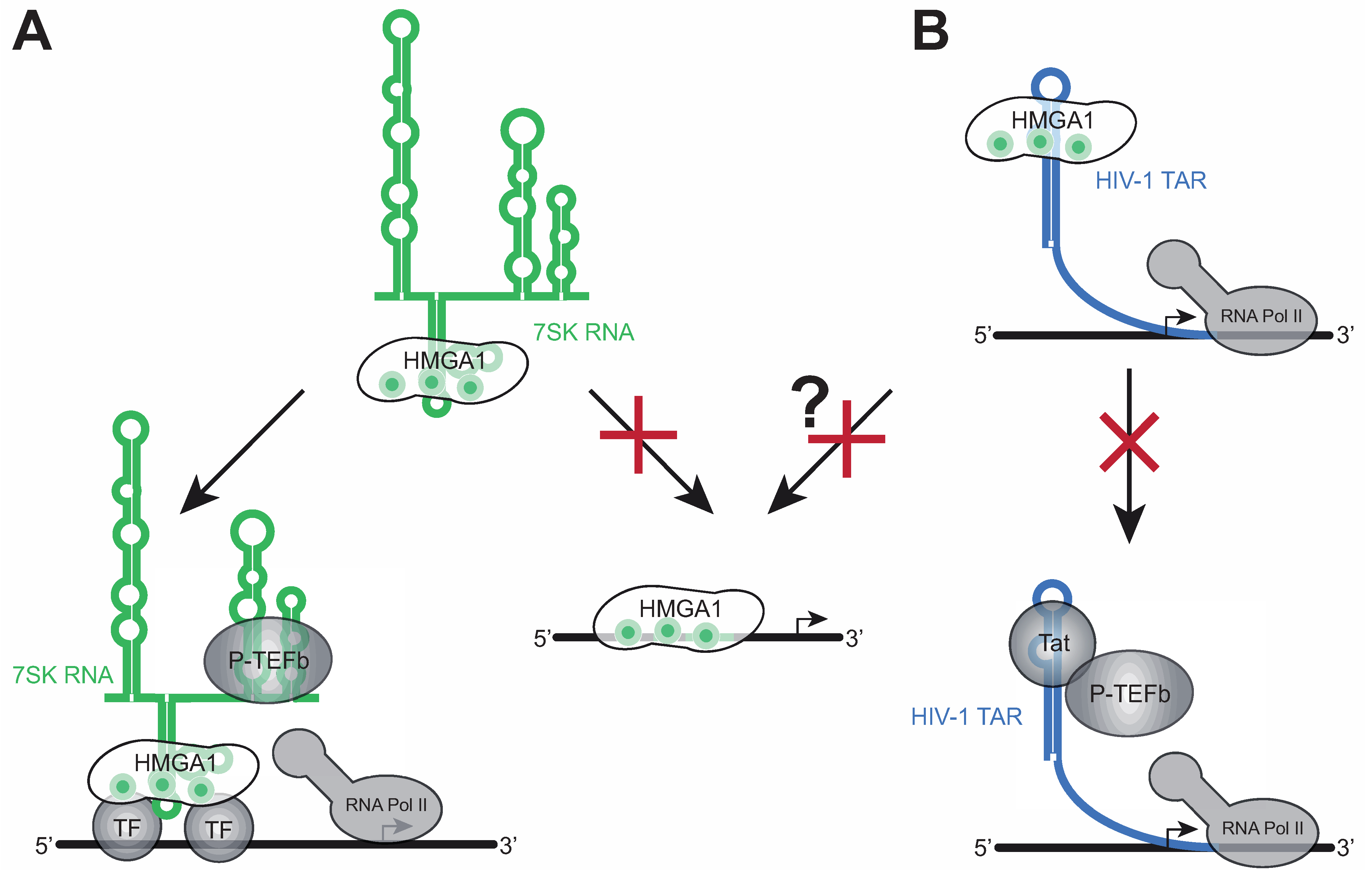
4. HMGA1 and 7SK-Dependent Transcription Elongation
5. HMGA1-RNA Complexes during HIV-1 Transcription
6. Conclusions
| Disease | HMGA1 Expression/Function | References |
|---|---|---|
| Bladder cancer | Overexpression | [54] |
| Breast cancer | Overexpression | [55,56,57] |
| Colorectal cancer | Overexpression; positively regulates Wnt/β-catenin signaling | [52,58] |
| Head and neck cancer | Overexpression | [59] |
| Leukemia | Overexpression; Cmyc target | [17,60,61] |
| Kidney cancer | Overexpression | [62] |
| Liver cancer | Overexpression | [63] |
| Lung cancer | Overexpression; promotes transformation | [64,65] |
| Glioblastoma/Neuroblastoma | Overexpression | [66,67,68,69] |
| Pancreatic cancer | Overexpression; promotes cellular invasiveness and metastatic potential | [70,71,72] |
| Prostate cancer | Overexpression; involved in chromosomal re-arrangements | [73,74] |
| Gastric cancer | Overexpression; let7-downregulation | [75,76] |
| Thyroid cancer | Overexpression; regulates expression of miR-603 and miR-10b | [77,78] |
| Cervix cancer | Overexpression | [79] |
| HIV infection | Co-factor for integration, transcription and spli-cing | [25,45,46,47,48,49,50] |
| Human papovavirus JC infection | Co-factor for transcription | [8] |
| Epstein Barr virus infection | Co-factor for transcription | [19] |
| Herpes Simplex virus 1 infection | Co-factor for transcription | [20,21] |
| Alzheimer’s disease | Involved in presenilin-2 pre-mRNA exon-skipping | [34] |
Acknowledgments
Conflicts of Interest
References
- Reeves, R.; Beckerbauer, L. HMGI/Y proteins: Flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 2001, 1519, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Huth, J.R.; Bewley, C.A.; Nissen, M.S.; Evans, J.N.; Reeves, R.; Gronenborn, A.M.; Clore, G.M. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 1997, 4, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Harrer, M.; Lührs, H.; Bustin, M.; Scheer, U.; Hock, R. Dynamic interaction of HMGA1a proteins with chromatin. J. Cell Sci. 2004, 117, 3459–3471. [Google Scholar] [CrossRef] [PubMed]
- Krech, A.B.; Wulff, D.; Grasser, K.D.; Feix, G. Plant chromosomal HMGI/Y proteins and histone H1 exhibit a protein domain of common origin. Gene 1999, 230, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Yie, J.; Liang, S.; Merika, M.; Thanos, D. Intra- and intermolecular cooperative binding of high-mobility-group protein I(Y) to the beta-interferon promoter. Mol. Cell. Biol. 1997, 17, 3649–3662. [Google Scholar] [PubMed]
- Currie, R.A. Functional interaction between the DNA binding subunit trimerization domain of NF-Y and the high mobility group protein HMG-I(Y). J. Biol. Chem. 1997, 272, 30880–30888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.M.; Verdine, G.L. A small region in HMG I(Y) is critical for cooperation with NF-κB on DNA. J. Biol. Chem. 1999, 274, 20235–20243. [Google Scholar] [CrossRef] [PubMed]
- Leger, H.; Sock, E.; Renner, K.; Grummt, F.; Wegner, M. Functional interaction between the POU domain protein Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol. Cell. Biol. 1995, 15, 3738–3747. [Google Scholar] [PubMed]
- Friedmann, M.; Holth, L.T.; Zoghbi, H.Y.; Reeves, R. Organization, inducible-expression and chromosome localization of the human HMG-I(Y) nonhistone protein gene. Nucleic Acids Res. 1993, 21, 4259–4267. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, S.; Ghosn, C.; DiSepio, D.; Molina, Y.; Sutter, M.; Klein, E.S.; Chandraratna, R.A. Retinoid-dependent recruitment of a histone H1 displacement activity by retinoic acid receptor. J. Biol. Chem. 1999, 274, 22563–22568. [Google Scholar] [CrossRef] [PubMed]
- Chiappetta, G.; Avantaggiato, V.; Visconti, R.; Fedele, M.; Battista, S.; Trapasso, F.; Merciai, B.M.; Fidanza, V.; Giancotti, V.; Santoro, M.; et al. High level expression of the HMGI (Y) gene during embryonic development. Oncogene 1996, 13, 2439–2446. [Google Scholar] [PubMed]
- Bustin, M.; Reeves, R. High-mobility-group chromosomal proteins: Architectural components that facilitate chromatin function. Prog. Nucleic Acids Res. Mol. Biol. 1996, 54, 35–100. [Google Scholar]
- Beaujean, N.; Bouniol-Baly, C.; Monod, C.; Kissa, K.; Jullien, D.; Aulner, N.; Amirand, C.; Debey, P.; Käs, E. Induction of early transcription in one-cell mouse embryos by microinjection of the nonhistone chromosomal protein HMG-I. Dev. Biol. 2000, 221, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Melillo, R.M.; Pierantoni, G.M.; Scala, S.; Battista, S.; Fedele, M.; Stella, A.; de Biasio, M.C.; Chiappetta, G.; Fidanza, V.; Condorelli, G.; et al. Critical role of the HMGI(Y) proteins in adipocytic cell growth and differentiation. Mol. Cell. Biol. 2001, 21, 2485–2495. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, K.; Karlson, J.R.; Ingebrigtsen, K.; Holtlund, J.; Lund, T.; Laland, S.G. On the presence of the chromosomal proteins HMG I and HMG Y in rat organs. Biochim. Biophys. Acta 1989, 1009, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Huso, T.H.; Resar, L. The high mobility group A1 molecular switch: Turning on cancer—Can we turn it off? Expert Opin. Ther. Targets 2014, 18, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.J.; Mukherjee, M.; Dolde, C.E.; Xu, Y.; Maher, J.F.; Bunton, T.E.; Williams, J.B.; Resar, L.M. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol. Cell. Biol. 2000, 20, 5490–5502. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.; Edberg, D.D.; Li, Y. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol. Cell. Biol. 2001, 21, 575–594. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, B.C.; Paulson, E.; Strominger, J.L.; Speck, S.H. Constitutive activation of Epstein-Barr virus (EBV) nuclear antigen 1 gene transcription by IRF1 and IRF2 during restricted EBV latency. Mol. Cell. Biol. 1997, 17, 873–886. [Google Scholar] [PubMed]
- Panagiotidis, C.A.; Silverstein, S.J. The host-cell architectural protein HMG I(Y) modulates binding of herpes simplex virus type 1 ICP4 to its cognate promoter. Virology 1999, 256, 64–74. [Google Scholar] [CrossRef] [PubMed]
- French, S.W.; Schmidt, M.C.; Glorioso, J.C. Involvement of a high-mobility-group protein in the transcriptional activity of herpes simplex virus latency-active promoter 2. Mol. Cell. Biol. 1996, 16, 5393–5399. [Google Scholar] [PubMed]
- Henderson, A.; Bunce, M.; Siddon, N.; Reeves, R.; Tremethick, D.J. High-mobility-group protein I can modulate binding of transcription factors to the U5 region of the human immunodeficiency virus type 1 proviral promoter. J. Virol. 2000, 74, 10523–10534. [Google Scholar] [CrossRef] [PubMed]
- Eilebrecht, S.; Brysbaert, G.; Wegert, T.; Urlaub, H.; Benecke, B.J.; Benecke, A. 7SK small nuclear RNA directly affects HMGA1 function in transcription regulation. Nucleic Acids Res. 2011, 39, 2057–2072. [Google Scholar] [CrossRef] [PubMed]
- Eilebrecht, S.; Bécavin, C.; Léger, H.; Benecke, B.J.; Benecke, A. HMGA1-dependent and independent 7SK RNA gene regulatory activity. RNA Biol. 2011, 8, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Eilebrecht, S.; Wilhelm, E.; Benecke, B.J.; Bell, B.; Benecke, A.G. HMGA1 directly interacts with TAR to modulate basal and Tat-dependent HIV transcription. RNA Biol. 2013, 10, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Kiss, T.; Michels, A.A.; Bensaude, O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 2001, 414, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhu, Q.; Luo, K.; Zhou, Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 2001, 414, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, T.; Price, D.H. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012, 81, 119–143. [Google Scholar] [CrossRef] [PubMed]
- Peterlin, B.M.; Brogie, J.E.; Price, D.H. 7SK snRNA: A noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 2012, 3, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Yik, J.H.; Chen, R.; Nishimura, R.; Jennings, J.L.; Link, A.J.; Zhou, Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 2003, 12, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Cherrier, T.; le Douce, V.; Eilebrecht, S.; Riclet, R.; Marban, C.; Dequiedt, F.; Goumon, Y.; Paillart, J.C.; Mericskay, M.; Parlakian, A.; et al. CTIP2 is a negative regulator of P-TEFb. Proc. Natl. Acad. Sci. USA 2013, 110, 12655–12660. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Ni, S.; Lindenberger, A.L.; Cho, J.; Tinch, S.L.; Kennedy, M.A. Characterization of the stoichiometry of HMGA1/DNA complexes. Open Biochem. J. 2013, 7, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Fonfría-Subirós, E.; Acosta-Reyes, F.; Saperas, N.; Pous, J.; Subirana, J.A.; Campos, J.L. Crystal structure of a complex of DNA with one AT-hook of HMGA1. PLoS ONE 2012, 7, e37120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manabe, T.; Ohe, K.; Katayama, T.; Matsuzaki, S.; Yanagita, T.; Okuda, H.; Bando, Y.; Imaizumi, K.; Reeves, R.; Tohyama, M.; et al. HMGA1a: Sequence-specific RNA-binding factor causing sporadic Alzheimer’s disease-linked exon skipping of presenilin-2 pre-mRNA. Genes Cells 2007, 12, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Gurney, T., Jr.; Eliceiri, G.L. Intracellular distribution of low molecular weight RNA species in HeLa cells. J. Cell Biol. 1980, 87, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.; Geyer, M.; Zhou, Q. The control of HIV transcription: Keeping RNA polymerase II on track. Cell Host Microbe 2011, 10, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Muniz, L.; Egloff, S.; Ughy, B.; Jády, B.E.; Kiss, T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 2010. [Google Scholar] [CrossRef]
- Berkhout, B.; Jeang, K.T. Trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: A quantitative analysis. J. Virol. 1989, 63, 5501–5504. [Google Scholar] [PubMed]
- Dumpelmann, E.; Mittendorf, H.; Benecke, B.J. Efficient transcription of the EBER2 gene depends on the structural integrity of the RNA. RNA 2003, 9, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Lerner, M.R.; Andrews, N.C.; Miller, G.; Steitz, J.A. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 1981, 78, 805–809. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.; Elton, T.S.; Nissen, M.S.; Lehn, D.; Johnson, K.R. Posttranscriptional gene regulation and specific binding of the nonhistone protein HMG-I by the 3' untranslated region of bovine interleukin 2 cDNA. Proc. Natl. Acad. Sci. USA 1987, 84, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.; Leonard, W.J.; Nissen, M.S. Binding of HMG-I(Y) imparts architectural specificity to a positioned nucleosome on the promoter of the human interleukin-2 receptor alpha gene. Mol. Cell. Biol. 2000, 20, 4666–4679. [Google Scholar] [CrossRef] [PubMed]
- Eilebrecht, S.; Benecke, B.J.; Benecke, A. 7SK snRNA-mediated, gene-specific cooperativity of HMGA1 and P-TEFb. RNA Biol. 2011, 8, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- D’Orso, I.; Frankel, A.D. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010, 17, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.; Holloway, A.; Reeves, R.; Tremethick, D.J. Recruitment of SWI/SNF to the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 2004, 24, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Eilebrecht, S.; le Douce, V.; Riclet, R.; Targat, B.; Hallay, H.; van Driessche, B.; Schwartz, C.; Robette, G.; van Lint, C.; Rohr, O.; et al. HMGA1 recruits CTIP2-repressed P-TEFb to the HIV-1 and cellular target promoters. Nucleic Acids Res. 2014, 42, 4962–4971. [Google Scholar] [CrossRef] [PubMed]
- Benecke, A. Genomic plasticity and information processing by transcription coregulators. Complexus 2003, 1, 65–76. [Google Scholar] [CrossRef]
- Tsuruno, C.; Ohe, K.; Kuramitsu, M.; Kohma, T.; Takahama, Y.; Hamaguchi, Y.; Hamaguchi, I.; Okuma, K. HMGA1a is involved in specific splice site regulation of human immunodeficiency virus type 1. Biochem. Biophys. Res. Commun. 2011, 406, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Gorelick, R.J.; Johnson, D.G.; Bushman, F. Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. J. Virol. 2003, 77, 1598–1603. [Google Scholar] [CrossRef] [PubMed]
- Van Maele, B.; Busschots, K.; Vandekerckhove, L.; Christ, F.; Debyser, Z. Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 2006, 31, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Fusco, A.; Fedele, M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer 2007, 7, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Belton, A.; Gabrovsky, A.; Bae, Y.K.; Reeves, R.; Iacobuzio-Donahue, C.; Huso, D.L.; Resar, L.M. HMGA1 induces intestinal polyposis in transgenic mice and drives tumor progression and stem cell properties in colon cancer cells. PloS ONE 2012, 7, e30034. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.N.; Resar, L.M. High mobility group A1 and cancer: Potential biomarker and therapeutic target. Histol. Histopathol. 2012, 27, 567–579. [Google Scholar] [PubMed]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.N.; Cope, L.; Poh, W.; Belton, A.; Roy, S.; Talbot, C.C., Jr.; Sukumar, S.; Huso, D.L.; Resar, L.M. HMGA1: A master regulator of tumor progression in triple-negative breast cancer cells. PloS ONE 2013, 8, e63419. [Google Scholar] [CrossRef] [PubMed]
- Dolde, C.E.; Mukherjee, M.; Cho, C.; Resar, L.M. HMG-I/Y in human breast cancer cell lines. Breast Cancer Res. Treat. 2002, 71, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Flohr, A.M.; Rogalla, P.; Bonk, U.; Puettmann, B.; Buerger, H.; Gohla, G.; Packeisen, J.; Wosniok, W.; Loeschke, S.; Bullerdiek, J. High mobility group protein HMGA1 expression in breast cancer reveals a positive correlation with tumour grade. Histol. Histopathol. 2003, 18, 999–1004. [Google Scholar] [PubMed]
- Xing, J.; Cao, G.; Fu, C. HMGA1 interacts with β-catenin to positively regulate Wnt/β-catenin signaling in colorectal cancer cells. Pathol. Oncol. Res. 2014, 20, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Rho, Y.S.; Lim, Y.C.; Park, I.S.; Kim, J.H.; Ahn, H.Y.; Cho, S.J.; Shin, H.S. High mobility group HMGI(Y) protein expression in head and neck squamous cell carcinoma. Acta Otolaryngol. 2007, 127, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Hillion, J.; Dhara, S.; Sumter, T.F.; Mukherjee, M.; di Cello, F.; Belton, A.; Turkson, J.; Jaganathan, S.; Cheng, L.; Ye, Z.; et al. The High-mobility group A1a/signal transducer and activator of transcription-3 Axis: An achilles heel for hematopoietic malignancies? Cancer Res. 2008, 68, 10121–10127. [Google Scholar] [CrossRef] [PubMed]
- Pierantoni, G.M.; Agosti, V.; Fedele, M.; Bond, H.; Caliendo, I.; Chiappetta, G.; lo Coco, F.; Pane, F.; Turco, M.C.; Morrone, G.; et al. High-mobility group A1 proteins are overexpressed in human leukaemias. Biochem. J. 2003, 372, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Takaha, N.; Sowa, Y.; Takeuchi, I.; Hongo, F.; Kawauchi, A.; Miki, T. Expression and role of HMGA1 in renal cell carcinoma. J. Urol. 2012, 187, 2215–2222. [Google Scholar] [CrossRef] [PubMed]
- Chuma, M.; Saeki, N.; Yamamoto, Y.; Ohta, T.; Asaka, M.; Hirohashi, S.; Sakamoto, M. Expression profiling in hepatocellular carcinoma with intrahepatic metastasis: Identification of high-mobility group I(Y) protein as a molecular marker of hepatocellular carcinoma metastasis. Keio J. Med. 2004, 53, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Sarhadi, V.K.; Wikman, H.; Salmenkivi, K.; Kuosma, E.; Sioris, T.; Salo, J.; Karjalainen, A.; Knuutila, S.; Anttila, S. Increased expression of high mobility group A proteins in lung cancer. J. Pathol. 2006, 209, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Hillion, J.; Wood, L.J.; Mukherjee, M.; Bhattacharya, R.; di Cello, F.; Kowalski, J.; Elbahloul, O.; Segal, J.; Poirier, J.; Rudin, C.M.; et al. Upregulation of MMP-2 by HMGA1 promotes transformation in undifferentiated, large-cell lung cancer. Mol. Cancer Res. 2009, 7, 1803–1812. [Google Scholar] [CrossRef] [PubMed]
- Pomeroy, S.L.; Tamayo, P.; Gaasenbeek, M.; Sturla, L.M.; Angelo, M.; McLaughlin, M.E.; Kim, J.Y.; Goumnerova, L.C.; Black, P.M.; Lau, C.; et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 2002, 415, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Giannini, G.; Cerignoli, F.; Mellone, M.; Massimi, I.; Ambrosi, C.; Rinaldi, C.; Dominici, C.; Frati, L.; Screpanti, I.; Gulino, A. High mobility group A1 is a molecular target for MYCN in human neuroblastoma. Cancer Res. 2005, 65, 8308–8316. [Google Scholar] [CrossRef] [PubMed]
- Giannini, G.; Cerignoli, F.; Mellone, M.; Massimi, I.; Ambrosi, C.; Rinaldi, C.; Gulino, A. Molecular mechanism of HMGA1 deregulation in human neuroblastoma. Cancer Lett. 2005, 228, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Cerignoli, F.; Ambrosi, C.; Mellone, M.; Assimi, I.; di Marcotullio, L.; Gulino, A.; Giannini, G. HMGA molecules in neuroblastic tumors. Ann. N Y Acad. Sci. 2004, 1028, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Liau, S.S.; Jazag, A.; Whang, E.E. HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res. 2006, 66, 11613–11622. [Google Scholar] [CrossRef] [PubMed]
- Abe, N.; Watanabe, T.; Masaki, T.; Mori, T.; Sugiyama, M.; Uchimura, H.; Fujioka, Y.; Chiappetta, G.; Fusco, A.; Atomi, Y. Pancreatic duct cell carcinomas express high levels of high mobility group I(Y) proteins. Cancer Res. 2000, 60, 3117–3122. [Google Scholar] [PubMed]
- Liau, S.S.; Rocha, F.; Matros, E.; Redston, M.; Whang, E. High mobility group AT-hook 1 (HMGA1) is an independent prognostic factor and novel therapeutic target in pancreatic adenocarcinoma. Cancer 2008, 113, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Takaha, N.; Hawkins, A.L.; Griffin, C.A.; Isaacs, W.B.; Coffey, D.S. High mobility group protein I(Y): A candidate architectural protein for chromosomal rearrangements in prostate cancer cells. Cancer Res. 2002, 62, 647–651. [Google Scholar] [PubMed]
- Takeuchi, I.; Takaha, N.; Nakamura, T.; Hongo, F.; Mikami, K.; Kamoi, K.; Okihara, K.; Kawauchi, A.; Miki, T. High mobility group protein AT-hook 1 (HMGA1) is associated with the development of androgen independence in prostate cancer cells. Prostate 2011, 72, 1124–1132. [Google Scholar] [PubMed]
- Rahman, M.M.; Qian, Z.R.; Wang, E.L.; Sultana, R.; Kudo, E.; Nakasono, M.; Hayashi, T.; Kakiuchi, S.; Sano, T. Frequent overexpression of HMGA1 and 2 in gastroenteropancreatic neuroendocrine tumours and its relationship to let-7 downregulation. Br. J. Cancer 2009, 100, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Akaboshi, S.; Watanabe, S.; Hino, Y.; Sekita, Y.; Xi, Y.; Araki, K.; Yamamura, K.; Oshima, M.; Ito, T.; Baba, H.; et al. HMGA1 is induced by Wnt/β-catenin pathway and maintains cell proliferation in gastric cancer. Am. J. Pathol. 2010, 175, 1675–1685. [Google Scholar]
- Chiappetta, G.; Bandiera, A.; Berlingieri, M.T.; Visconti, R.; Manfioletti, G.; Battista, S.; Martinez-Tello, F.J.; Santoro, M.; Giancotti, V.; Fusco, A. The expression of the high mobility group HMGI (Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene 1995, 10, 1307–1314. [Google Scholar] [PubMed]
- Mussnich, P.; D’Angelo, D.; Leone, V.; Croce, C.M.; Fusco, A. The high mobility group A proteins contribute to thyroid cell transformation by regulating miR-603 and miR-10b expression. Mol. Oncol. 2013, 7, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, A.; Bonifacio, D.; Manfioletti, G.; Mantovani, F.; Rustighi, A.; Zanconati, F.; Fusco, A.; di Bonito, L.; Giancotti, V. Expression of HMGI(Y) proteins in squamous intraepithelial and invasive lesions of the uterine cervix. Cancer Res. 1998, 58, 426–431. [Google Scholar] [PubMed]
- Rajski, S.R.; Williams, R.M. Observations on the covalent cross-linking of the binding domain (BD) of the high mobility group I/Y (HMG I/Y) proteins to DNA by FR66979. Bioorganice Med. Chem. 2000, 8, 1331–1342. [Google Scholar] [CrossRef]
- Beckerbauer, L.; Tepe, J.J.; Cullison, J.; Reeves, R.; Williams, R.M. FR900482 class of anti-tumor drugs cross-links oncoprotein HMG I/Y to DNA in vivo. Chem. Biol. 2000, 7, 805–812. [Google Scholar] [CrossRef]
- Beckerbauer, L.; Tepe, J.J.; Eastman, R.A.; Mixter, P.F.; Williams, R.M.; Reeves, R. Differential effects of FR900482 and FK317 on apoptosis, IL-2 gene expression, and induction of vascular leak syndrome. Chem. Biol. 2002, 9, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Baron, R.M.; Lopez-Guzman, S.; Riascos, D.F.; Macias, A.A.; Layne, M.D.; Cheng, G.; Harris, C.; Chung, S.W.; Reeves, R.; von Andrian, U.H.; et al. Distamycin A inhibits HMGA1-binding to the P-selectin promoter and attenuates lung and liver inflammation during murine endotoxemia. PloS ONE 2010, 5, e10656. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.A.; Baron, R.M.; Macias, A.A.; Layne, M.D.; Perrella, M.A.; Rigby, A.C. Netropsin improves survival from endotoxaemia by disrupting HMGA1 binding to the NOS2 promoter. Biochem. J. 2009, 418, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Maasch, C.; Vater, A.; Buchner, K.; Purschke, W.G.; Eulberg, D.; Vonhoff, S.; Klussmann, S. Polyetheylenimine-polyplexes of spiegelmer NOX-A50 directed against intracellular high mobility group protein A1 (HMGA1) reduce tumor growth in Vivo. J. Biol. Chem. 2010, 285, 40012–40018. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Sheriff, S.; Lewis, K.B.; Tinch, S.L.; Cho, J.; Balasubramaniam, A.; Kennedy, M.A. HMGA-targeted phosphorothioate DNA aptamers increase sensitivity to gemcitabine chemotherapy in human pancreatic cancer cell lines. Cancer Lett. 2012, 315, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Eilebrecht, S.; Pellay, F.; Odenwälder, P.; Brysbaert, G.; Benecke, B.; Benecke, A. EBER2 RNA-induced transcriptome changes identify cellular processes likely targeted during Epstein Barr virus infection. BMC Res. Notes 2008. [Google Scholar] [CrossRef]
- Sun, J.; Lu, H.; Wang, X.; Jin, H. MicroRNAs in hepatocellular carcinoma: Regulation, function, and clinical implications. Sci. World J. 2013. [Google Scholar] [CrossRef]
- Fujita, Y.; Kuwano, K.; Ochiya, T. Development of small RNA delivery systems for lung cancer therapy. IJMS 2015, 16, 5254–5270. [Google Scholar] [CrossRef] [PubMed]
- Fedele, M.; Fidanza, V.; Battista, S.; Pentimalli, F.; Klein-Szanto, A.J.; Visone, R.; de Martino, I.; Curcio, A.; Morisco, C.; del Vecchio, L.; et al. Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res. 2006, 66, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, I.M.; Baron, R.M.; Perrella, M.A. High-mobility group-I/Y proteins: Potential role in the pathophysiology of critical illnesses. Crit. Care Med. 2002, 30, S36–S42. [Google Scholar] [CrossRef] [PubMed]
- Cleynen, I.; van de Ven, W.J. The HMGA proteins: A myriad of functions (Review). Int. J. Oncol. 2008, 32, 289–305. [Google Scholar] [PubMed]
- Foti, D.; Chiefari, E.; Fedele, M.; Iuliano, R.; Brunetti, L.; Paonessa, F.; Manfioletti, G.; Barbetti, F.; Brunetti, A.; Croce, C.M.; et al. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nat. Med. 2005, 11, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.; Thum, T. Novel non-coding RNA-based therapeutic approaches to prevent statin-induced liver damage. EMBO Mol. Med. 2012, 4, 863–865. [Google Scholar] [CrossRef] [PubMed]
- Archin, N.M.; Sung, J.M.; Garrido, C.; Soriano-Sarabia, N.; Margolis, D.M. Eradicating HIV-1 infection: Seeking to clear a persistent pathogen. Nat. Publ. Group 2014, 12, 750–764. [Google Scholar]
- Battistini, A.; Sgarbanti, M. HIV-1 latency: An update of molecular mechanisms and therapeutic strategies. Viruses 2014, 6, 1715–1758. [Google Scholar] [CrossRef] [PubMed]
- Chiefari, E.; Nevolo, M.T.; Arcidiacono, B.; Maurizio, E.; Nocera, A.; Iiritano, S.; Sgarra, R.; Possidente, K.; Palmieri, C.; Paonessa, F.; et al. HMGA1 is a novel downstream nuclear target of the insulin receptor signaling pathway. Sci. Rep. 2012. [Google Scholar] [CrossRef]
- Arcidiacono, B.; Liritano, S.; Chiefari, E.; Brunetti, S.F.; Gu, G.; Foti, D.P.; Brunetti, A. Cooperation between HMGA1, PDX-1, and MafA is essential for glucose-induced insulin transcription in pancreatic β cells. Front. Endocrinol. Lausanne 2015. [Google Scholar] [CrossRef]
- Garriga, J.; Graña, X. CDK9 inhibition strategy defines distinct sets of target genes. BMC Res. Notes. 2014. [Google Scholar] [CrossRef]
- Esposito, F.; de Martino, M.; Forzati, F.; Fusco, A. HMGA1-pseudogene overexpression contributes to cancer progression. Cell Cycle 2014, 13, 3636–3639. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benecke, A.G.; Eilebrecht, S. RNA-Mediated Regulation of HMGA1 Function. Biomolecules 2015, 5, 943-957. https://doi.org/10.3390/biom5020943
Benecke AG, Eilebrecht S. RNA-Mediated Regulation of HMGA1 Function. Biomolecules. 2015; 5(2):943-957. https://doi.org/10.3390/biom5020943
Chicago/Turabian StyleBenecke, Arndt G., and Sebastian Eilebrecht. 2015. "RNA-Mediated Regulation of HMGA1 Function" Biomolecules 5, no. 2: 943-957. https://doi.org/10.3390/biom5020943





