Mass Spectrometry-Based Methods for Identifying Oxidized Proteins in Disease: Advances and Challenges
Abstract
:1. Introduction to Protein Oxidation

2. Overview of Mass Spectrometry Methods for Protein Oxidation Analysis

2.1. Sample Preparation and Digestion
2.2. Enrichment and Separation
2.3. Intact Protein and Top-Down Analysis
2.4. Bottom-Up Analysis
3. Untargeted Mass Spectrometry and “Discovery” Approaches
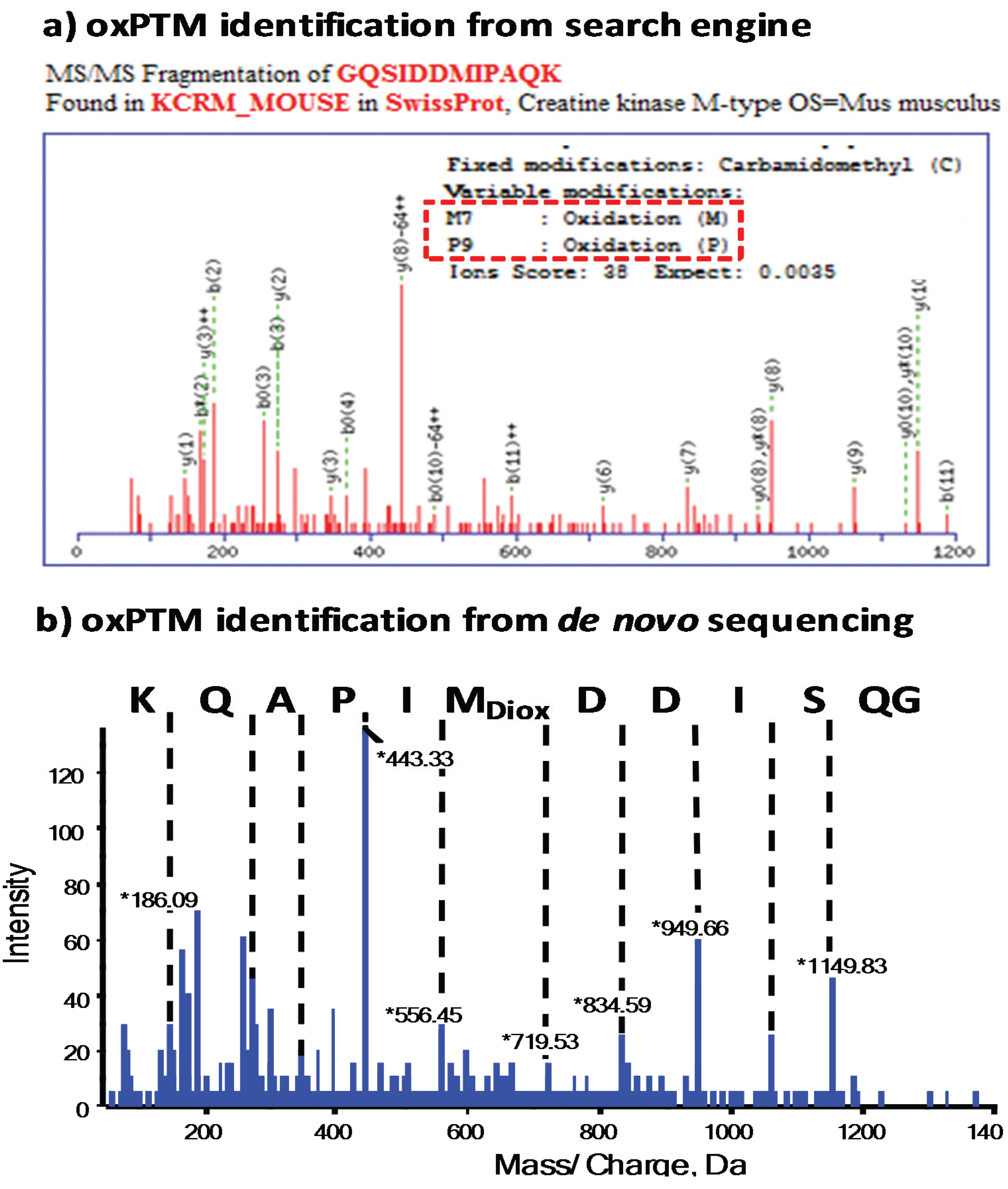
3.1. Use of Search Engines for MS Data and Analysis of oxPTMs
| Search Engine | Method | Advantages | Disadvantages |
|---|---|---|---|
| Mascot | Uses a probability modelling algorithm and protein database searching. Matches experimental peptide and fragment ion masses to ones generated in silico from databases. | User-friendly interface. Provides an error-tolerant search facility. Sophisticated but complex data export possibilities. | Very reliant on user input for correct identification of oxPTMs, otherwise false positives and negatives occur. |
| Sequest | Uses an algorithm based on a cross correlation function, plus protein data base searching. Matches experimental peptide and fragment ion masses to ones generated in silico from databases. | User-friendly interface. Provides an error-tolerant search facility. | Very reliant on user input for correct identification of oxPTMs, otherwise false positives and negatives occur. |
| ProteinPilot | Uses a sequence tag method plus protein database searching. | User-friendly interface. Potentially better at identifying unsuspected modifications. | If the initial sequence tag is incorrectly identified, the experimental peptide will not be matched to the correct peptide. Long analysis run times. |
| pMatch | Spectral library searching against experimentally-derived data. | Has been reported to be better at identifying PTMs, and specifically at coping with the unusual fragmentation of peptides caused by PTMs. | Since this method uses a spectral library, the peptide will only be identified if the spectra are available in the spectral library. |
| MS Amanda | Based on a binomial distribution function. Protein data base searching. Matches experimental peptide and fragment ion masses to ones generated in silico from databases. | Reported to be better at identifying peptides of higher m/z than Mascot and Sequest. | Very reliant on user input for correct identification of oxPTMs, otherwise false positives and negatives occur. |
3.2. The Importance of Data Validation
4. Reporter Ion-Based Methodologies
4.1. Semi-Targeted MS/MS Analysis
4.2. Narrow-Window Extracted Ion Chromatograms
4.3. Targeted Methods of Analysis
5. Quantification of (ox)PTMs
5.1. Label-Free Methods of Quantification
5.2. Label-Dependent Methods of Quantification
6. Applications in Vivo and in Disease
6.1. Considerations for Clinical Sample Type in oxPTM Analysis
6.2. MS Analysis of Protein Oxidation in Disease
| Modification Type | Disease | Method | Sample Type | Protein Type | Oxidation Sites Identified? | Reference |
|---|---|---|---|---|---|---|
| Carbonylation | Alzheimer’s disease | DNPH, MALDI-TOF/MS | Blood (human) | Fibrinogen γ-chain precursor protein, α-1-Antitrypsin precursor | no | Choi et al., 2002 [150] |
| Carbonylation | Aging | Avidin affinity, LC-MS/MS | Brain tissue (mouse) | Brain proteins | yes | Soreghan et al., 2003 [151] |
| Carbonylation | Aging | FTCl-labeling; 2DE-MS | Liver tissue (mouse) | Cytosolic liver proteins | no | Chaudhuri et al., 2006 [152] |
| Carbonylation | Aging | ITRAQ/LC-MS/MS | Skeletal muscle (rat) | Mitochondrial muscle proteins | no | Feng et al., 2008 [153] |
| Carbonylation | Mild Cognitive impairment and Early Alzheimer’s disease | DNPH, MALDI-TOF/MS | inferior parietal lobule (human) | CA II, Syntaxin binding protein I, Hsp70, MAPK kinase I, FBA-C, PM-1, GFAP | no | Sultana et al., 2010 [154] |
| Carbonylation | Aging | ARP-labeling, MS/MS | Heart (rat) | Cardiac mitochondrial proteins | yes | Chavez et al., 2011 [155] |
| Carbonylation | Diabetes | ITRAQ/LS-MS/MS(SRM) | Plasma (rat) | Plasma proteins | yes | Madian et al., 2011 [156] |
| Carbonylation | Obesity-induced diabetes mellitus | ARP-labeling RPC-MS/MS | Plasma (human) | Plasma proteins | yes | Bollineni et al., 2014 [157] |
| Carbonylation | Breast cancer | iTRAQ | Plasma (human) | Plasma proteins | yes | Madian & Regnier, 2010 [29] |
| Carbonylation | Ischemia/reperfusion | 2D-PAGE-MALDI-TOF/TOF/MS/MS, | Hippocampus (monkey) | Hsp70-1, DRP2 isoform 2, GFAP, β-actin | yes | Oikawa et al., 2009 [158] |
| Carbonylation, cysteic acid, MetO, MetO2 | Alzheimer’s disease, Parkinson’s disease | 2D-PAGE, MALDI-TOF/MS MALDI-TOF/TOF/MS/MS, HPLC-ESI/MS/MS MALDI-MS/MS | Brain (human) | DJ-1 | yes | Choi et al., 2006 [137] |
| 3-NO2Y | Cancer | NTAC-based MALDI–LTQ MS/MS | Non-functional pituitary adenoma tissue (human) | NTAC-enriched proteins | yes | Zhan & Desiderio, 2006 [120] |
| 3-NO2Y, 3-Cl-Y | Influenza | LC-MS/MS | Serum (mouse) | Serum proteins | yes | Kumar et al., 2014 [159] |
7. Conclusions and Perspectives
Acknowledgments
Author Contributions
Abbreviations
| DNPH | 2,4-dinitrophenylhydrazine |
| DTT | dithiotreitol |
| ESI | electrospray ionization |
| HDL | high density lipoprotein |
| HPLC | high performance liquid chromatography |
| ICAT | isotope coded affinity tags |
| iTRAQ | isobaric tags for relative and absolute quantification |
| LC-MS or LC-MS/MS | Liquid chromatography coupled to mass spectrometry or tandem mass spectrometry |
| LPS | lipopolysaccharide |
| MALDI | matrix-assisted laser desorption/ionisation |
| MRM | multiple reaction monitoring |
| MS | mass spectrometry |
| MS/MS | tandem mass spectrometry |
| oxPTM | oxidative post-translational modification |
| PTM | post-translational modification |
| SLE | systemic lupus erythematosus |
| SNO | S-nitrosothiol |
| XIC | extracted ion chromatogram |
Conflicts of Interest
References
- Droge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [PubMed]
- Spickett, C.M.; Pitt, A.R. Protein oxidation: Role in signalling and detection by mass spectrometry. Amino Acids 2012, 42, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Shacter, E. Quantification and significance of protein oxidation in biological samples. Drug Metab. Rev. 2000, 32, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Domingues, R.M.; Domingues, P.; Melo, T.; Perez-Sala, D.; Reis, A.; Spickett, C.M. Lipoxidation adducts with peptides and proteins: Deleterious modifications or signaling mechanisms? J. Proteomics. 2013, 92, 110–131. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Thornalley, P.J. Quantitative screening of protein biomarkers of early glycation, advanced glycation, oxidation and nitrosation in cellular and extracellular proteins by tandem mass spectrometry multiple reaction monitoring. Biochem. Soc. Trans. 2003, 31, 1417–1422. [Google Scholar] [CrossRef] [PubMed]
- Perdivara, I.; Deterding, L.J.; Przybylski, M.; Tomer, K.B. Mass spectrometric identification of oxidative modifications of tryptophan residues in proteins: Chemical artifact or post-translational modification? J. Am. Soc. Mass Spectrom. 2010, 21, 1114–1117. [Google Scholar] [CrossRef] [PubMed]
- Auclair, J.R.; Salisbury, J.P.; Johnson, J.L.; Petsko, G.A.; Ringe, D.; Bosco, D.A.; Agar, N.Y.R.; Santagata, S.; Durham, H.D.; Agar, J.N. Artifacts to avoid while taking advantage of top-down mass spectrometry based detection of protein S-thiolation. Proteomics 2014, 14, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, G.; Wu, S.L.; Shieh, P.; Hancock, W.S. Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J. Proteome Res. 2003, 2, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.; Lorenzen, K.; Kerkhoven, R.; van Breukelen, B.; Vannini, A.; Cramer, P.; Heck, A.J. Multiplexed proteomics mapping of yeast RNA polymerase II and III allows near-complete sequence coverage and reveals several novel phosphorylation sites. Anal. Chem. 2008, 80, 3584–3592. [Google Scholar] [CrossRef] [PubMed]
- Swaney, D.L.; Wenger, C.D.; Coon, J.J. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 2010, 9, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Table 2. List of proteases commonly used for fragmenting proteins. Cold Spring Harbor Protocols 2007. [CrossRef]
- Moulaei, T.; Stuchlik, O.; Reed, M.; Yuan, W.; Pohl, J.; Lu, W.; Haugh-Krumpe, L.; O’Keefe, B.R.; Wlodawer, A. Topology of the disulfide bonds in the antiviral lectin scytovirin. Protein Sci. 2010, 19, 1649–1661. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Lv, X.; Wang, S.; Iqbal, J.; Qing, H.; Li, Q.; Deng, Y. An aptamer-based trypsin reactor for on-line protein digestion with electrospray ionization tandem mass spectrometry. Anal. Biochem. 2013, 441, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wojcik, R.; Dovichi, N.J. A replaceable microreactor for on-line protein digestion in a two-dimensional capillary electrophoresis system with tandem mass spectrometry detection. J. Chromatogr. A 2011, 1218, 2007–2011. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Young, R.; Straubinger, R.M.; Page, B.; Cao, J.; Wang, H.; Yu, H.; Canty, J.M.; Qu, J. A straightforward and highly efficient precipitation/on-pellet digestion procedure coupled with a long gradient nano-lc separation and orbitrap mass spectrometry for label-free expression profiling of the swine heart mitochondrial proteome. J. Proteome Res. 2009, 8, 2838–2850. [Google Scholar] [CrossRef] [PubMed]
- Saveliev, S.V.; Woodroofe, C.C.; Sabat, G.; Adams, C.M.; Klaubert, D.; Wood, K.; Urh, M. Mass spectrometry compatible surfactant for optimized in-gel protein digestion. Anal. Chem. 2013, 85, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.Q.; Gilar, M.; Kaska, J.; Gebler, J.C. A rapid sample preparation method for mass spectrometric characterization of n-linked glycans. Rapid Commun. Mass Spectrom. 2005, 19, 2331–2336. [Google Scholar] [CrossRef] [PubMed]
- Luque-Garcia, J.L.; Neubert, T.A. On-membrane tryptic digestion of proteins for mass spectrometry analysis. Methods Mol. Biol. 2009, 536, 331–341. [Google Scholar] [PubMed]
- Su, D.; Gaffrey, M.J.; Guo, J.; Hatchell, K.E.; Chu, R.K.; Clauss, T.R.; Aldrich, J.T.; Wu, S.; Purvine, S.; Camp, D.G.; et al. Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radic. Biol. Med. 2014, 67, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Gaffrey, M.J.; Su, D.; Liu, T.; Camp, D.G., 2nd; Smith, R.D.; Qian, W.J. Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications. Nat. Protoc. 2014, 9, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Rees, J.S.; Lilley, K.S. Method for suppressing non-specific protein interactions observed with affinity resins. Methods 2011, 54, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Trinkle-Mulcahy, L.; Boulon, S.; Lam, Y.W.; Urcia, R.; Boisvert, F.M.; Vandermoere, F.; Morrice, N.A.; Swift, S.; Rothbauer, U.; Leonhardt, H.; et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell. Biol. 2008, 183, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Pignatelli, B.; Li, C.Q.; Boffetta, P.; Chen, Q.; Ahrens, W.; Nyberg, F.; Mukeria, A.; Bruske-Hohlfeld, I.; Fortes, C.; Constantinescu, V.; et al. Nitrated and oxidized plasma proteins in smokers and lung cancer patients. Cancer Res. 2001, 61, 778–784. [Google Scholar] [PubMed]
- Kim, J.K.; Lee, J.R.; Kang, J.W.; Lee, S.J.; Shin, G.C.; Yeo, W.S.; Kim, K.H.; Park, H.S.; Kim, K.P. Selective enrichment and mass spectrometric identification of nitrated peptides using fluorinated carbon tags. Anal. Chem. 2011, 83, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, J.R.; Eaton, P.; Enrique, C.; Lester, P. Chapter 15—A rapid approach for the detection, quantification, and discovery of novel sulfenic acid or S-nitrosothiol modified proteins using a biotin-switch method. In Methods in Enzymology; Academic Press: Waltham, MA, USA, 2010; Volume 473, pp. 281–303. [Google Scholar]
- Jaffrey, S.R.; Snyder, S.H. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001. [Google Scholar] [CrossRef]
- Qu, Z.; Meng, F.; Bomgarden, R.D.; Viner, R.I.; Li, J.; Rogers, J.C.; Cheng, J.; Greenlief, C.M.; Cui, J.; Lubahn, D.B.; et al. Proteomic quantification and site-mapping of S-nitrosylated proteins using isobaric iodotmt reagents. J. Proteome Res. 2014, 13, 3200–3211. [Google Scholar] [CrossRef] [PubMed]
- Doulias, P.T.; Raju, K.; Greene, J.L.; Tenopoulou, M.; Ischiropoulos, H. Mass spectrometry-based identification of S-nitrosocysteine in vivo using organic mercury assisted enrichment. Methods 2013, 62, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Madian, A.G.; Regnier, F.E. Profiling carbonylated proteins in human plasma. J. Proteome Res. 2010, 9, 1330–1343. [Google Scholar] [CrossRef] [PubMed]
- Bollineni, R.; Hoffmann, R.; Fedorova, M. Identification of protein carbonylation sites by two-dimensional liquid chromatography in combination with MALDI- and ESI-MS. J. Proteomics 2011, 74, 2338–2350. [Google Scholar] [CrossRef] [PubMed]
- Bollineni, R.C.; Hoffmann, R.; Fedorova, M. Proteome-wide profiling of carbonylated proteins and carbonylation sites in HeLa cells under mild oxidative stress conditions. Free Radic. Biol. Med. 2014, 68, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Qian, W.J.; Knyushko, T.V.; Clauss, T.R.; Purvine, S.O.; Moore, R.J.; Sacksteder, C.A.; Chin, M.H.; Smith, D.J.; Camp, D.G., 2nd; et al. A method for selective enrichment and analysis of nitrotyrosine-containing peptides in complex proteome samples. J. Proteome Res. 2007, 6, 2257–2268. [Google Scholar] [CrossRef] [PubMed]
- Amoresano, A.; Chiappetta, G.; Pucci, P.; D'Ischia, M.; Marino, G. Bidimensional tandem mass spectrometry for selective identification of nitration sites in proteins. Anal. Chem. 2007, 79, 2109–2117. [Google Scholar] [CrossRef] [PubMed]
- Houée-Lévin, C.; Bobrowski, K.; Horakova, L.; Karademir, B.; Schöneich, C.; Davies, M.J.; Spickett, C.M. Exploring oxidative modifications of tyrosine: An update on mechanisms of formation, advances in analysis and biological consequences. Free Radic. Res. 2015, 49. [Google Scholar] [CrossRef]
- Tran, J.C.; Doucette, A.A. Gel-eluted liquid fraction entrapment electrophoresis: An electrophoretic method for broad molecular weight range proteome separation. Anal. Chem. 2008, 80, 1568–1573. [Google Scholar] [CrossRef] [PubMed]
- Ghesquiere, B.; Colaert, N.; Helsens, K.; Dejager, L.; Vanhaute, C.; Verleysen, K.; Kas, K.; Timmerman, E.; Goethals, M.; Libert, C.; et al. In vitro and in vivo protein-bound tyrosine nitration characterized by diagonal chromatography. Mol. Cell. Proteomics 2009, 8, 2642–2652. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, Y.; Aslanian, A.; Fonslow, B.; Graczyk, B.; Davis, T.N.; Yates, J.R., 3rd. In-line separation by capillary electrophoresis prior to analysis by top-down mass spectrometry enables sensitive characterization of protein complexes. J. Proteome Res. 2014, 13, 6078–6086. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Compton, P.D.; Tran, J.C.; Ntai, I.; Kelleher, N.L. Optimizing capillary electrophoresis for top-down proteomics of 30–80 kda proteins. Proteomics 2014, 14, 1158–1164. [Google Scholar] [CrossRef] [PubMed]
- Chait, B.T.; Kent, S.B. Weighing naked proteins: Practical, high-accuracy mass measurement of peptides and proteins. Science 1992, 257, 1885–1894. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, P.V.; Second, T.P.; Zabrouskov, V.; Makarov, A.A.; Zhang, Z. Mass measurement and top-down hplc/ms analysis of intact monoclonal antibodies on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. J. Am. Soc. Mass Spectrom. 2009, 20, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Heck, A.J.; van den Heuvel, R.H. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 2004, 23, 368–389. [Google Scholar] [CrossRef] [PubMed]
- Mouls, L.; Silajdzic, E.; Haroune, N.; Spickett, C.M.; Pitt, A.R. Development of novel mass spectrometric methods for identifying HOCL-induced modifications to proteins. Proteomics 2009, 9, 1617–1631. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Uehara, H.; Shacter, E. Taurine chloramine-induced inactivation of cofilin protein through methionine oxidation. Free Radic. Biol. Med. 2014, 75, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Ehrmann, D.C.; Rose, K.; Calcutt, M.W.; Beller, A.B.; Hill, S.; Rogers, T.J.; Steele, S.D.; Hachey, D.L.; Aschner, J.L. Glutathionylated gammag and gammaa subunits of hemoglobin F: A novel post-translational modification found in extremely premature infants by LC-MS and nanoLC-MS/MS. J. Mass Spectrom. 2014, 49, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Carini, M.; Regazzoni, L.; Aldini, G. Mass spectrometric strategies and their applications for molecular mass determination of recombinant therapeutic proteins. Curr. Pharm. Biotechnol. 2011, 12, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Ansong, C.; Wu, S.; Meng, D.; Liu, X.W.; Brewer, H.M.; Kaiser, B.L.D.; Nakayasu, E.S.; Cort, J.R.; Pevzner, P.; Smith, R.D.; et al. Top-down proteomics reveals a unique protein S-thiolation switch in salmonella typhimurium in response to infection-like conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 10153–10158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ge, Y. Comprehensive analysis of protein modifications by top-down mass spectrometry. Circ. Cardiovasc. Genet. 2011. [Google Scholar] [CrossRef]
- Scotcher, J.; Clarke, D.J.; Mackay, C.L.; Hupp, T.; Sadler, P.J.; Langridge-Smith, P.R.R. Redox regulation of tumour suppressor protein p53: Identification of the sites of hydrogen peroxide oxidation and glutathionylation. Chem. Sci. 2013, 4, 1257–1269. [Google Scholar] [CrossRef]
- Holzmann, J.; Hausberger, A.; Rupprechter, A.; Toll, H. Top-down ms for rapid methionine oxidation site assignment in filgrastim. Anal. Bioanal. Chem. 2013, 405, 6667–6674. [Google Scholar] [CrossRef] [PubMed]
- Lourette, N.; Smallwood, H.; Wu, S.; Robinson, E.W.; Squier, T.C.; Smith, R.D.; Pasa-Tolic, L. A top-down LC-fticr MS-based strategy for characterizing oxidized calmodulin in activated macrophages. J. Am. Soc. Mass Spectrom. 2010, 21, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Aebersold, R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 2010, 28, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Stevens, S.M., Jr.; Prokai-Tatrai, K.; Prokai, L. Factors that contribute to the misidentification of tyrosine nitration by shotgun proteomics. Mol. Cell. Proteomics. 2008, 7, 2442–2451. [Google Scholar] [CrossRef] [PubMed]
- Craig, R.; Beavis, R.C. Tandem: Matching proteins with tandem mass spectra. Bioinformatics 2004, 20, 1466–1467. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef] [PubMed]
- Geer, L.Y.; Markey, S.P.; Kowalak, J.A.; Wagner, L.; Xu, M.; Maynard, D.M.; Yang, X.; Shi, W.; Bryant, S.H. Open mass spectrometry search algorithm. J. Proteome Res. 2004, 3, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. Peaks: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.M.N.; Vitorino, R.; Domingues, M.R.M.; Spickett, C.M.; Domingues, P. Post-translational modifications and mass spectrometry detection. Free Radic. Biol. Med. 2013, 65, 925–941. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [PubMed]
- Spickett, C.M.; Reis, A.; Pitt, A.R. Use of narrow mass-window, high-resolution extracted product ion chromatograms for the sensitive and selective identification of protein modifications. Anal. Chem. 2013, 85, 4621–4627. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, R.; Wilson, J.; Bridgewater, J.D.; Numbers, J.R.; Lim, J.; Olbris, M.R.; Kettani, A.; Vachet, R.W. Improved sequencing of oxidized cysteine and methionine containing peptides using electron transfer dissociation. J. Am. Soc. Mass Spectrom. 2007, 18, 1499–1506. [Google Scholar] [CrossRef] [PubMed]
- Shilov, I.V.; Seymour, S.L.; Patel, A.A.; Loboda, A.; Tang, W.H.; Keating, S.P.; Hunter, C.L.; Nuwaysir, L.M.; Schaeffer, D.A. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 2007, 6, 1638–1655. [Google Scholar] [CrossRef] [PubMed]
- Dorfer, V.; Pichler, P.; Stranzl, T.; Stadlmann, J.; Taus, T.; Winkler, S.; Mechtler, K. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J. Proteome Res. 2014, 13, 3679–3684. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Fu, Y.; Sun, R.X.; Wang, H.P.; Yuan, Z.F.; Chi, H.; He, S.M. Open MS/MS spectral library search to identify unanticipated post-translational modifications and increase spectral identification rate. Bioinformatics 2010, 26, i399–i406. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J. Detection and localization of methionine sulfoxide residues of specific proteins in brain tissue. Protein Pept. Lett. 2014, 21, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Steen, H.; Mann, M. The abc’s (and xyz’s) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 2004, 5, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Medzihradszky, K.F.; Chalkley, R.J. Lessons in de novo peptide sequencing by tandem mass spectrometry. Mass Spectrom. Rev. 2013, 34, 43–63. [Google Scholar] [CrossRef]
- Curran, T.G.; Bryson, B.D.; Reigelhaupt, M.; Johnson, H.; White, F.M. Computer aided manual validation of mass spectrometry-based proteomic data. Methods 2013, 61, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.; Kim, S.; Pevzner, P.A. Uninovo: A universal tool for de novo peptide sequencing. Bioinformatics 2013, 29, 1953–1962. [Google Scholar] [CrossRef] [PubMed]
- Muth, T.; Weilnbock, L.; Rapp, E.; Huber, C.G.; Martens, L.; Vaudel, M.; Barsnes, H. Denovogui: An open source graphical user interface for de novo sequencing of tandem mass spectra. J. Proteome Res. 2014, 13, 1143–1146. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Dekker, L.J.; Wu, S.; Vanduijn, M.M.; Luider, T.M.; Tolic, N.; Kou, Q.; Dvorkin, M.; Alexandrova, S.; Vyatkina, K.; et al. De novo protein sequencing by combining top-down and bottom-up tandem mass spectra. J. Proteome Res. 2014, 13, 3241–3248. [Google Scholar] [CrossRef] [PubMed]
- Petersson, A.S.; Steen, H.; Kalume, D.E.; Caidahl, K.; Roepstorff, P. Investigation of tyrosine nitration in proteins by mass spectrometry. J. Mass Spectrom. 2001, 36, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Abello, N.; Kerstjens, H.A.; Postma, D.S.; Bischoff, R. Protein tyrosine nitration: Selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 2009, 8, 3222–3238. [Google Scholar] [CrossRef] [PubMed]
- Tveen-Jensen, K.; Reis, A.; Mouls, L.; Pitt, A.R.; Spickett, C.M. Reporter ion-based mass spectrometry approaches for the detection of non-enzymatic protein modifications in biological samples. J. Proteomics 2013, 92, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Held, J.M.; Schilling, B.; Danielson, S.R.; Gibson, B.W. Confident identification of 3-nitrotyrosine modifications in mass spectral data across multiple mass spectrometry platforms. J. Proteomics 2011, 74, 2510–2521. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.Q.; Yates, N.A.; Bakhtiar, R. Detection and characterization of methionine oxidation in peptides by collision-induced dissociation and electron capture dissociation. J. Am. Soc. Mass Spectrom. 2003, 14, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Galeva, N.A.; Esch, S.W.; Williams, T.D.; Markille, L.M.; Squier, T.C. Rapid method for quantifying the extent of methionine oxidation in intact calmodulin. J. Am. Soc. Mass Spectrom. 2005, 16, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Rauniyar, N.; Prokai, L. Isotope-coded dimethyl tagging for differential quantification of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. J. Mass Spectrom. 2011, 46, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.M.; Zhong, F.; Du, M.; Duchoslav, E.; Sakuma, T.; McDermott, J.C. Multiple reaction monitoring as a method for identifying protein posttranslational modifications. J. Biomol. Technol. 2005, 16, 83–90. [Google Scholar]
- Held, J.M.; Danielson, S.R.; Behring, J.B.; Atsriku, C.; Britton, D.J.; Puckett, R.L.; Schilling, B.; Campisi, J.; Benz, C.C.; Gibson, B.W. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol. Cell Proteomics 2010, 9, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Lam, H.; Aebersold, R. Peptideatlas: A resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008, 9, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Valim, L.R.; Davies, J.A.; Jensen, K.T.; Guo, R.; Willison, K.R.; Spickett, C.M.; Pitt, A.R.; Klug, D.R. Identification and relative quantification of tyrosine nitration in a model peptide using two-dimensional infrared spectroscopy. J. Phys. Chem. B 2014, 118, 12855–12864. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.A.; Rush, J.; Stemman, O.; Kirschner, M.W.; Gygi, S.P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem ms. Proc. Natl. Acad. Sci .USA 2003, 100, 6940–6945. [Google Scholar] [CrossRef] [PubMed]
- Nahnsen, S.; Bielow, C.; Reinert, K.; Kohlbacher, O. Tools for label-free peptide quantification. Mol. Cell Proteomics 2013, 12, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Turk, R.; Piras, C.; Kovacic, M.; Samardzija, M.; Ahmed, H.; de Canio, M.; Urbani, A.; Mestric, Z.F.; Soggiu, A.; Bonizzi, L.; et al. Proteomics of inflammatory and oxidative stress response in cows with subclinical and clinical mastitis. J. Proteomics 2012, 75, 4412–4428. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Silva, G.M.; Marcotte, E.M. Protein expression regulation under oxidative stress. Mol. Cell Proteomics 2011. [Google Scholar] [CrossRef]
- May, D.; Fitzgibbon, M.; Liu, Y.; Holzman, T.; Eng, J.; Kemp, C.J.; Whiteaker, J.; Paulovich, A.; McIntosh, M. A platform for accurate mass and time analyses of mass spectrometry data. J. Proteome Res. 2007, 6, 2685–2694. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. Maxquant enables high peptide identification rates, individualized p.P.B.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Sturm, M.; Bertsch, A.; Gropl, C.; Hildebrandt, A.; Hussong, R.; Lange, E.; Pfeifer, N.; Schulz-Trieglaff, O.; Zerck, A.; Reinert, K.; et al. Openms—An open-source software framework for mass spectrometry. BMC Bioinform. 2008. [Google Scholar] [CrossRef]
- Mueller, L.N.; Rinner, O.; Schmidt, A.; Letarte, S.; Bodenmiller, B.; Brusniak, M.-Y.; Vitek, O.; Aebersold, R.; Müller, M. Superhirn—A novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 2007, 7, 3470–3480. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Trudgian, D.C.; Wright, C.; Thomas, G.; Bradbury, L.A.; Brown, M.A.; Bowness, P.; Kessler, B.M. Discovery of candidate serum proteomic and metabolomic biomarkers in ankylosing spondylitis. Mol. Cell Proteomics 2012. [Google Scholar] [CrossRef]
- Zaccarin, M.; Falda, M.; Roveri, A.; Bosello-Travain, V.; Bordin, L.; Maiorino, M.; Ursini, F.; Toppo, S. Quantitative label-free redox proteomics of reversible cysteine oxidation in red blood cell membranes. Free Radic. Biol. Med. 2014, 71, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.E.; Carroll, K.S. Chemical “omics” approaches for understanding protein cysteine oxidation in biology. Curr. Opin. Chem. Biol. 2011, 15, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Sethuraman, M.; McComb, M.E.; Huang, H.; Huang, S.; Heibeck, T.; Costello, C.E.; Cohen, R.A. Isotope-coded affinity tag (ICAT) approach to redox proteomics: Identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J. Proteome Res. 2004, 3, 1228–1233. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lee, W.N.; Xiao, G.G. Quantitative proteomics and biomarker discovery in human cancer. Expert Rev. Proteomics 2009, 6, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Scotcher, J.; Bythell, B.J.; Marshall, A.G. Unequivocal determination of site-specific protein disulfide bond reduction potentials by top-down FTICR MS: Characterization of the N- and C-terminal redox-active sites in human thioredoxin 1. Anal. Chem. 2013, 85, 9164–9172. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.T.; Chen, Y.Y.; Pu, T.H.; Chao, Y.S.; Yang, C.Y.; Bomgarden, R.D.; Rogers, J.C.; Meng, T.C.; Khoo, K.H. Mass spectrometry-based quantitative proteomics for dissecting multiplexed redox cysteine modifications in nitric oxide-protected cardiomyocyte under hypoxia. Antioxid. Redox Signal. 2014, 20, 1365–1381. [Google Scholar] [CrossRef] [PubMed]
- Wojdyla, K.; Williamson, J.; Roepstorff, P.; Rogowska-Wrzesinska, A. The SNO/SOH TMT strategy for combinatorial analysis of reversible cysteine oxidations. J. Proteomics 2015, 113, 415–434. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, B.; Martinez-Acedo, P.; Vazquez, J.; Padilla, C.A.; Sheehan, D.; Barcena, J.A. Application of iTRAQ reagents to relatively quantify the reversible redox state of cysteine residues. Int. J. Proteomics 2012, 2012, 514847. [Google Scholar] [CrossRef] [PubMed]
- Palmese, A.; de Rosa, C.; Chiappetta, G.; Marino, G.; Amoresano, A. Novel method to investigate protein carbonylation by iTRAQ strategy. Anal. Bioanal. Chem. 2012, 404, 1631–1635. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.A.; Evans, A.R. Enhanced sample multiplexing for nitrotyrosine-modified proteins using combined precursor isotopic labeling and isobaric tagging. Anal. Chem. 2012, 84, 4677–4686. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ponniah, G.; Neill, A.; Patel, R.; Andrien, B. Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis. Anal. Chem. 2013, 85, 11705–11709. [Google Scholar] [CrossRef] [PubMed]
- Fahlman, R.P.; Chen, W.; Overall, C.M. Absolute proteomic quantification of the activity state of proteases and proteolytic cleavages using proteolytic signature peptides and isobaric tags. J. Proteomics 2014, 100, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Knoefler, D.; Leicher, L.I.O.; Thamsen, M.; Cremers, C.M.; Reichmann, D.; Gray, M.J.; Wholey, W.Y.; Jakob, U. About the dangers, costs and benefits of living an aerobic lifestyle. Biochem. Soc. Trans. 2014, 42, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Ursini, F.; Maiorino, M. An overview of mechanisms of redox signaling. J. Mol. Cell. Cardiol. 2014, 73, 2–9. [Google Scholar] [CrossRef]
- Wani, R.; Nagata, A.; Murray, B.W. Protein redox chemistry: Post-translational cysteine modifications that regulate signal transduction and drug pharmacology. Front. Pharmacol. 2014. [Google Scholar] [CrossRef]
- Forman, H.J.; Fukuto, J.M.; Torres, M. Redox signaling: Thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am. J. Physiol. Cell. Physiol. 2004, 287, C246–C256. [Google Scholar] [CrossRef] [PubMed]
- Karplus, P.A. A primer on peroxiredoxin biochemistry. Free Radic. Biol. Med. 2015, 80, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Venereau, E.; Casalgrandi, M.; Schiraldi, M.; Antoine, D.J.; Cattaneo, A.; De Marchis, F.; Liu, J.; Antonelli, A.; Preti, A.; Raeli, L.; et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J. Exp. Med. 2012, 209, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Enescu, M.; Kassim, R.; Ramseyer, C.; Cardey, B. Theoretical insights into the mechanism of redox switch in heat shock protein Hsp33. J. Biol. Inorg. Chem. 2015. [Google Scholar] [CrossRef]
- Iyer, A.K.; Rojanasakul, Y.; Azad, N. Nitrosothiol signaling and protein nitrosation in cell death. Nitric Oxide 2014, 42, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Methner, C.; Nadtochiy, S.M.; Logan, A.; Pell, V.R.; Ding, S.; James, A.M.; Cocheme, H.M.; Reinhold, J.; Lilley, K.S.; et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat. Med. 2013, 19, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Bottari, S.P. Protein tyrosine nitration: A signaling mechanism conserved from yeast to man. Proteomics 2015, 15, 185–187. [Google Scholar] [CrossRef] [PubMed]
- Yeo, W.S.; Kim, Y.J.; Kabir, M.H.; Kang, J.W.; Kim, K.P. Mass spectrometric analysis of protein tyrosine nitration in aging and neurodegenerative diseases. Mass Spectrom. Rev. 2015, 34, 166–183. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; Kohr, M.; Menazza, S.; Nguyen, T.; Evangelista, A.; Sun, J.; Steenbergen, C. Signaling by S-nitrosylation in the heart. J. Mol. Cell Cardiol. 2014, 73, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Torta, F.; Usuelli, V.; Malgaroli, A.; Bachi, A. Proteomic analysis of protein S-nitrosylation. Proteomics 2008, 8, 4484–4494. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J.; Rabbani, N. Detection of oxidized and glycated proteins in clinical samples using mass spectrometry—A user’s perspective. Biochim. Biophys. Acta 2014, 1840, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Sacksteder, C.A.; Qian, W.J.; Knyushko, T.V.; Wang, H.X.; Chin, M.H.; Lacan, G.; Melega, W.P.; Camp, D.G.; Smith, R.D.; Smith, D.J.; et al. Endogenously nitrated proteins in mouse brain: Links to neurodegenerative disease. Biochemistry 2006, 45, 8009–8022. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Wehr, N.B. Protein carbonylation: Avoiding pitfalls in the 2,4-dinitrophenylhydrazine assay. Redox Rep. 2009, 14, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Augustyniak, E.; Adam, A.; Wojdyla, K.; Rogowska-Wrzesinska, A.; Willetts, R.; Korkmaz, A.; Atalay, M.; Weber, D.; Grune, T.; Borsa, C.; et al. Validation of protein carbonyl measurement: A multi-centre study. Redox Biol. 2014, 4, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Desiderio, D.M. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal. Biochem. 2006, 354, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Conrad, D.H.; Goyette, J.; Thomas, P.S. Proteomics as a method for early detection of cancer: A review of proteomics, exhaled breath condensate, and lung cancer screening. J. Gen. Int. Med. 2008, 23, 78–84. [Google Scholar] [CrossRef]
- Larstad, M.; Soderling, A.S.; Caidahl, K.; Olin, A.C. Selective quantification of free 3-nitrotyrosine in exhaled breath condensate in asthma using gas chromatography/tandem mass spectrometry. Nitric Oxide 2005, 13, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D. Analytical methods for 3-nitrotyrosine quantification in biological samples: The unique role of tandem mass spectrometry. Amino Acids 2012, 42, 45–63. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.L.; Anderson, N.G. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell Proteomics 2002, 1, 845–867. [Google Scholar] [CrossRef]
- Colombo, G.; Clerici, M.; Giustarini, D.; Rossi, R.; Milzani, A.; Dalle-Donne, I. Redox albuminomics: Oxidized albumin in human diseases. Antioxid. Redox Signal. 2012, 17, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Weisel, J.W.; Ischiropoulos, H. Functional impact of oxidative posttranslational modifications on fibrinogen and fibrin clots. Free Radic. Biol. Med. 2013, 65, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Wang, H.; Liu, J.J.; Zhang, Z.P.; McLuckey, M.N.; Ouyang, Z. Analysis of biological samples using paper spray mass spectrometry: An investigation of impacts by the substrates, solvents and elution methods. Chromatographia 2013, 76, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Ahmed, U.; Thornalley, P.J.; Hager, K.; Fleischer, G.; Munch, G. Protein glycation, oxidation and nitration adduct residues and free adducts of cerebrospinal fluid in Alzheimer’s disease and link to cognitive impairment. J. Neurochem. 2005, 92, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Korolainen, M.A.; Nyman, T.A.; Nyyssonen, P.; Hartikainen, E.S.; Pirttila, T. Multiplexed proteomic analysis of oxidation and concentrations of cerebrospinal fluid proteins in Alzheimer’s disease. Clin. Chem. 2007, 53, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Radabaugh, M.R.; Nemirovskiy, O.V.; Misko, T.P.; Aggarwal, P.; Mathews, W.R. Immunoaffinity liquid chromatography-tandem mass spectrometry detection of nitrotyrosine in biological fluids: Development of a clinically translatable biomarker. Anal. Biochem. 2008, 380, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Aydemir, B.; Onaran, I.; Kiziler, A.R.; Alici, B.; Akyolcu, M.C. The influence of oxidative damage on viscosity of seminal fluid in infertile men. J. Androl. 2008, 29, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Perluigi, M.; di Domenico, F.; Fiorini, A.; Cocciolo, A.; Giorgi, A.; Foppoli, C.; Butterfield, D.A.; Giorlandino, M.; Giorlandino, C.; Schinina, M.E.; et al. Oxidative stress occurs early in down syndrome pregnancy: A redox proteomics analysis of amniotic fluid. Proteomics Clin. Appl. 2011, 5, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Bahar, G.; Feinmesser, R.; Shpitzer, T.; Popovtzer, A.; Nagler, R.M. Salivary analysis in oral cancer patients: DNA and protein oxidation, reactive nitrogen species, and antioxidant profile. Cancer 2007, 109, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Reyzer, M.L.; Choi, I.J.; Kim, C.G.; Kim, H.S.; Oshima, A.; Chertov, O.; Colantonio, S.; Fisher, R.J.; Allen, J.L.; et al. Gastric cancer-specific protein profile identified using endoscopic biopsy samples via maldi mass spectrometry. J. Proteome Res. 2010, 9, 4123–4130. [Google Scholar] [CrossRef] [PubMed]
- Diamond, D.L.; Jacobs, J.M.; Paeper, B.; Proll, S.C.; Gritsenko, M.A.; Carithers, R.L., Jr.; Larson, A.M.; Yeh, M.M.; Camp, D.G., II; Smith, R.D.; et al. Proteomic profiling of human liver biopsies: Hepatitis C virus-induced fibrosis and mitochondrial dysfunction. Hepatology 2007, 46, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Canton, M.; Menazza, S.; Sheeran, F.L.; Polverino de Laureto, P.; di Lisa, F.; Pepe, S. Oxidation of myofibrillar proteins in human heart failure. J. Am. Coll. Cardiol. 2011, 57, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Sullards, M.C.; Olzmann, J.A.; Rees, H.D.; Weintraub, S.T.; Bostwick, D.E.; Gearing, M.; Levey, A.I.; Chin, L.S.; Li, L. Oxidative damage of DJ-1 is linked to sporadic parkinson and Alzheimer diseases. J. Biol. Chem. 2006, 281, 10816–10824. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.P.; Biswas, S.; Merchant, A.S.; Satoskar, A.; Taslim, C.; Lin, S.L.; Rovin, B.H.; Sen, C.K.; Roy, S.; Freitas, M.A. A quantitative proteomic workflow for characterization of frozen clinical biopsies: Laser capture microdissection coupled with label-free mass spectrometry. J. Proteomics 2012, 77, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Djidja, M.C.; Claude, E.; Snel, M.F.; Francese, S.; Scriven, P.; Carolan, V.; Clench, M.R. Novel molecular tumour classification using MALDI-mass spectrometry imaging of tissue micro-array. Anal. Bioanal. Chem. 2010, 397, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol Med 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Dozaki, N.; Nakamura, T.; Kitamoto, N.; Yoshida, A.; Naito, M.; Kitamura, M.; Osawa, T. Quantification of modified tyrosines in healthy and diabetic human urine using liquid chromatography/tandem mass spectrometry. J. Clin. Biochem. Nutr. 2009, 44, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Svatikova, A.; Wolk, R. Circulating free nitrotyrosine in obstructive sleep apnea. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004, 55905, 284–287. [Google Scholar] [CrossRef]
- Oriolli, M.; Aldini, G.; Benfatto, M.C.; Facino, R.M.; Carini, M. Hne michael adducts to histidine and histidine-containing peptides as biomarkers of lipid-derived carbonyl stress in urines: Lc-MS/NIS profiling in zucker obese rats. Anal. Chem. 2007, 79, 9174–9184. [Google Scholar] [CrossRef] [PubMed]
- Hui, Y.; Wong, M.; Zhao, S.S.; Love, J.A.; Ansley, D.M.; Chen, D.D. A simple and robust LC-MS/MS method for quantification of free 3-nitrotyrosine in human plasma from patients receiving on-pump CABG surgery. Electrophoresis 2012, 33, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Kuligowski, J.; Torres-Cuevas, I.; Quintas, G.; Rook, D.; van Goudoever, J.B.; Cubells, E.; Asensi, M.; Lliso, I.; Nunez, A.; Vento, M.; et al. Assessment of oxidative damage to proteins and DNA in urine of newborn infants by a validated upLC-MS/MS approach. PLOS ONE 2014, 9, e93703. [Google Scholar] [CrossRef] [PubMed]
- Nemirovskiy, O.V.; Radabaugh, M.R.; Aggarwal, P.; Funckes-Shippy, C.L.; Mnich, S.J.; Meyer, D.M.; Sunyer, T.; Rodney Mathews, W.; Misko, T.P. Plasma 3-nitrotyrosine is a biomarker in animal models of arthritis: Pharmacological dissection of iNOS’ role in disease. Nitric Oxide 2009, 20, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.M.; Strobel, F.H.; Reed, M.; Pohl, J.; Jones, D.P. A rapid LC-FTMS method for the analysis of cysteine, cystine and cysteine/cystine steady-state redox potential in human plasma. Clin. Chim. Acta 2008, 396, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Murdaugh, L.S.; Wang, Z.; Del Priore, L.V.; Dillon, J.; Gaillard, E.R. Age-related accumulation of 3-nitrotyrosine and nitro-A2E in human Bruch’s membrane. Exp. Eye Res. 2010, 90, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Winyard, P.G.; Ryan, B.; Eggleton, P.; Nissim, A.; Taylor, E.; lo Faro, M.L.; Burkholz, T.; Szabo-Taylor, K.E.; Fox, B.; Viner, N.; et al. Measurement and meaning of markers of reactive species of oxygen, nitrogen and sulfur in healthy human subjects and patients with inflammatory joint disease. Biochem. Soc. Trans. 2011, 39, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Malakowsky, C.A.; Talent, J.M.; Conrad, C.C.; Gracy, R.W. Identification of oxidized plasma proteins in Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2002, 293, 1566–1570. [Google Scholar] [CrossRef] [PubMed]
- Soreghan, B.A.; Yang, F.; Thomas, S.N.; Hsu, J.; Yang, A.J. High-throughput proteomic-based identification of oxidatively induced protein carbonylation in mouse brain. Pharm. Res. 2003, 20, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.R.; de Waal, E.M.; Pierce, A.; van Remmen, H.; Ward, W.F.; Richardson, A. Detection of protein carbonyls in aging liver tissue: A fluorescence-based proteomic approach. Mech. Ageing Dev. 2006, 127, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Xie, H.; Meany, D.L.; Thompson, L.V.; Arriaga, E.A.; Griffin, T.J. Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J. Gerontol. A Biol. Sci. Med. Sci. 2008, 63, 1137–1152. [Google Scholar] [CrossRef] [PubMed]
- Sultana, R.; Perluigi, M.; Newman, S.F.; Pierce, W.M.; Cini, C.; Coccia, R.; Butterfield, D.A. Redox proteomic analysis of carbonylated brain proteins in mild cognitive impairment and early Alzheimer’s disease. Antioxid. Redox Signal. 2010, 12, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Chavez, J.D.; Wu, J.; Bisson, W.; Maier, C.S. Site-specific proteomic analysis of lipoxidation adducts in cardiac mitochondria reveals chemical diversity of 2-alkenal adduction. J. Proteomics 2011, 74, 2417–2429. [Google Scholar] [CrossRef] [PubMed]
- Madian, A.G.; Myracle, A.D.; Diaz-Maldonado, N.; Rochelle, N.S.; Janle, E.M.; Regnier, F.E. Differential carbonylation of proteins as a function of in vivo oxidative stress. J. Proteome Res. 2011, 10, 3959–3972. [Google Scholar] [CrossRef] [PubMed]
- Bollineni, R.C.; Fedorova, M.; Bluher, M.; Hoffmann, R. Carbonylated plasma proteins as potential biomarkers of obesity induced type 2 diabetes mellitus. J. Proteome Res. 2014, 13, 5081–5093. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, S.; Yamada, T.; Minohata, T.; Kobayashi, H.; Furukawa, A.; Tada-Oikawa, S.; Hiraku, Y.; Murata, M.; Kikuchi, M.; Yamashima, T. Proteomic identification of carbonylated proteins in the monkey hippocampus after ischemia-reperfusion. Free Radic. Biol. Med. 2009, 46, 1472–1477. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Liang, C.; Limmon, G.V.; Liang, L.; Engelward, B.P.; Ooi, E.E.; Chen, J.; Tannenbaum, S.R. Molecular analysis of serum and bronchoalveolar lavage in a mouse model of influenza reveals markers of disease severity that can be clinically useful in humans. PLOS ONE 2014, 9, e86912. [Google Scholar] [CrossRef] [PubMed]
- Dalle-Donne, I.; Aldini, G.; Carini, M.; Colombo, R.; Rossi, R.; Milzani, A. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med. 2006, 10, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Rees, H.D.; Weintraub, S.T.; Levey, A.I.; Chin, L.S.; Li, L. Oxidative modifications and aggregation of Cu,Zn-superoxide dismutase associated with Alzheimer and Parkinson diseases. J. Biol. Chem. 2005, 280, 11648–11655. [Google Scholar] [CrossRef] [PubMed]
- Madian, A.G.; Diaz-Maldonado, N.; Gao, Q.; Regnier, F.E. Oxidative stress induced carbonylation in human plasma. J. Proteomics 2011, 74, 2395–2416. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.M.; Jones, D.P. Cysteine/cystine redox signaling in cardiovascular disease. Free Radic. Biol. Med. 2011, 50, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Sanders, L.H.; Greenamyre, J.T. Oxidative damage to macromolecules in human parkinson disease and the rotenone model. Free Radic. Biol. Med. 2013, 62, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Sharov, V.S.; Dremina, E.S.; Galeva, N.A.; Williams, T.D.; Schoneich, C. Quantitative mapping of oxidation-sensitive cysteine residues in SERCA in vivo and in vitro by HPLC-electrospray-tandem MS: Selective protein oxidation during biological aging. Biochem. J. 2006, 394, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Levey, A.I.; Weintraub, S.T.; Rees, H.D.; Gearing, M.; Chin, L.S.; Li, L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J. Biol. Chem. 2004, 279, 13256–13264. [Google Scholar] [CrossRef] [PubMed]
- Riederer, I.M.; Schiffrin, M.; Kovari, E.; Bouras, C.; Riederer, B.M. Ubiquitination and cysteine nitrosylation during aging and Alzheimer’s disease. Brain Res. Bull. 2009, 80, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Kohr, M.J.; Aponte, A.; Sun, J.; Gucek, M.; Steenbergen, C.; Murphy, E. Measurement of S-nitrosylation occupancy in the myocardium with cysteine-reactive tandem mass tags: Short communication. Circ. Res. 2012, 111, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kleffmann, T.; Hampton, M.B.; Cannell, M.B.; Winterbourn, C.C. Redox proteomics of thiol proteins in mouse heart during ischemia/reperfusion using ICAT reagents and mass spectrometry. Free Radic. Biol. Med. 2013, 58, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.M.; Roede, J.R.; Orr, M.; Liang, Y.; Jones, D.P. Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of CD toxicity. Toxicol. Sci. 2014, 139, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Schoneich, C. Methionine oxidation by reactive oxygen species: Reaction mechanisms and relevance to Alzheimer’s disease. Biochim. Biophys. Acta 2005, 1703, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Glaser, C.B.; Yamin, G.; Uversky, V.N.; Fink, A.L. Methionine oxidation, alpha-synuclein and Parkinson’s disease. Biochim. Biophys. Acta 2005, 1703, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Brock, J.W.; Jenkins, A.J.; Lyons, T.J.; Klein, R.L.; Yim, E.; Lopes-Virella, M.; Carter, R.E.; Research, G.; Thorpe, S.R.; Baynes, J.W. Increased methionine sulfoxide content of ApoA-I in type 1 diabetes. J. Lipid Res. 2008, 49, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Tsimikas, S. In vivo markers of oxidative stress and therapeutic interventions. Am. J. Cardiol. 2008, 101, 34D–42D. [Google Scholar] [CrossRef] [PubMed]
- Castegna, A.; Thongboonkerd, V.; Klein, J.B.; Lynn, B.; Markesbery, W.R.; Butterfield, D.A. Proteomic identification of nitrated proteins in Alzheimer’s disease brain. J. Neurochem. 2003, 85, 1394–1401. [Google Scholar] [CrossRef] [PubMed]
- Knutson, C.G.; Mangerich, A.; Zeng, Y.; Raczynski, A.R.; Liberman, R.G.; Kang, P.; Ye, W.; Prestwich, E.G.; Lu, K.; Wishnok, J.S.; et al. Chemical and cytokine features of innate immunity characterize serum and tissue profiles in inflammatory bowel disease. Proc. Natl. Acad. Sci. USA 2013, 110, E2332–E2341. [Google Scholar] [CrossRef] [PubMed]
- Pennathur, S.; Bergt, C.; Shao, B.; Byun, J.; Kassim, S.Y.; Singh, P.; Green, P.S.; McDonald, T.O.; Brunzell, J.; Chait, A.; et al. Human atherosclerotic intima and blood of patients with established coronary artery disease contain high density lipoprotein damaged by reactive nitrogen species. J. Biol. Chem. 2004, 279, 42977–42983. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.K.; Vivekanandan-Giri, A.; Tang, C.; Knight, J.S.; Mathew, A.; Padilla, R.L.; Gillespie, B.W.; Carmona-Rivera, C.; Liu, X.; Subramanian, V.; et al. Neutrophil extracellular trap-derived enzymes oxidize high-density lipoprotein: An additional proatherogenic mechanism in systemic lupus erythematosus. Arthritis Rheumatol. 2014, 66, 2532–2544. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.; Pennathur, S.; Heinecke, J.W. Myeloperoxidase targets apolipoprotein A–I, the major high density lipoprotein protein, for site-specific oxidation in human atherosclerotic lesions. J. Biol. Chem. 2012, 287, 6375–6386. [Google Scholar] [CrossRef] [PubMed]
- Paton, L.N.; Mocatta, T.J.; Richards, A.M.; Winterbourn, C.C. Increased thrombin-induced polymerization of fibrinogen associated with high protein carbonyl levels in plasma from patients post myocardial infarction. Free Radic. Biol. Med. 2010, 48, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Sparvero, L.J.; Amoscato, A.A.; Kochanek, P.M.; Pitt, B.R.; Kagan, V.E.; Bayir, H. Mass-spectrometry based oxidative lipidomics and lipid imaging: Applications in traumatic brain injury. J. Neurochem. 2010, 115, 1322–1336. [Google Scholar] [CrossRef] [PubMed]
- Zanivan, S.; Krueger, M.; Mann, M. In vivo quantitative proteomics: The silac mouse. Methods Mol. Biol. 2012, 757, 435–450. [Google Scholar] [PubMed]
- McClatchy, D.B.; Liao, L.; Park, S.K.; Xu, T.; Lu, B.; Yates III, J.R. Differential proteomic analysis of mammalian tissues using silam. PLOS ONE 2011, 6, e16039. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.S.; Xiong, Y.; Ho, D.S.; Gao, J.; Chua, B.H.; Pai, H.; Mieyal, J.J. Targeted disruption of the glutaredoxin 1 gene does not sensitize adult mice to tissue injury induced by ischemia/reperfusion and hyperoxia. Free Radic. Biol. Med. 2007, 43, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Putker, M.; Vos, H.R.; van Dorenmalen, K.; de Ruiter, H.; Duran, A.G.; Snel, B.; Burgering, B.M.; Vermeulen, M.; Dansen, T.B. Evolutionary acquisition of cysteines determines FOXO paralog-specific redox signaling. Antioxid. Redox Signal. 2015, 22, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Rivera, J.; Sobey, C.G.; Walduck, A.K.; Drummond, G.R. Nox isoforms in vascular pathophysiology: Insights from transgenic and knockout mouse models. Redox Rep. 2010, 15, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Adimora, N.J.; Jones, D.P.; Kemp, M.L. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxid. Redox Signal. 2010, 13, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Coon, J.J.; Zurbig, P.; Dakna, M.; Dominiczak, A.F.; Decramer, S.; Fliser, D.; Frommberger, M.; Golovko, I.; Good, D.M.; Herget-Rosenthal, S.; et al. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clin. Appl. 2008, 2, 964–973. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verrastro, I.; Pasha, S.; Jensen, K.T.; Pitt, A.R.; Spickett, C.M. Mass Spectrometry-Based Methods for Identifying Oxidized Proteins in Disease: Advances and Challenges. Biomolecules 2015, 5, 378-411. https://doi.org/10.3390/biom5020378
Verrastro I, Pasha S, Jensen KT, Pitt AR, Spickett CM. Mass Spectrometry-Based Methods for Identifying Oxidized Proteins in Disease: Advances and Challenges. Biomolecules. 2015; 5(2):378-411. https://doi.org/10.3390/biom5020378
Chicago/Turabian StyleVerrastro, Ivan, Sabah Pasha, Karina Tveen Jensen, Andrew R. Pitt, and Corinne M. Spickett. 2015. "Mass Spectrometry-Based Methods for Identifying Oxidized Proteins in Disease: Advances and Challenges" Biomolecules 5, no. 2: 378-411. https://doi.org/10.3390/biom5020378
APA StyleVerrastro, I., Pasha, S., Jensen, K. T., Pitt, A. R., & Spickett, C. M. (2015). Mass Spectrometry-Based Methods for Identifying Oxidized Proteins in Disease: Advances and Challenges. Biomolecules, 5(2), 378-411. https://doi.org/10.3390/biom5020378