Regulation of AU-Rich Element RNA Binding Proteins by Phosphorylation and the Prolyl Isomerase Pin1
Abstract
:1. Introduction
| AUBP | Phosphorylation site * | Kinase | Interaction w/Pin1 | mRNA stability affected by AUBP | mRNA stability/expression affected by Pin1 |
|---|---|---|---|---|---|
| AUF1 | Ser83, Ser87, Thr91 | CK1, GSK3β, PKA [27,28,29] | Yes [30,31] | c-myc, c-fos, Cyclin D1, GM-CSF, iNOS, IL-1β, IL-2, IL-3, IL-6, IL-10, p21, PTH, TNF-α [32,33,34,35] | Cyclin D1, GM-CSF, IL-1β, IL-2, IL-6, PTH, TNF-α [30,36,37,38,39,40,41] |
| BRF1 | Ser54, Ser92, Ser203 | AKT, ERK2 [42,43,44] | N/D ** | GM-CSF, IL-3, TNF-α [8] | GM-CSF, TNF-α [30,38,40] |
| DAZAP1 | Thr269, Thr315 | ERK2 [45,46] | N/D | Regulates RNA splicing and translation [45,46,47] | |
| hnRNP C | N/D | N/D | No [30,31,36] | APP, GM-CSF, TGF-β, Urokinase receptor [30,36,48,49,50] | GM-CSF, TGF-β [30,36,38] |
| HuR | Ser88, Ser100, Thr118, Ser158, Tyr200, Ser202, Ser221, Ser242, Ser318 | AMPK, MAPKs, CDK1, CHK2, JAK3, PKCs [51,52,53,54,55,56,57,58,59,60,61,62,63] | Yes [30,31,36] | AFT-2, C/EBP-β, Cyclin A/B1/D1, Cox-2, cPLA2α, CXCL8, CXCL1/5, c-fos, Dll1, DNMT3B, GATA3, GM-CSF, iNOS, IL-3, IL-8, MyoD, Myogenin, Musashi1, NPM, p21, PEPCK, RGS4, SIRT1, SMN, Survivin, TNF-α, VEGF, VHL, XIAP [8,50,56,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78] | Cyclin D1, Cox-2, GM-CSF, iNOS, IL-8, TNF-α, VEGF [30,38,40,79,80] |
| KSRP | Ser193, Thr692 | AKT, p38 MAPK [81,82] | Yes [37] | β-catenin, c-fos, c-jun, GAP43, IL-2, iNOS, MyoD, Myogenin, p21, TNF-α [8,64,81,82,83] | IL-2, iNOS, β-catenin, TNF-α [40,84,85,86,87] |
| La (SSB) | Thr301, Ser366, Thr389 | AKT, CK2 [88,89,90,91] | N/D | Regulates RNA translation [88,90,91] | |
| NF90 | Ser482, Ser647 | AKT, PKC-β [92,93,94] | N/D | Cyclin E1, IL-2, MKP-1 [95,96,97] | IL-2 [84] |
| Nucleolin | Thr641, Thr707 | CDK1,CDC2,CK2,GSK3β,PI3K [98,99,100,101,102] | No [30] | Bcl-xL, β-globin, CD154, GM-CSF, Gadd45a, Gastrin, HIF1α, IL-2, p53 [18,102,103,104,105,106,107,108,109,110] | IL-2, GM-CSF [30,38,84] |
| SLBP | Ser60, Thr62, Thr171 | N/D [111,112,113] | Yes [26,31] | Histone [26] | Histone [26] |
| TIA-1/R | N/D | FASTK [114,115,116] | No [30,31,36] | Regulates RNA translation [13] | |
| TTP | Ser52, Ser66, Ser88, Thr92, Ser93, Ser169, Ser178, Ser186, Ser197, Ser218, Ser220, Ser228, Ser245, Thr250, Ser276, Ser296 | AKT, GSK3β, MK2, MAPKs, PKA, PKC [117,118,119,120,121,122,123,124,125] | N/D | Bdp1, Claudin-1, Cyclin D1, c-fos, Cox-2, GM-CSF, IL-1α, IL-2, IL-3, IL-6, IL-8, IL-10, IL-23, ler3, IFN-γ, iNOS, LIF, Mllt11, c-myc, Pim3, PLK3, PHLDA1, PAI-2, Pitx2, Rusc2, TNF-α, VEGF [32,126,127] | Cox-2, GM-CSF, IL-2, IL-8, IFN-γ, iNOS, TNF-α, VEGF [30,38,40,79,84,85] |
| YB1 | Ser316 | AKT, ERK2, GSK3β,JNK [103,128,129,130] | No [30] | IL-2 [32,103,128] | IL-2 [84] |
2. Post-translational Regulation of AUBPs by Phosphorylation
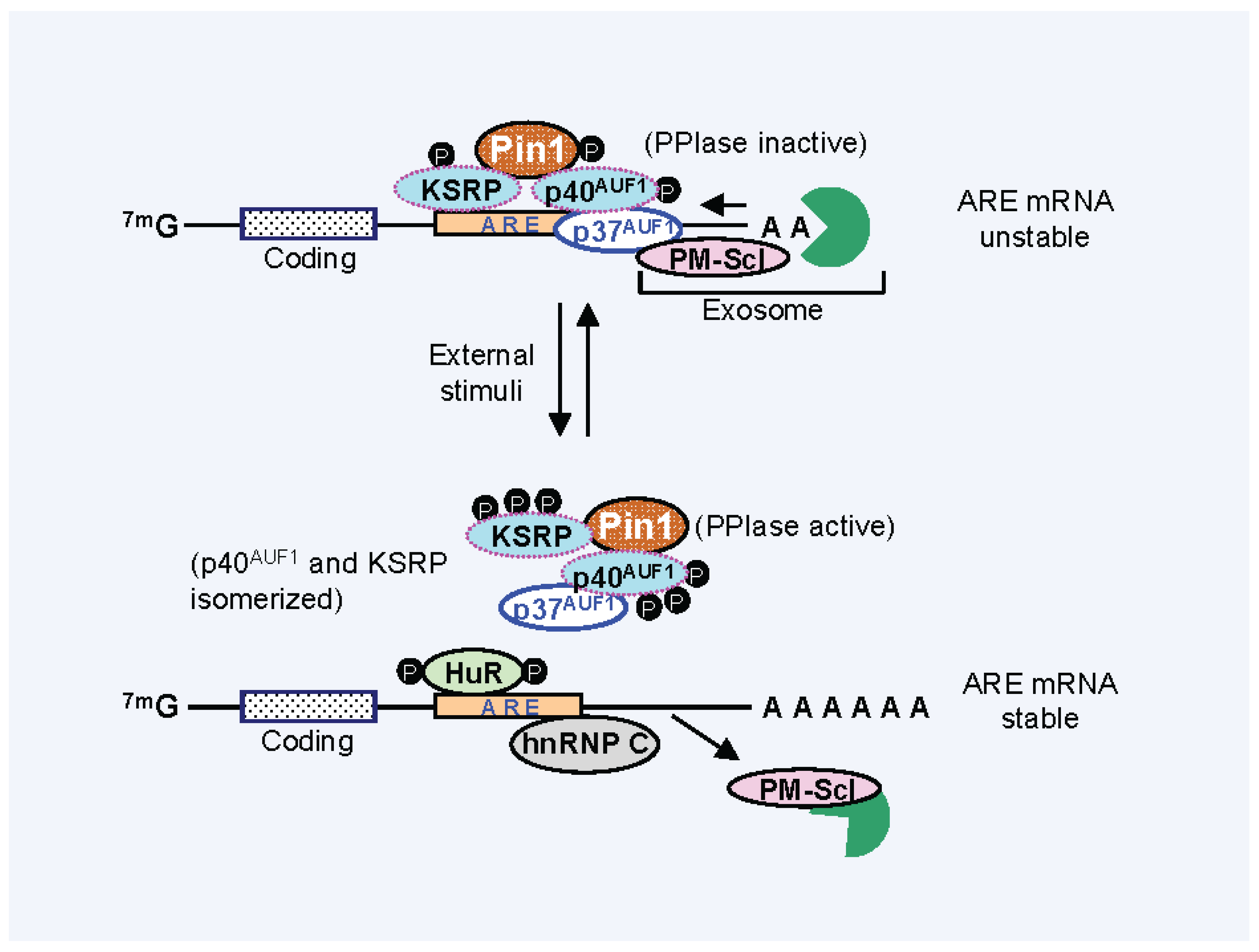
3. Pin1 Is Associated with AUBPs and Regulates Cytokine mRNA Stability
4. Effect of Pin1 Prolyl Directed, Cis-Trans Isomerization Activity on mRNA-AUBP Complex Remodeling
5. Pin1 and Immune Disorders
6. Conclusions
Acknowledgments
Conflicts of interest
References
- Schoenberg, D.R.; Maquat, L.E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012, 13, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ortín, J.E.; Alepuz, P.; Chávez, S.; Choder, M. Eukaryotic mRNA decay: Methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 2013, 425, 3750–3775. [Google Scholar] [CrossRef] [PubMed]
- Bakheet, T.; Williams, B.R.; Khabar, K.S. ARED 2.0: An update of AU-rich element mRNA database. Nucleic Acids Res. 2003, 31, 421–423. [Google Scholar]
- Halees, A.S.; El-Badrawi, R.; Khabar, K.S. ARED Organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008, 36, D137–D140. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; van Nimwegen, E.; Zavolan, M.; Rajewsky, N.; Schroeder, M.; Magnasco, M.; Darnell, J.E., Jr. Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 2003, 13, 1863–1872. [Google Scholar] [CrossRef] [PubMed]
- Gingerich, T.J.; Feige, J.J.; LaMarre, J. AU-rich elements and the control of gene expression through regulated mRNA stability. Anim. Health Res. Rev. 2004, 5, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Malter, J.S. Identification of an AUUUA-specific messenger RNA binding protein. Science 1989, 246, 664–666. [Google Scholar] [CrossRef] [PubMed]
- Barreau, C.; Paillard, L.; Osborne, H.B. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2006, 33, 7138–7150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoecklin, G.; Anderson, P. Posttranscriptional mechanisms regulating the inflammatory response. Adv. Immunol. 2006, 89, 1–37. [Google Scholar] [PubMed]
- Shaw, G.; Kamen, R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 1986, 46, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Stoecklin, G.; Tenenbaum, S.A.; Mayo, T.; Chittur, S.V.; George, A.D.; Baroni, T.E.; Blackshear, P.J.; Anderson, P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J. Biol. Chem. 2008, 283, 11689–11699. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.S.; Parker, J.S.; Grissom, S.F.; Stumpo, D.J.; Blackshear, P.J. Novel mRNA argets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell Biol. 2006, 26, 9196–9208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Kruys, V.; Huez, G.; Gueydan, C. AU-rich element-mediated translational control: Complexity and multiple activities of trans-activating factors. Biochem. Soc. Trans. 2002, 30, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Thapar, R.; Denmon, A.P. Signaling pathways that control mRNA turnover. Cell Signal 2013, 25, 1699–1710. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Shyu, A.B. Emerging mechanisms of mRNP remodeling regulation. Wiley Interdiscip. Rev. RNA 2014, 5, 713–722. [Google Scholar] [PubMed]
- Liu, Q.; Dreyfuss, G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell Biol. 1995, 15, 2800–2808. [Google Scholar] [PubMed]
- Eberhardt, W.; Doller, A.; Pfeilschifter, J. Regulation of the mRNA-binding protein HuR by posttranslational modification: Spotlight on phosphorylation. Curr. Protein Pept. Sci. 2012, 13, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liang, Y.; Xie, Q.; Gao, G.; Wei, J.; Huang, H.; Li, J.; Gao, J.; Huang, C. A novel post-translational modification of nucleolin, SUMOylation at K294, mediates Arsenite-induced cell death by regulating GADD45α mRNA stability. J. Biol. Chem. 2015. [Google Scholar] [CrossRef]
- Laroia, G.; Sarkar, B.; Schneider, R.J. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl. Acad. Sci. USA 2002, 99, 1842–1846. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.P.; Hanes, S.D.; Hunter, T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 1996, 380, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, L.; Sun, A.; Lu, P.J.; Zhou, X.Z.; Balastik, M.; Finn, G.; Wulf, G.; Lim, J.; Li, S.H.; Li, X.; et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature 2006, 440, 528–534. [Google Scholar]
- Esnault, S.; Shen, Z.J.; Malter, J.S. Pinning down signaling in the immune system: The role of the peptidyl-prolyl isomerase Pin1 in immune cell function. Crit. Rev. Immunol. 2008, 28, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, R.; Lu, K.P.; Hunter, T.; Noel, J.P. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 1997, 89, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Verdecia, M.A.; Bowman, M.E.; Lu, K.P.; Hunter, T.; Noel, J.P. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 2000, 7, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Daum, S.; Wildemann, D.; Zhou, X.Z.; Verdecia, M.A.; Bowman, M.E.; Lücke, C.; Hunter, T.; Lu, K.P.; Fischer, G.; et al. Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem. Biol. 2007, 2, 320–328. [Google Scholar]
- Krishnan, N.; Lam, T.T.; Fritz, A.; Rempinski, D.; O’Loughlin, K.; Minderman, H.; Berezney, R.; Marzluff, W.F.; Thapar, R. The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol. Cell Biol. 2012, 32, 4306–4322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wagner, B.J.; Ehrenman, K.; Schaefer, A.W.; DeMaria, C.T.; Crater, D.; DeHaven, K.; Long, L.; Brewer, G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell Biol. 1993, 13, 7652–7665. [Google Scholar] [PubMed]
- Wilson, G.M.; Lu, J.; Sutphen, K.; Sun, Y.; Huynh, Y.; Brewer, G. Regulation of A + U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 2003, 278, 33029–33038. [Google Scholar] [CrossRef] [PubMed]
- Kreegipuu, A.; Blom, N.; Brunak, S. PhosphoBase, a database of phosphorylation sites: Release 2.0. Nucleic Acids Res. 1999, 27, 237–239. [Google Scholar] [CrossRef]
- Shen, Z.J.; Esnault, S.; Malter, J.S. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat. Immunol. 2005, 6, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, N.; Titus, M.A.; Thapar, R. The prolyl isomerase pin1 regulates mRNA levels of genes with short half-lives by targeting specific RNA binding proteins. PLoS ONE 2014, 9, e85427. [Google Scholar] [CrossRef] [PubMed]
- Stumpo, D.J.; Lai, W.S.; Blackshear, P.J. Inflammation: Cytokines and RNA-based regulation. Wiley Interdiscip. Rev. RNA 2010, 1, 60–80. [Google Scholar] [CrossRef] [PubMed]
- Paschoud, S.; Dogar, A.M.; Kuntz, C.; Grisoni-Neupert, B.; Richman, L.; Kühn, L.C. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol. Cell Biol. 2006, 26, 8228–8241. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Sinsimer, K.S.; Foster, R.L.; Brewer, G.; Pestka, S. AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes. J. Interferon Cytokine Res. 2008, 28, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, D.; Zuraw, L.; Ramalingam, S.; Sengupta, T.K.; Bandyopadhyay, S.; Reuben, A.; Fernandes, D.J.; Spicer, E.K. Mechanism of regulation of Bcl-2 mRNA by nucleolin and A + U-rich element-binding factor 1 (AUF1). J. Biol. Chem. 2010, 285, 27182–27191. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.J.; Esnault, S.; Rosenthal, L.A.; Szakaly, R.J.; Sorkness, R.L.; Westmark, P.R.; Sandor, M.; Malter, J.S. Pin1 regulates TGF-beta1 production by activated human and murine eosinophils and contributes to allergic lung fibrosis. J. Clin. Invest. 2008, 118, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Nechama, M.; Uchida, T.; Mor Yosef-Levi, I.; Silver, J.; Naveh-Many, T. The peptidyl-prolyl isomerase Pin1 determines parathyroid hormone mRNA levels and stability in rat models of secondary hyperparathyroidism. J. Clin. Invest. 2009, 119, 3102–3114. [Google Scholar] [CrossRef] [PubMed]
- Esnault, S.; Shen, Z.J.; Whitesel, E.; Malter, J.S. The peptidyl-prolyl isomerase Pin1 regulates granulocyte-macrophage colony-stimulating factor mRNA stability in T lymphocytes. J. Immunol. 2006, 177, 6999–7006. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Misawa, T.; Ono, M.; Uchida, C.; Uchida, T. Prolyl isomerase Pin1 protects mice from endotoxin shock. PLoS ONE 2011, 6, e14656. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Schneider, R.A.; Lee, N.Y.; Hoyt, D.G. Peptidylprolyl cis/trans isomerase, NIMA-interacting 1 (PIN1) regulates pulmonary effects of endotoxin and tumor necrosis factor-α in mice. Biochem. Biophys. Res. Commun. 2014, 452, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, H.; Uchida, T.; Mori, S.; Echigo, S.; Motegi, K. Expression status of Pin1 and cyclins in oral squamous cell carcinoma: Pin1 correlates with Cyclin D1 mRNA expression and clinical significance of cyclins. Oncol. Rep. 2003, 10, 1045–1048. [Google Scholar] [PubMed]
- Maitra, S.; Chou, C.F.; Luber, C.A.; Lee, K.Y.; Mann, M.; Chen, C.Y. The AU-rich element mRNA decay-promoting activity of BRF1 is regulated by mitogen-activated protein kinase-activated protein kinase 2. RNA 2008, 14, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.; Schmidlin, M.; Min, L.; Gross, B.; Moroni, C. BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol. Cell Biol. 2006, 26, 9497–9507. [Google Scholar] [CrossRef] [PubMed]
- Schmidlin, M.; Lu, M.; Leuenberger, S.A.; Stoecklin, G.; Mallaun, M.; Gross, B.; Gherzi, R.; Hess, D.; Hemmings, B.A.; Moroni, C. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004, 23, 4760–4769. [Google Scholar] [CrossRef] [PubMed]
- Morton, S.; Yang, H.T.; Moleleki, N.; Campbell, D.G.; Cohen, P.; Rousseau, S. Phosphorylation of the ARE-binding protein DAZAP1 by ERK2 induces its dissociation from DAZ. Biochem. J. 2006, 399, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.W.; Anderson, R.C.; Smith, J.W.; Brook, M.; Richardson, W.A.; Gray, N.K. DAZAP1, an RNA-binding protein required for development and spermatogenesis, can regulate mRNA translation. RNA 2011, 17, 1282–1295. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Yu, Y.H.; Yen, P.H. DAZAP1 regulates the splicing of Crem, Crisp2 and Pot1a transcripts. Nucleic Acids Res. 2013, 41, 9858–9869. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, L.E.; Westmark, C.J.; Jarzembowski, J.A.; Malter, J.S. hnRNP C increases amyloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res. 1998, 26, 3418–3423. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S. Regulation of urokinase receptor mRNA stability by hnRNP C in lung epithelial cells. Mol. Cell Biochem. 2005, 272, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Esnault, S.; Malter, J.S. Hyaluronic acid or TNF-alpha plus fibronectin triggers granulocyte macrophage-colony-stimulating factor mRNA stabilization in eosinophils yet engages differential intracellular pathways and mRNA binding proteins. J. Immunol. 2003, 171, 6780–6787. [Google Scholar] [CrossRef] [PubMed]
- Doller, A.; Schlepckow, K.; Schwalbe, H.; Pfeilschifter, J.; Eberhardt, W. Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol. Cell Biol. 2010, 30, 1397–1410. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Yang, X.; Kuwano, Y.; Gorospe, M. Modification at HuR(S242) alters HuR localization and proliferative influence. Cell Cycle 2008, 7, 3371–3377. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Pullmann, R., Jr.; Lal, A.; Kim, H.H.; Galban, S.; Yang, X.; Blethrow, J.D.; Walker, M.; Shubert, J.; Gillespie, D.A.; et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007, 25, 543–557. [Google Scholar]
- Kim, H.H.; Abdelmohsen, K.; Lal, A.; Pullmann, R., Jr.; Yang, X.; Galban, S.; Srikantan, S.; Martindale, J.L.; Blethrow, J.; Shokat, K.M.; et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008, 22, 1804–1815. [Google Scholar]
- Doller, A.; Huwiler, A.; Müller, R.; Radeke, H.H.; Pfeilschifter, J.; Eberhardt, W. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: Implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007, 18, 2137–2148. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Guo, R.; Yang, X.; Martindale, J.L.; Gorospe, M. Tyrosine phosphorylation of HuR by JAK3 triggers dissociation and degradation of HuR target mRNAs. Nucleic Acids Res. 2014, 42, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Akaike, Y.; Masuda, K.; Kuwano, Y.; Nishida, K.; Kajita, K.; Kurokawa, K.; Satake, Y.; Shoda, K.; Imoto, I.; Rokutan, K. HuR regulates alternative splicing of the TRA2β gene in human colon cancer cells under oxidative stress. Mol. Cell Biol. 2014, 34, 2857–2873. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Doller, A.; Pendini, N.R.; Wilce, J.A.; Pfeilschifter, J.; Eberhardt, W. Domain-specific phosphomimetic mutation allows dissection of different protein kinase C (PKC) isotype-triggered activities of the RNA binding protein HuR. Cell Signal. 2013, 25, 2485–2495. [Google Scholar] [CrossRef] [PubMed]
- Doller, A.; Schulz, S.; Pfeilschifter, J.; Eberhardt, W. RNA-dependent association with myosin IIA promotes F-actin-guided trafficking of the ELAV-like protein HuR to polysomes. Nucleic Acids Res. 2013, 41, 9152–9167. [Google Scholar] [CrossRef] [PubMed]
- Filippova, N.; Yang, X.; King, P.; Nabors, L.B. Phosphoregulation of the RNA-binding protein Hu antigen R (HuR) by Cdk5 affects centrosome function. J. Biol. Chem. 2012, 287, 32277–32287. [Google Scholar] [CrossRef] [PubMed]
- Doller, A.; Winkler, C.; Azrilian, I.; Schulz, S.; Hartmann, S.; Pfeilschifter, J.; Eberhardt, W. High-constitutive HuR phosphorylation at Ser 318 by PKCδ propagates tumor relevant functions in colon carcinoma cells. Carcinogenesis 2011, 32, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Chantar, M.L.; Vázquez-Chantada, M.; Garnacho, M.; Latasa, M.U.; Varela-Rey, M.; Dotor, J.; Santamaria, M.; Martínez-Cruz, L.A.; Parada, L.A.; Lu, S.C.; et al. S-adenosylmethionine regulates cytoplasmic HuR via AMP-activated kinase. Gastroenterology 2006, 131, 223–232. [Google Scholar]
- Lafarga, V.; Cuadrado, A.; Lopez de Silanes, I.; Bengoechea, R.; Fernandez-Capetillo, O.; Nebreda, A.R. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21Cip1 mRNA mediates the G1/S checkpoint. Mol. Cell Biol. 2009, 29, 4341–4351. [Google Scholar] [CrossRef] [PubMed]
- Cammas, A.; Sanchez, B.J.; Lian, X.J.; Dormoy-Raclet, V.; van der Giessen, K.; de Silanes, I.L.; Ma, J.; Wilusz, C.; Richardson, J.; Gorospe, M.; et al. Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation. Nat. Commun. 2014. [Google Scholar] [CrossRef]
- González-Feliciano, J.A.; Hernández-Pérez, M.; Estrella, L.A.; Colón-López, D.D.; López, A.; Martínez, M.; Maurás-Rivera, K.R.; Lasalde, C.; Martínez, D.; Araujo-Pérez, F.; et al. The role of HuR in the post-transcriptional regulation of interleukin-3 in T cells. PLOS ONE 2014, 9, e92457. [Google Scholar]
- Zhang, X.; Zou, T.; Rao, J.N.; Liu, L.; Xiao, L.; Wang, P.Y.; Cui, Y.H.; Gorospe, M.; Wang, J.Y. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 2009, 37, 7623–7637. [Google Scholar] [CrossRef] [PubMed]
- Hudy, M.H.; Proud, D. Cigarette smoke enhances human rhinovirus-induced CXCL8 production via HuR-mediated mRNA stabilization in human airway epithelial cells. Respir. Res. 2013. [Google Scholar] [CrossRef]
- Herjan, T.; Yao, P.; Qian, W.; Li, X.; Liu, C.; Bulek, K.; Sun, D.; Yang, W.P.; Zhu, J.; He, A.; et al. HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J. Immunol. 2013, 191, 640–649. [Google Scholar]
- Gummadi, L.; Taylor, L.; Curthoys, N.P. Concurrent binding and modifications of AUF1 and HuR mediate the pH-responsive stabilization of phosphoenolpyruvate carboxykinase mRNA in kidney cells. Am. J. Physiol. Renal Physiol. 2012, 303, F1545–F1554. [Google Scholar] [CrossRef] [PubMed]
- Vo, D.T.; Abdelmohsen, K.; Martindale, J.L.; Qiao, M.; Tominaga, K.; Burton, T.L.; Gelfond, J.A.; Brenner, A.J.; Patel, V.; Trageser, D.; et al. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol. Cancer Res. 2012, 10, 143–155. [Google Scholar]
- Liao, W.L.; Wang, W.C.; Chang, W.C.; Tseng, J.T. The RNA-binding protein HuR stabilizes cytosolic phospholipase A2α mRNA under interleukin-1β treatment in non-small cell lung cancer A549 Cells. J. Biol. Chem. 2011, 286, 35499–35508. [Google Scholar] [CrossRef] [PubMed]
- Donahue, J.M.; Chang, E.T.; Xiao, L.; Wang, P.Y.; Rao, J.N.; Turner, D.J.; Wang, J.Y.; Battafarano, R.J. The RNA-binding protein HuR stabilizes survivin mRNA in human oesophageal epithelial cells. Biochem. J. 2011, 437, 89–96. [Google Scholar] [CrossRef] [PubMed]
- García-Domínguez, D.J.; Morello, D.; Cisneros, E.; Kontoyiannis, D.L.; Frade, J.M. Stabilization of Dll1 mRNA by Elavl1/HuR in neuroepithelial cells undergoing mitosis. Mol. Biol. Cell. 2011, 22, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Bergalet, J.; Fawal, M.; Lopez, C.; Desjobert, C.; Lamant, L.; Delsol, G.; Morello, D.; Espinos, E. HuR-mediated control of C/EBPbeta mRNA stability and translation in ALK-positive anaplastic large cell lymphomas. Mol. Cancer Res. 2011, 9, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hu, D.Y.; Liu, S.; Mahavadi, S.; Yen, W.; Murthy, K.S.; Khalili, K.; Hu, W. RNA-binding protein HuR regulates RGS4 mRNA stability in rabbit colonic smooth muscle cells. Am. J. Physiol. Cell Physiol. 2010, 299, C1418–C1429. [Google Scholar] [CrossRef] [PubMed]
- Licata, L.A.; Hostetter, C.L.; Crismale, J.; Sheth, A.; Keen, J.C. The RNA-binding protein HuR regulates GATA3 mRNA stability in human breast cancer cell lines. Breast Cancer Res. Treat. 2010, 122, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Farooq, F.; Balabanian, S.; Liu, X.; Holcik, M.; MacKenzie, A. p38 Mitogen-activated protein kinase stabilizes SMN mRNA through RNA binding protein HuR. Hum. Mol. Genet. 2009, 18, 4035–4045. [Google Scholar] [CrossRef] [PubMed]
- López de Silanes, I.; Gorospe, M.; Taniguchi, H.; Abdelmohsen, K.; Srikantan, S.; Alaminos, M.; Berdasco, M.; Urdinguio, R.G.; Fraga, M.F.; Jacinto, F.V.; et al. The RNA-binding protein HuR regulates DNA methylation through stabilization of DNMT3b mRNA. Nucleic Acids Res. 2009, 37, 2658–2671. [Google Scholar]
- Atkinson, G.P.; Nozell, S.E.; Harrison, D.K.; Stonecypher, M.S.; Chen, D.; Benveniste, E.N. The prolyl isomerase Pin1 regulates the NF-κB signaling pathway and interleukin-8 expression in glioblastoma. Oncogene 2009, 28, 3735–3745. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.R.; Choi, H.S.; Heo, T.H.; Hwang, S.W.; Kang, K.W. Induction of vascular endothelial growth factor by peptidyl-prolyl isomerase Pin1 in breast cancer cells. Biochem. Biophys. Res. Commun. 2008, 369, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Briata, P.; Forcales, S.V.; Ponassi, M.; Corte, G.; Chen, C.Y.; Karin, M.; Puri, P.L.; Gherzi, R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell. 2005, 20, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Gherzi, R.; Trabucchi, M.; Ponassi, M.; Ruggiero, T.; Corte, G.; Moroni, C.; Chen, C.Y.; Khabar, K.S.; Andersen, J.S.; Briata, P. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 2006, 5, e5. [Google Scholar] [CrossRef] [PubMed]
- Bird, C.W.; Gardiner, A.S.; Bolognani, F.; Tanner, D.C.; Chen, C.Y.; Lin, W.J.; Yoo, S.; Twiss, J.L.; Perrone-Bizzozero, N. KSRP modulation of GAP-43 mRNA stability restricts axonal outgrowth in embryonic hippocampal neurons. PLoS ONE 2013, 8, e79255. [Google Scholar] [CrossRef] [PubMed]
- Esnault, S.; Braun, R.K.; Shen, Z.J.; Xiang, Z.; Heninger, E.; Love, R.B.; Sandor, M.; Malter, J.S. Pin1 modulates the type 1 immune response. PLoS ONE 2007, 2, e226. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.A.; Jue, S.S.; Bae, W.J.; Heo, S.H.; Shin, S.I.; Kwon, I.K.; Lee, S.C.; Kim, E.C. PIN1 inhibition suppresses osteoclast differentiation and inflammatory responses. J. Dent Res. 2015, 94, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.W.; Tsai, C.H.; Hsiao, M.; Tseng, C.J.; Ger, L.P.; Lee, K.H.; Lu, P.J. Pin1 overexpression is associated with poor differentiation and survival in oral squamous cell carcinoma. Oncol. Rep. 2009, 21, 1097–1104. [Google Scholar] [PubMed]
- Lin, F.C.; Lee, Y.C.; Goan, Y.G.; Tsai, C.H.; Yao, Y.C.; Cheng, H.C.; Lai, W.W.; Wang, Y.C.; Sheu, B.S.; Lu, P.J. Pin1 positively affects tumorigenesis of esophageal squamous cell carcinoma and correlates with poor survival of patients. J. Biomed. Sci. 2014. [Google Scholar] [CrossRef]
- Schwartz, E.I.; Intine, R.V.; Maraia, R.J. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5'-terminal oligopyrimidine mRNA metabolism. Mol. Cell Biol. 2004, 24, 9580–9591. [Google Scholar] [CrossRef] [PubMed]
- Terzoglou, A.G.; Routsias, J.G.; Avrameas, S.; Moutsopoulos, H.M.; Tzioufas, A.G. Preferential recognition of the phosphorylated major linear B-cell epitope of La/SSB 349–368 aa by anti-La/SSB autoantibodies from patients with systemic autoimmune diseases. Clin. Exp. Immunol. 2006, 144, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Brenet, F.; Socci, N.D.; Sonenberg, N.; Holland, E.C. Akt phosphorylation of La regulates specific mRNA translation in glial progenitors. Oncogene 2009, 28, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Kuehnert, J.; Sommer, G.; Zierk, A.W.; Fedarovich, A.; Brock, A.; Fedarovich, D.; Heise, T. Novel RNA chaperone domain of RNA-binding protein La is regulated by AKT phosphorylation. Nucleic Acids Res. 2015, 43, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Zhu, P.; Dang, Y.; Wu, J.; Yang, X.; Wan, B.; Liu, J.O.; Yi, Q.; Yu, L. Nuclear export of NF90 to stabilize IL-2 mRNA is mediated by AKT-dependent phosphorylation at Ser647 in response to CD28 costimulation. J. Immunol. 2008, 180, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.L.; Miskimins, W.K. Phosphorylation at serine 482 affects stability of NF90 and its functional role in mitosis. Cell Prolif. 2011, 44, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Jiang, W.; Cao, L.; Yu, W.; Pei, Y.; Yang, X.; Wan, B.; Liu, J.O.; Yi, Q.; Yu, L. IL-2 mRNA stabilization upon PMA stimulation is dependent on NF90-Ser647 phosphorylation by protein kinase CbetaI. J. Immunol. 2010, 185, 5140–5149. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Huang, H.; Ding, L.; Zhu, P.; Saiyin, H.; Ji, G.; Zuo, J.; Han, D.; Pan, Y.; Ding, D.; et al. Regulation of cell cycle of hepatocellular carcinoma by NF90 through modulation of cyclin E1 mRNA stability. Oncogene 2014. [Google Scholar] [CrossRef]
- Kuwano, Y.; Kim, H.H.; Abdelmohsen, K.; Pullmann, R., Jr.; Martindale, J.L.; Yang, X.; Gorospe, M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell Biol. 2008, 28, 4562–4575. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.; Lim, H.; R Yates, J.; Karin, M. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell 2002, 10, 1331–1344. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, H.; Filhol, O.; Truchet, I.; Brethenou, P.; Cochet, C.; Amalric, F.; Bouche, G. Fibroblast growth factor-2 binds to the regulatory beta subunit of CK2 and directly stimulates CK2 activity toward nucleolin. J. Biol. Chem. 1996, 271, 24781–24787. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.A.; Li, H.Y.; Hsu, T.I.; Chen, S.H.; Wu, C.J.; Chang, W.C.; Hung, J.J. Heat shock protein 90 stabilizes nucleolin to increase mRNA stability in mitosis. J. Biol. Chem. 2011, 286, 43816–43829. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.M.; Zhang, P.; Liu, R.Y.; Sang, Y.X.; Zhou, C.; Xu, G.C.; Yang, J.L.; Tong, A.P.; Wang, C.T. Phosphorylation and changes in the distribution of nucleolin promote tumor metastasis via the PI3K/Akt pathway in colorectal carcinoma. FEBS Lett. 2014, 588, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Dranovsky, A.; Vincent, I.; Gregori, L.; Schwarzman, A.; Colflesh, D.; Enghild, J.; Strittmatter, W.; Davies, P.; Goldgaber, D. Cdc2 phosphorylation of nucleolin demarcates mitotic stages and Alzheimer’s disease pathology. Neurobiol. Aging 2001, 22, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.D.; Zhao, H.G.; Yang, Y.S.; Hu, T.; Yang, Q.C. GSK3β negatively regulates HIF1α mRNA stability via nucleolin in the MG63 osteosarcoma cell line. Biochem. Biophys. Res. Commun. 2014, 443, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Gherzi, R.; Andersen, J.S.; Gaietta, G.; Jürchott, K.; Royer, H.D.; Mann, M.; Karin, M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000, 14, 1236–1248. [Google Scholar] [PubMed]
- Woo, H.H.; Baker, T.; Laszlo, C.; Chambers, S.K. Nucleolin mediates microRNA-directed CSF-1 mRNA deadenylation but increases translation of CSF-1 mRNA. Mol. Cell. Proteomics 2013, 12, 1661–1677. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Childress, M.O.; Geahlen, R.L. Syk interacts with and phosphorylates nucleolin to stabilize Bcl-xL mRNA and promote cell survival. Mol. Cell Biol. 2014, 34, 3788–3799. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.T.; Liao, P.C.; Chang, W.C.; Tseng, J.T. Epidermal growth factor increases the interaction between nucleolin and heterogeneous nuclear ribonucleoprotein K/poly(C) binding protein 1 complex to regulate the gastrin mRNA turnover. Mol. Biol. Cell. 2007, 18, 5004–5013. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Xu, X.S.; Russell, J.E. A nucleolin-binding 3' untranslated region element stabilizes beta-globin mRNA in vivo. Mol. Cell Biol. 2006, 26, 2419–2429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bhatia, D.; Xia, H.; Castranova, V.; Shi, X.; Chen, F. Nucleolin links to arsenic-induced stabilization of GADD45α mRNA. Nucleic Acids Res. 2006, 34, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Laughlin, J.; Kosinski, P.A.; Covey, L.R. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J. Immunol. 2004, 173, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, T.K.; Bandyopadhyay, S.; Fernandes, D.J.; Spicer, E.K. Identification of nucleolin as an AU-rich element binding protein involved in Bcl-2 mRNA stabilization. J. Biol. Chem. 2004, 279, 10855–10863. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Dominski, Z.; Yang, X.C.; Elms, P.; Raska, C.S.; Borchers, C.H.; Marzluff, W.F. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell Biol. 2003, 23, 1590–1601. [Google Scholar] [CrossRef] [PubMed]
- Bansal, N.; Zhang, M.; Bhaskar, A.; Itotia, P.; Lee, E.; Shlyakhtenko, L.S.; Lam, T.T.; Fritz, A.; Berezney, R.; Lyubchenko, Y.L.; et al. Assembly of the SLIP1-SLBP complex on histone mRNA requires heterodimerization and sequential binding of SLBP followed by SLIP1. Biochemistry 2013, 52, 520–536. [Google Scholar]
- Zhang, M.; Lam, T.T.; Tonelli, M.; Marzluff, W.F.; Thapar, R. Interaction of the histone mRNA hairpin with stem-loop binding protein (SLBP) and regulation of the SLBP-RNA complex by phosphorylation and proline isomerization. Biochemistry 2012, 51, 3215–3231. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Taupin, J.; Elledge, S.; Robertson, M.; Anderson, P. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J. Exp. Med. 1995, 182, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, J.M.; Valcárcel, J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J. Biol. Chem. 2007, 282, 1539–1543. [Google Scholar] [CrossRef] [PubMed]
- Gottschald, O.R.; Malec, V.; Krasteva, G.; Hasan, D.; Kamlah, F.; Herold, S.; Rose, F.; Seeger, W.; Hänze, J. TIAR and TIA-1 mRNA-binding proteins co-aggregate under conditions of rapid oxygen decline and extreme hypoxia and suppress the HIF-1α pathway. J. Mol. Cell Biol. 2010, 2, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Chrestensen, C.A.; Schroeder, M.J.; Shabanowitz, J.; Hunt, D.F.; Pelo, J.W.; Worthington, M.T.; Sturgill, T.W. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14–3-3 binding. J. Biol. Chem. 2004, 279, 10176–10184. [Google Scholar] [CrossRef] [PubMed]
- Stoecklin, G.; Stubbs, T.; Kedersha, N.; Wax, S.; Rigby, W.F.; Blackwell, T.K.; Anderson, P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004, 23, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.A.; Thompson, M.J.; Lai, W.S.; Blackshear, P.J. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J. Biol. Chem. 1995, 270, 13341–13347. [Google Scholar] [CrossRef] [PubMed]
- Hitti, E.; Iakovleva, T.; Brook, M.; Deppenmeier, S.; Gruber, A.D.; Radzioch, D.; Clark, A.R.; Blackshear, P.J.; Kotlyarov, A.; Gaestel, M. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell Biol. 2006, 26, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Brook, M.; Tchen, C.R.; Santalucia, T.; McIlrath, J.; Arthur, J.S.; Saklatvala, J.; Clark, A.R. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell Biol. 2006, 26, 2408–2418. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Tokuhara, M.; Morita, E.H.; Abe, S. TTP at Ser245 phosphorylation by AKT is required for binding to 14-3-3. J. Biochem. 2009, 145, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Kedar, V.P.; Darby, M.K.; Williams, J.G.; Blackshear, P.J. Phosphorylation of human tristetraprolin in response to its interaction with the Cbl interacting protein CIN85. PLoS ONE 2010, 5, e9588. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Lin, R. Phosphorylation of recombinant tristetraprolin in vitro. Protein J. 2008, 27, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Deterding, L.J.; Venable, J.D.; Kennington, E.A.; Yates, J.R., III; Tomer, K.B.; Blackshear, P.J. Identification of the anti-inflammatory protein tristetraprolin as a hyperphosphorylated protein by mass spectrometry and site-directed mutagenesis. Biochem. J. 2006, 394, 285–297. [Google Scholar]
- Arma, A.; Bhat, A.A.; Krishnan, M.; Singh, A.B.; Dhawan, P. Trichostatin-A modulates claudin-1 mRNA stability through the modulation of Hu antigen R and tristetraprolin in colon cancer cells. Carcinogenesis 2013, 34, 2610–2621. [Google Scholar] [CrossRef] [PubMed]
- Molle, C.; Zhang, T.; Ysebrant de Lendonck, L.; Gueydan, C.; Andrianne, M.; Sherer, F.; van Simaeys, G.; Blackshear, P.J.; Leo, O.; Goriely, S. Tristetraprolin regulation of interleukin 23 mRNA stability prevents a spontaneous inflammatory disease. J. Exp. Med. 2013, 210, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Evdokimova, V.; Ruzanov, P.; Anglesio, M.S.; Sorokin, A.V.; Ovchinnikov, L.P.; Buckley, J.; Triche, T.J.; Sonenberg, N.; Sorensen, P.H. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol. Cell Biol. 2006, 26, 277–292. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, B.W.; Kucab, J.; Wu, J.; Lee, C.; Cheang, M.C.; Yorida, E.; Turbin, D.; Dedhar, S.; Nelson, C.; Pollak, M.; et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene 2005, 24, 4281–4292. [Google Scholar]
- Coles, L.S.; Lambrusco, L.; Burrows, J.; Hunter, J.; Diamond, P.; Bert, A.G.; Vadas, M.A.; Goodall, G.J. Phosphorylation of cold shock domain/Y-box proteins by ERK2 and GSK3β and repression of the human VEGF promoter. FEBS Lett. 2005, 579, 5372–5378. [Google Scholar] [CrossRef] [PubMed]
- Khabar, K.S. Post-transcriptional control during chronic inflammation and cancer: A focus on AU-rich elements. Cell Mol. Life Sci. 2010, 67, 2937–2955. [Google Scholar] [CrossRef] [PubMed]
- Venigalla, R.K.; Turner, M. RNA-binding proteins as a point of convergence of the PI3K and p38 MAPK pathways. Front Immunol. 2012. [Google Scholar] [CrossRef]
- Wilson, G.M.; Lu, J.; Sutphen, K.; Suarez, Y.; Sinha, S.; Brewer, B.; Villanueva-Feliciano, E.C.; Ysla, R.M.; Charles, S.; Brewer, G. Phosphorylation of p40AUF1 regulates binding to A + U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J. Biol. Chem. 2003, 278, 33039–33048. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Stehn, J.R.; Yaffe, M.B.; Blackwell, T.K. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J. Biol. Chem. 2002, 277, 18029–18036. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.A.; Thompson, M.J.; Lai, W.S.; Blackshear, P.J. Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor. Mol. Endocrinol. 1996, 10, 140–146. [Google Scholar] [PubMed]
- Gringhuis, S.I.; García-Vallejo, J.J.; van Het Hof, B.; van Dijk, W. Convergent actions of I kappa B kinase beta and protein kinase C delta modulate mRNA stability through phosphorylation of 14-3-3 beta complexed with tristetraprolin. Mol. Cell Biol. 2005, 25, 6454–6463. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Stoecklin, G.; van Way, S.; Hinkovska-Galcheva, V.; Guo, R.F.; Anderson, P.; Shanley, T.P. Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-alpha mRNA. J. Biol. Chem. 2007, 282, 3766–3777. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, B.; Lu, J.Y.; Schneider, R.J. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 2003, 278, 20700–20707. [Google Scholar] [CrossRef] [PubMed]
- Wagner, B.J.; DeMaria, C.T.; Sun, Y.; Wilson, G.M.; Brewer, G. Structure and genomic organization of the human AUF1 gene: Alternative pre-mRNA splicing generates four protein isoforms. Genomics 1998, 48, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Misquitta, C.M.; Iyer, V.R.; Werstiuk, E.S.; Grover, A.K. The role of 3'-untranslated region (3'-UTR) mediated mRNA stability in cardiovascular pathophysiology. Mol. Cell Biochem. 2001, 224, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Fellows, A.; Deng, B.; Mierke, D.F.; Robey, R.B.; Nichols, R.C. Peptides modeled on the RGG domain of AUF1/hnRNP-D regulate 3' UTR-dependent gene expression. Int. Immunopharmacol. 2013, 17, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Li, M.L.; Defren, J.; Brewer, G. Hsp27 and F-box protein β-TrCP promote degradation of mRNA decay factor AUF1. Mol. Cell Biol. 2013, 33, 2315–2326. [Google Scholar] [CrossRef] [PubMed]
- Loflin, P.; Chen, C.Y.; Shyu, A.B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999, 13, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Tolnay, M.; Baranyi, L.; Tsokos, G.C. Heterogeneous nuclear ribonucleoprotein D0 contains transactivator and DNA-binding domains. Biochem. J. 2000, 348, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Fawal, M.; Armstrong, F.; Ollier, S.; Dupont, H.; Touriol, C.; Monsarrat, B.; Delsol, G.; Payrastre, B.; Morello, D. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood 2006, 108, 2780–2788. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.M.; Steitz, J.A. HuR and mRNA stability. Cell Mol. Life Sci. 2001, 58, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Liu, L.; Rao, J.N.; Marasa, B.S.; Chen, J.; Xiao, L.; Zhou, H.; Gorospe, M.; Wang, J.Y. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1. Biochem. J. 2008, 409, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Winzen, R.; Kracht, M.; Ritter, B.; Wilhelm, A.; Chen, C.Y.; Shyu, A.B.; Müller, M.; Gaestel, M.; Resch, K.; Holtmann, H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999, 18, 4969–4980. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.F.; Stoecklin, G.; Lu, M.; Looser, R.; Moroni, C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell Biol. 2001, 21, 5778–5789. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Caldwell, M.C.; Lin, S.; Furneaux, H.; Gorospe, M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000, 19, 2340–2350. [Google Scholar] [CrossRef] [PubMed]
- Doller, A.; Pfeilschifter, J.; Eberhardt, W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal. 2008, 20, 2165–2173. [Google Scholar] [CrossRef] [PubMed]
- Pascale, A.; Amadio, M.; Scapagnini, G.; Lanni, C.; Racchi, M.; Provenzani, A.; Govoni, S.; Alkon, D.L.; Quattrone, A. Neuronal ELAV proteins enhance mRNA stability by a PKCalpha-dependent pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 12065–12070. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.X.; Wang, P.Y.; Rao, J.N.; Zou, T.; Liu, L.; Xiao, L.; Gorospe, M.; Wang, J.Y. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res. 2011, 39, 8472–8487. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Rao, J.N.; Zou, T.; Xiao, L.; Wang, P.Y.; Turner, D.J.; Gorospe, M.; Wang, J.Y. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol. Biol. Cell. 2009, 20, 4885–4898. [Google Scholar] [CrossRef] [PubMed]
- Briata, P.; Ilengo, C.; Corte, G.; Moroni, C.; Rosenfeld, M.G.; Chen, C.Y.; Gherzi, R. The Wnt/beta-catenin-Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol. Cell. 2003, 12, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.J.; Malter, J.S.; Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA. Direct interaction of phosphorylated AUF1 with ARE mRNA. Unpublished work. 2014. [Google Scholar]
- Shen, Z.J.; Braun, R.K.; Hu, J.; Xie, Q.; Chu, H.; Love, R.B.; Stodola, L.A.; Rosenthal, L.A.; Szakaly, R.J.; Sorkness, R.L.; et al. Pin1 protein regulates Smad protein signaling and pulmonary fibrosis. J. Biol. Chem. 2012, 287, 23294–23305. [Google Scholar]
- Esnault, S.; Rosenthal, L.A.; Shen, Z.J.; Sedgwick, J.B.; Szakaly, R.J.; Sorkness, R.L.; Malter, J.S. A critical role for Pin1 in allergic pulmonary eosinophilia in rats. J. Allergy Clin. Immunol. 2007, 120, 1082–1088. [Google Scholar] [CrossRef] [PubMed]
- Rajbhandari, P.; Schalper, K.A.; Solodin, N.M.; Ellison-Zelski, S.J.; Ping Lu, K.; Rimm, D.L.; Alarid, E.T. Pin1 modulates ERα levels in breast cancer through inhibition of phosphorylation-dependent ubiquitination and degradation. Oncogene 2014, 33, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- Innes, B.T.; Bailey, M.L.; Brandl, C.J.; Shilton, B.H.; Litchfield, D.W. Non-catalytic participation of the Pin1 peptidyl-prolyl isomerase domain in target binding. Front Physiol. 2013. [Google Scholar] [CrossRef]
- Lu, P.J.; Zhou, X.Z.; Shen, M.; Lu, K.P. Function of WW domains as phosphoserine-or phosphothreonine-binding modules. Science 1999, 283, 1325–1328. [Google Scholar] [CrossRef] [PubMed]
- Smet, C.; Wieruszeski, J.M.; Buée, L.; Landrieu, I.; Lippens, G. Regulation of Pin1 peptidyl-prolyl cis/trans isomerase activity by its WW binding module on a multi-phosphorylated peptide of Tau protein. FEBS Lett. 2005, 579, 4159–4164. [Google Scholar] [CrossRef] [PubMed]
- Khanal, P.; Yun, H.J.; Lim, S.C.; Ahn, S.G.; Yoon, H.E.; Kang, K.W.; Hong, R.; Choi, H.S. Proyl isomerase Pin1 facilitates ubiquitin-mediated degradation of cyclin-dependent kinase 10 to induce tamoxifen resistance in breast cancer cells. Oncogene 2012, 31, 3845–3856. [Google Scholar] [CrossRef] [PubMed]
- Valley, C.C.; Métivier, R.; Solodin, N.M.; Fowler, A.M.; Mashek, M.T.; Hill, L.; Alarid, E.T. Differential regulation of estrogen-inducible proteolysis and transcription by the estrogen receptor alpha N terminus. Mol. Cell Biol. 2005, 25, 5417–5428. [Google Scholar] [CrossRef] [PubMed]
- Min, S.H.; Lau, A.W.; Lee, T.H.; Inuzuka, H.; Wei, S.; Huang, P.; Shaik, S.; Lee, D.Y.; Finn, G.; Balastik, M.; et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol. Cell. 2012, 46, 771–783. [Google Scholar]
- Epis, M.R.; Barker, A.; Giles, K.M.; Beveridge, D.J.; Leedman, P.J. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J. Biol. Chem. 2011, 286, 41442–41454. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.N.; Habermacher, R.; Martine, U.; Closs, E.I.; Filipowicz, W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant Biol. 2006, 71, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Kuwano, Y.; Srikantan, S.; Lee, E.K.; Martindale, J.L.; Gorospe, M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009, 23, 1743–1748. [Google Scholar] [CrossRef] [PubMed]
- Holbert, M.A.; Sikorski, T.; Carten, J.; Snowflack, D.; Hodawadekar, S.; Marmorstein, R. The human monocytic leukemia zinc finger histone acetyltransferase domain contains DNA-binding activity implicated in chromatin targeting. J. Biol. Chem. 2007, 282, 36603–36613. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Shimojo, H.; Hayashi, A.; Kawaguchi, R.; Ohtani, Y.; Uegaki, K.; Nishimura, Y.; Nakayama, J. Intrinsic nucleic acid-binding activity of Chp1 chromodomain is required for heterochromatic gene silencing. Mol. Cell. 2012, 47, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Seko, Y.; Cole, S.; Kasprzak, W.; Shapiro, B.A.; Ragheb, J.A. The role of cytokine mRNA stability in the pathogenesis of autoimmune disease. Autoimmun. Rev. 2006, 5, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Carter, B.Z.; Malter, J.S. Regulation of mRNA stability and its relevance to disease. Lab. Investig. 1991, 65, 610–621. [Google Scholar] [PubMed]
- Manganaro, L.; Lusic, M.; Gutierrez, M.I.; Cereseto, A.; Del Sal, G.; Giacca, M. Concerted action of cellular JNK and Pin1 restricts HIV-1 genome integration to activated CD4+ T lymphocytes. Nat. Med. 2010, 16, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, T.; Tun-Kyi, A.; Ryo, A.; Yamamoto, M.; Finn, G.; Fujita, T.; Akira, S.; Yamamoto, N.; Lu, K.P.; Yamaoka, S. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 2006, 7, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Lang, R.A.; Metcalf, D.; Cuthbertson, R.A.; Lyons, I.; Stanley, E.; Kelso, A.; Kannourakis, G.; Williamson, D.J.; Klintworth, G.K.; Gonda, T.J.; et al. Transgenic mice expressing a hemopoietic growth factor gene (GM-CSF) develop accumulations of macrophages, blindness, and a fatal syndrome of tissue damage. Cell 1987, 51, 675–686. [Google Scholar]
- Biondo, M.; Nasa, Z.; Marshall, A.; Toh, B.H.; Alderuccio, F. Local transgenic expression of granulocyte macrophage-colony stimulating factor initiates autoimmunity. J. Immunol. 2001, 166, 2090–2099. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.R.; Gonda, T.J.; Metcalf, D.; Hariharan, I.K.; Cory, S. A lethal myeloproliferative syndrome in mice transplanted with bone marrow cells infected with a retrovirus expressing granulocyte-macrophage colony stimulating factor. EMBO J. 1989, 8, 441–448. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Z.-J.; Malter, J.S. Regulation of AU-Rich Element RNA Binding Proteins by Phosphorylation and the Prolyl Isomerase Pin1. Biomolecules 2015, 5, 412-434. https://doi.org/10.3390/biom5020412
Shen Z-J, Malter JS. Regulation of AU-Rich Element RNA Binding Proteins by Phosphorylation and the Prolyl Isomerase Pin1. Biomolecules. 2015; 5(2):412-434. https://doi.org/10.3390/biom5020412
Chicago/Turabian StyleShen, Zhong-Jian, and James S. Malter. 2015. "Regulation of AU-Rich Element RNA Binding Proteins by Phosphorylation and the Prolyl Isomerase Pin1" Biomolecules 5, no. 2: 412-434. https://doi.org/10.3390/biom5020412





