SUMO Wrestles with Recombination
Abstract
:1. Introduction
1.1. Double-Strand Break Repair

1.2. SUMO
2. SUMO in Recombination
2.1. SUMO in HR
| Pathway | Yeast | Human | Function | Effect of sumoylation | Reference |
|---|---|---|---|---|---|
| NHEJ | Ku70 | KU70 | subunit of Ku complex, protection of DNA ends, recruitment of other NHEJ factors | unknown | [25,64] |
| Ku80 | KU80 | subunit of Ku complex, protection of DNA ends, recruitment of other NHEJ factors | unknown | [38,65] | |
| Lif1 | XRCC4 | DNA ligation | intracellular localization (human) | [38,66] | |
| HR | Mre11 1 | MRE11 | subunit of MRX complex (DSB resection) | unknown | [38] |
| Rad50 1 | RAD50 | subunit of MRX complex (DSB resection) | unknown | [38] | |
| Xrs2 1 | NBS1 | subunit of MRX complex (DSB resection) | unknown | [38] | |
| Sae2 | CtIP | DSB resection | unknown | [38] | |
| Rad52 2 | RAD52 | recombination mediator | inhibition of biochemical activities, intranuclear localization, protein stability (yeast) | [42,43] | |
| subcellular localization (human) | |||||
| RPA 2 | RPA | binding resected DNA tails | recruitment of RAD51 to initiate HR (human) | [48,67] | |
| Rad59 2 | stabilization of Rad51 filament, ssDNA annealing | unknown | [67] | ||
| Sgs1 | BLM | RecQ-like helicase, resolution of dHJ | Sgs1 sumoylation stimulates recombination at telomeres | [34,68] | |
| BLM sumoylation promotes Rad51-dependent recombination | |||||
| WRN | RecQ-like helicase, resolution of dHJ | WRN sumoylation affects its nuclear localization | [69,70] | ||
| Srs2 | helicase, disruption of Rad51 filament, promoting SDSA | unscheduled sumoylation in | [54] | ||
| non-phosphorylatable Srs2 causes recombinational repair defects | |||||
| SSA | Rad1 | XPF | subunit of Rad1–Rad10 complex (nuclease activity) | unknown | [38] |
2.1.1. DNA End Resection
2.1.2. Presynaptic Filament Formation
2.1.3. Synaptic Phase
2.1.4. Post-Synaptic Phase
2.2. SUMO in NHEJ
3. The Interplay of HR and SUMO at the Repetitive Sequences
3.1. SUMO and the rDNA
3.2. SUMO and the Telomeres
4. Meiotic Recombination
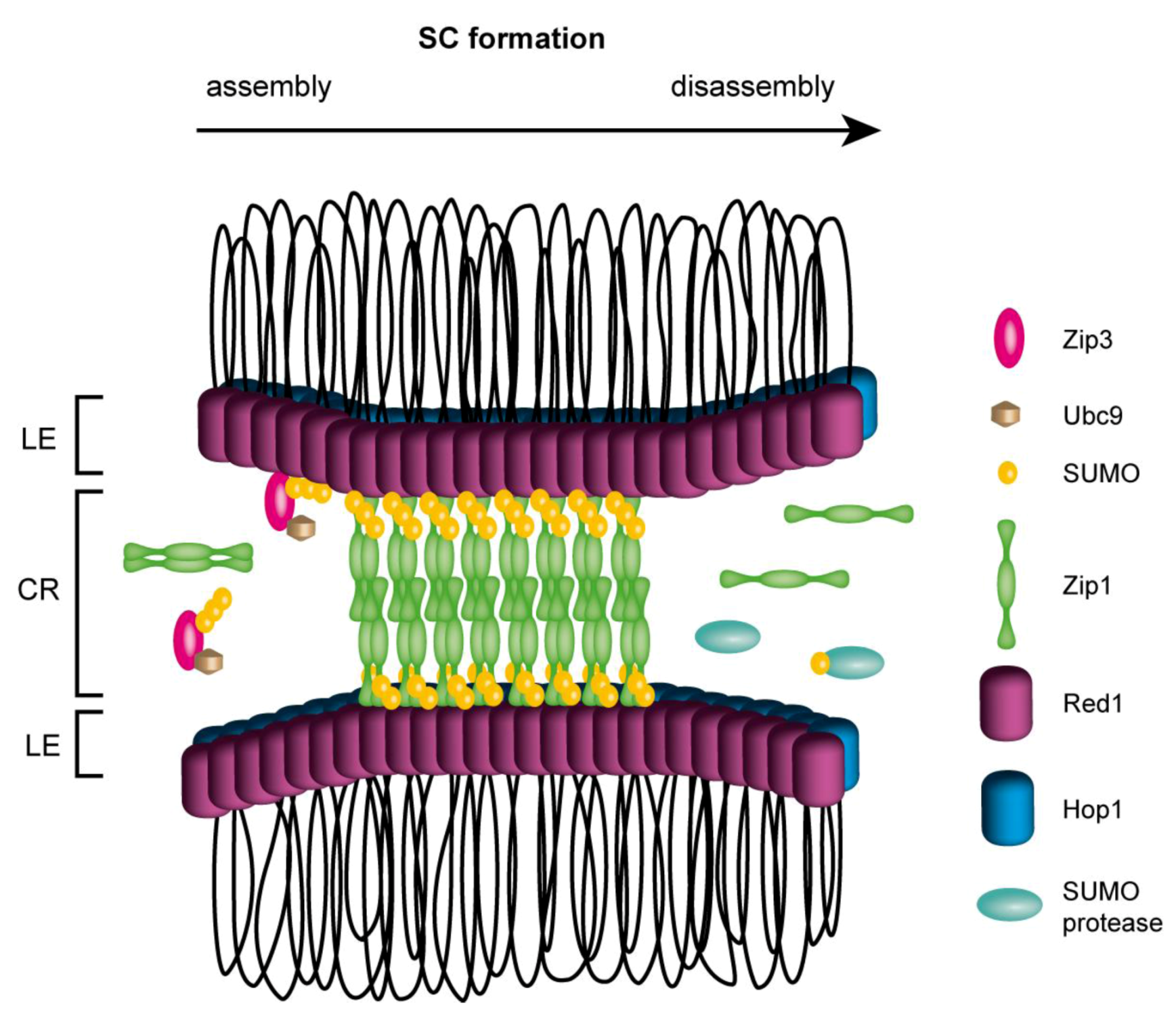
5. Interplay of Post-Translational Modifications
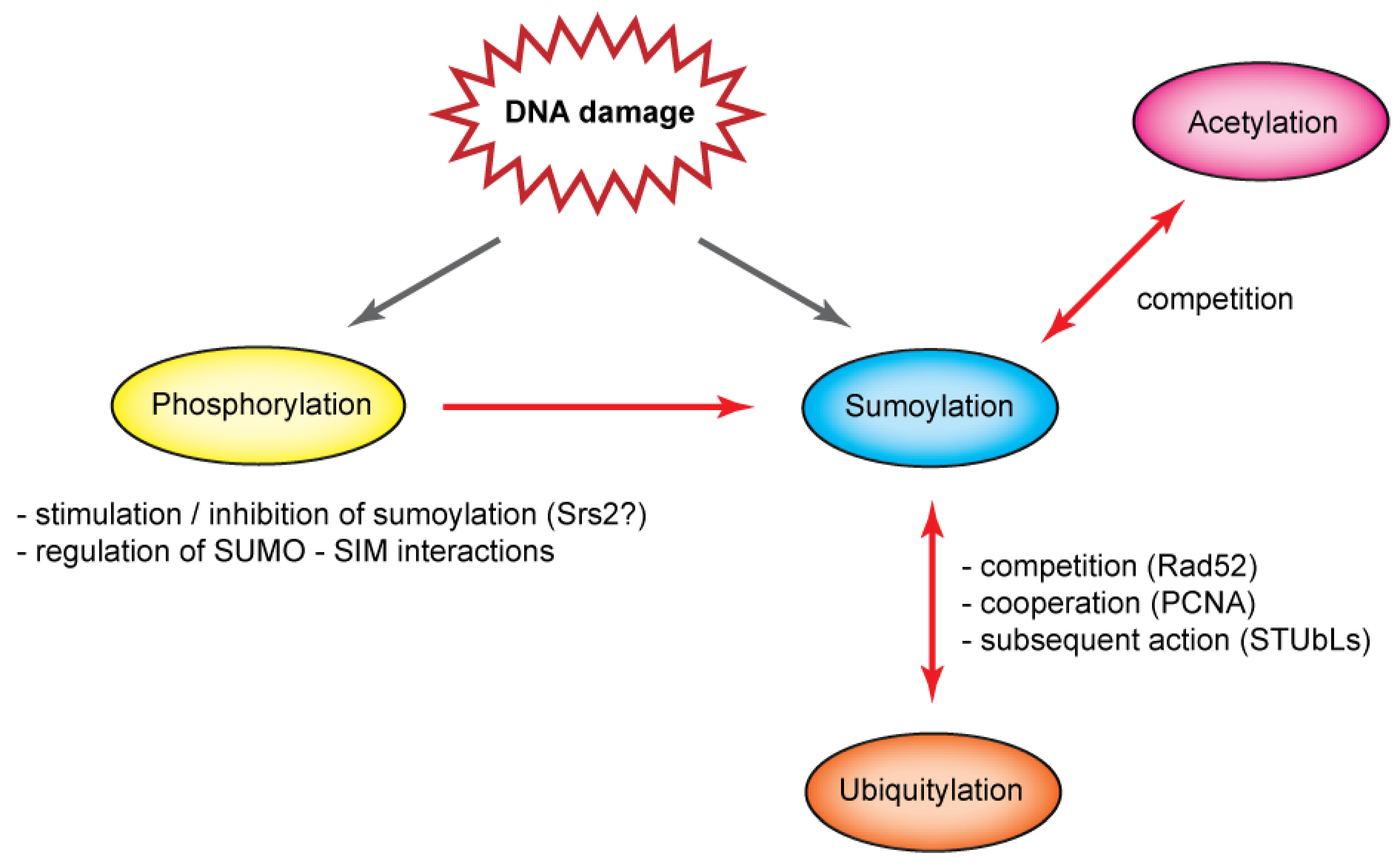
6. Conclusions
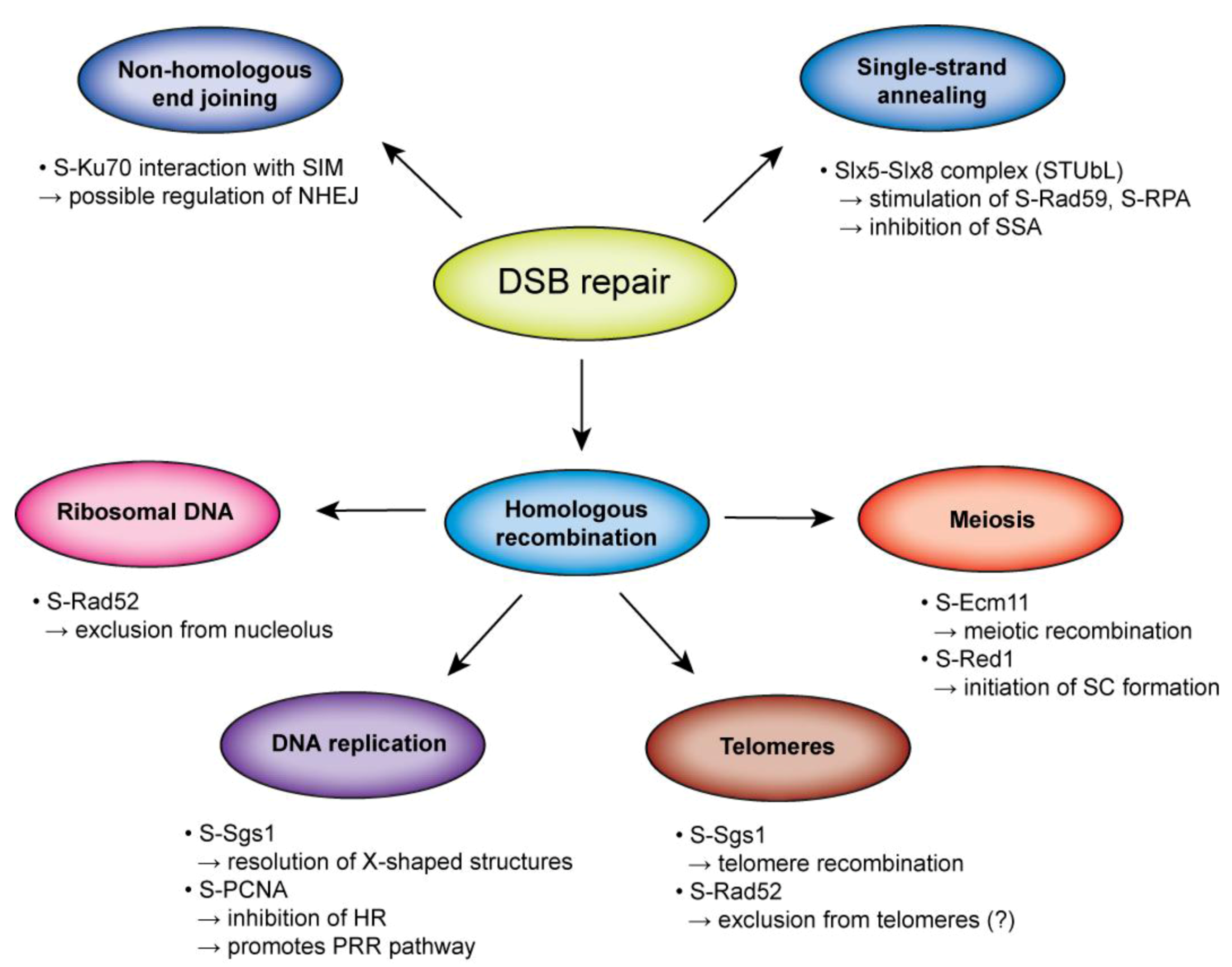
Acknowledgments
References
- Aylon, Y.; Liefshitz, B.; Kupiec, M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004, 23, 4868–4875. [Google Scholar] [CrossRef]
- Ira, G.; Pellicioli, A.; Balijja, A.; Wang, X.; Fiorani, S.; Carotenuto, W.; Liberi, G.; Bressan, D.; Wan, L.; Hollingsworth, N.M.; Haber, J.E.; Foiani, M. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 2004, 431, 1011–1017. [Google Scholar]
- Shrivastav, M.; de Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef]
- Chen, X.; Niu, H.; Chung, W.H.; Zhu, Z.; Papusha, A.; Shim, E.Y.; Lee, S.E.; Sung, P.; Ira, G. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat. Struct. Mol. Biol. 2011, 18, 1015–1019. [Google Scholar] [CrossRef]
- Huertas, P.; Cortes-Ledesma, F.; Sartori, A.A.; Aguilera, A.; Jackson, S.P. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 2008, 455, 689–692. [Google Scholar] [CrossRef]
- Saleh-Gohari, N.; Helleday, T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004, 32, 3683–3688. [Google Scholar] [CrossRef]
- Yun, M.H.; Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 2009, 459, 460–463. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Mimitou, E.P.; Symington, L.S. DNA end resection--unraveling the tail. DNA Repair (Amst) 2011, 10, 344–348. [Google Scholar]
- Sung, P.; Krejci, L.; van Komen, S.; Sehorn, M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003, 278, 42729–42732. [Google Scholar]
- Mazin, A.V.; Mazina, O.M.; Bugreev, D.V.; Rossi, M.J. Rad54, the motor of homologous recombination. DNA Repair (Amst) 2010, 9, 286–302. [Google Scholar]
- Szostak, J.W.; Orr-Weaver, T.L.; Rothstein, R.J.; Stahl, F.W. The double-strand-break repair model for recombination. Cell 1983, 33, 25–35. [Google Scholar] [CrossRef]
- Nassif, N.; Penney, J.; Pal, S.; Engels, W.R.; Gloor, G.B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell Biol. 1994, 14, 1613–1625. [Google Scholar]
- Malkova, A.; Ivanov, E.L.; Haber, J.E. Double-strand break repair in the absence of RAD51 in yeast: A possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 1996, 93, 7131–7136. [Google Scholar] [CrossRef]
- Heyer, W.D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet 2010, 44, 113–139. [Google Scholar] [CrossRef]
- Krejci, L.; Altmannova, V.; Spirek, M.; Zhao, X. Homologous recombination and its regulation. Nucleic Acids Res 2012. [Google Scholar]
- Krogh, B.O.; Symington, L.S. Recombination proteins in yeast. Annu. Rev. Genet 2004, 38, 233–271. [Google Scholar] [CrossRef]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef]
- Hay, R.T. SUMO: A history of modification. Mol. Cell 2005, 18, 1–12. [Google Scholar] [CrossRef]
- Johnson, E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004, 73, 355–382. [Google Scholar] [CrossRef]
- Ulrich, H.D. The SUMO system: An overview. Methods Mol. Biol. 2009, 497, 3–16. [Google Scholar] [CrossRef]
- Zhao, J. Sumoylation regulates diverse biological processes. Cell Mol. Life Sci. 2007, 64, 3017–3033. [Google Scholar] [CrossRef]
- Takahashi, Y.; Kahyo, T.; Toh, E.A.; Yasuda, H.; Kikuchi, Y. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 2001, 276, 48973–48977. [Google Scholar]
- Johnson, E.S.; Gupta, A.A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 2001, 106, 735–744. [Google Scholar] [CrossRef]
- Zhao, X.; Blobel, G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 2005, 102, 4777–4782. [Google Scholar] [CrossRef]
- Cheng, C.H.; Lo, Y.H.; Liang, S.S.; Ti, S.C.; Lin, F.M.; Yeh, C.H.; Huang, H.Y.; Wang, T.F. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006, 20, 2067–2081. [Google Scholar] [CrossRef]
- Kerscher, O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007, 8, 550–555. [Google Scholar] [CrossRef]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef]
- Hannich, J.T.; Lewis, A.; Kroetz, M.B.; Li, S.J.; Heide, H.; Emili, A.; Hochstrasser, M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 4102–4110. [Google Scholar]
- Hecker, C.M.; Rabiller, M.; Haglund, K.; Bayer, P.; Dikic, I. Specification of SUMO1-and SUMO2-interacting motifs. J. Biol. Chem. 2006, 281, 16117–16127. [Google Scholar]
- Song, J.; Durrin, L.K.; Wilkinson, T.A.; Krontiris, T.G.; Chen, Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 14373–14378. [Google Scholar]
- Chang, C.C.; Naik, M.T.; Huang, Y.S.; Jeng, J.C.; Liao, P.H.; Kuo, H.Y.; Ho, C.C.; Hsieh, Y.L.; Lin, C.H.; Huang, N.J.; et al. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 2011, 42, 62–74. [Google Scholar] [CrossRef]
- Stehmeier, P.; Muller, S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell 2009, 33, 400–409. [Google Scholar] [CrossRef]
- Branzei, D.; Sollier, J.; Liberi, G.; Zhao, X.; Maeda, D.; Seki, M.; Enomoto, T.; Ohta, K.; Foiani, M. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 2006, 127, 509–522. [Google Scholar] [CrossRef]
- Maeda, D.; Seki, M.; Onoda, F.; Branzei, D.; Kawabe, Y.; Enomoto, T. Ubc9 is required for damage-tolerance and damage-induced interchromosomal homologous recombination in S. cerevisiae. DNA Repair (Amst) 2004, 3, 335–341. [Google Scholar]
- Potts, P.R.; Yu, H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell Biol. 2005, 25, 7021–7032. [Google Scholar] [CrossRef]
- Soustelle, C.; Vernis, L.; Fréon, K.; Reynaud-Angelin, A.; Chanet, R.; Fabre, F.; Heude, M. A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Mol. Cell Biol. 2004, 24, 5130–5143. [Google Scholar] [CrossRef]
- Cremona, C.A.; Sarangi, P.; Yang, Y.; Hang, L.E.; Rahman, S.; Zhao, X. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol. Cell 2012, 45, 422–432. [Google Scholar] [CrossRef]
- Choi, J.H.; Lindsey-Boltz, L.A.; Kemp, M.; Mason, A.C.; Wold, M.S.; Sancar, A. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 13660–13665. [Google Scholar]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar]
- Ho, J.C.; Warr, N.J.; Shimizu, H.; Watts, F.Z. SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52. Nucleic Acids Res. 2001, 29, 4179–4186. [Google Scholar] [CrossRef]
- Sacher, M.; Pfander, B.; Hoege, C.; Jentsch, S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 2006, 8, 1284–1290. [Google Scholar] [CrossRef]
- Saito, K.; Kagawa, W.; Suzuki, T.; Suzuki, H.; Yokoyama, S.; Saitoh, H.; Tashiro, S.; Dohmae, N.; Kurumizaka, H. The putative nuclear localization signal of the human RAD52 protein is a potential sumoylation site. J. Biochem. 2010, 147, 833–842. [Google Scholar] [CrossRef]
- Altmannova, V.; Eckert-Boulet, N.; Arneric, M.; Kolesar, P.; Chaloupkova, R.; Damborsky, J.; Sung, P.; Zhao, X.; Lisby, M.; Krejci, L. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 2010, 38, 4708–4721. [Google Scholar]
- Ohuchi, T.; Seki, M.; Branzei, D.; Maeda, D.; Ui, A.; Ogiwara, H.; Tada, S.; Enomoto, T. Rad52 sumoylation and its involvement in the efficient induction of homologous recombination. DNA Repair (Amst) 2008, 7, 879–889. [Google Scholar]
- Xie, Y.; Kerscher, O.; Kroetz, M.B.; McConchie, H.F.; Sung, P.; Hochstrasser, M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007, 282, 34176–34184. [Google Scholar] [CrossRef]
- Mullen, J.R.; Brill, S.J. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J. Biol. Chem. 2008, 283, 19912–19921. [Google Scholar] [CrossRef]
- Dou, H.; Huang, C.; Singh, M.; Carpenter, P.B.; Yeh, E.T. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 2010, 39, 333–345. [Google Scholar] [CrossRef]
- Kovalenko, O.V.; Plug, A.W.; Haaf, T.; Gonda, D.K.; Ashley, T.; Ward, D.C.; Radding, C.M.; Golub, E.I. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 1996, 93, 2958–2963. [Google Scholar]
- Shen, Z.; Pardington-Purtymun, P.E.; Comeaux, J.C.; Moyzis, R.K.; Chen, D.J. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics 1996, 37, 183–186. [Google Scholar] [CrossRef]
- Krejci, L.; van Komen, S.; Li, Y.; Villemain, J.; Reddy, M.S.; Klein, H.; Ellenberger, T.; Sung, P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 2003, 423, 305–309. [Google Scholar]
- Veaute, X.; Jeusset, J.; Soustelle, C.; Kowalczykowski, S.C.; Le Cam, E.; Fabre, F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 2003, 423, 309–312. [Google Scholar]
- Kolesar, P.; Sarangi, P.; Altmannova, V.; Zhao, X.; Krejci, L. Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res. 2012. [Google Scholar]
- Saponaro, M.; Callahan, D.; Zheng, X.; Krejci, L.; Haber, J.E.; Klein, H.L.; Liberi, G. Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet 2010, 6. [Google Scholar]
- Heyer, W.D.; Li, X.; Rolfsmeier, M.; Zhang, X.P. Rad54: The Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006, 34, 4115–4125. [Google Scholar] [CrossRef]
- Krejci, L.; Altmannova, V.; Spirek, M.; Zhao, X. Homologous recombination and its regulation. 2012. [Google Scholar]
- Uzunova, K.; Göttsche, K.; Miteva, M.; Weisshaar, S.R.; Glanemann, C.; Schnellhardt, M.; Niessen, M.; Scheel, H.; Hofmann, K.; Johnson, E.S.; Praefcke, G.J.; Dohmen, R.J. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007, 282, 34167–34175. [Google Scholar]
- Shah, P.P.; Zheng, X.; Epshtein, A.; Carey, J.N.; Bishop, D.K.; Klein, H.L. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol. Cell 2010, 39, 862–872. [Google Scholar] [CrossRef]
- Cal-Bakowska, M.; Litwin, I.; Bocer, T.; Wysocki, R.; Dziadkowiec, D. The Swi2-Snf2-like protein Uls1 is involved in replication stress response. Nucleic Acids Res. 2011. [Google Scholar]
- Lu, C.Y.; Tsai, C.H.; Brill, S.J.; Teng, S.C. Sumoylation of the BLM ortholog, Sgs1, promotes telomere-telomere recombination in budding yeast. Nucleic Acids Res. 2010, 38, 488–498. [Google Scholar] [CrossRef]
- Rog, O.; Miller, K.M.; Ferreira, M.G.; Cooper, J.P. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol. Cell 2009, 33, 559–569. [Google Scholar] [CrossRef]
- Ouyang, K.J.; Woo, L.L.; Zhu, J.; Huo, D.; Matunis, M.J.; Ellis, N.A. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 2009, 7. [Google Scholar] [CrossRef]
- Li, Y.J.; Stark, J.M.; Chen, D.J.; Ann, D.K.; Chen, Y. Role of SUMO:SIM-mediated protein-protein interaction in non-homologous end joining. Oncogene 2010, 29, 3509–3518. [Google Scholar] [CrossRef]
- Yurchenko, V.; Xue, Z.; Gama, V.; Matsuyama, S.; Sadofsky, M.J. Ku70 is stabilized by increased cellular SUMO. Biochem. Biophys. Res. Commun. 2008, 366, 263–268. [Google Scholar] [CrossRef]
- Gocke, C.B.; Yu, H.; Kang, J. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J. Biol. Chem. 2005, 280, 5004–5012. [Google Scholar]
- Yurchenko, V.; Xue, Z.; Sadofsky, M.J. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol. Cell Biol. 2006, 26, 1786–1794. [Google Scholar] [CrossRef]
- Burgess, R.C.; Rahman, S.; Lisby, M.; Rothstein, R.; Zhao, X. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell Biol. 2007, 27, 6153–6162. [Google Scholar] [CrossRef]
- Eladad, S.; Ye, T.Z.; Hu, P.; Leversha, M.; Beresten, S.; Matunis, M.J.; Ellis, N.A. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet 2005, 14, 1351–1365. [Google Scholar] [CrossRef]
- Kawabe, Y.; Seki, M.; Seki, T.; Wang, W.S.; Imamura, O.; Furuichi, Y.; Saitoh, H.; Enomoto, T. Covalent modification of the Werner’s syndrome gene product with the ubiquitin-related protein, SUMO-1. J. Biol. Chem. 2000, 275, 20963–20966. [Google Scholar]
- Woods, Y.L.; Xirodimas, D.P.; Prescott, A.R.; Sparks, A.; Lane, D.P.; Saville, M.K. p14 Arf promotes small ubiquitin-like modifier conjugation of Werners helicase. J. Biol. Chem. 2004, 279, 50157–50166. [Google Scholar]
- Takahashi, Y.; Dulev, S.; Liu, X.; Hiller, N.J.; Zhao, X.; Strunnikov, A. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet 2008, 4. [Google Scholar] [CrossRef]
- Torres-Rosell, J.; Sunjevaric, I.; de Piccoli, G.; Sacher, M.; Eckert-Boulet, N.; Reid, R.; Jentsch, S.; Rothstein, R.; Aragón, L.; Lisby, M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007, 9, 923–931. [Google Scholar] [CrossRef]
- Takahashi, Y.; Strunnikov, A. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma 2008, 117, 189–198. [Google Scholar] [CrossRef]
- De Piccoli, G.; Torres-Rosell, J.; Aragon, L. The unnamed complex: What do we know about Smc5-Smc6? Chromosome Res. 2009, 17, 251–263. [Google Scholar] [CrossRef]
- Stephan, A.K.; Kliszczak, M.; Morrison, C.G. The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett. 2011, 585, 2907–2913. [Google Scholar] [CrossRef]
- Fujioka, Y.; Kimata, Y.; Nomaguchi, K.; Watanabe, K.; Kohno, K. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J. Biol. Chem. 2002, 277, 21585–21591. [Google Scholar]
- Grandin, N.; Charbonneau, M. Protection against chromosome degradation at the telomeres. Biochimie 2008, 90, 41–59. [Google Scholar] [CrossRef]
- Hang, L.E.; Liu, X.; Cheung, I.; Yang, Y.; Zhao, X. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat. Struct. Mol. Biol. 2011, 18, 920–926. [Google Scholar] [CrossRef]
- Tanaka, K.; Nishide, J.; Okazaki, K.; Kato, H.; Niwa, O.; Nakagawa, T.; Matsuda, H.; Kawamukai, M.; Murakami, Y. Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregati. Mol. Cell Biol. 1999, 19, 8660–8672. [Google Scholar]
- Xhemalce, B.; Riising, E.M.; Baumann, P.; Dejean, A.; Arcangioli, B.; Seeler, J.S. Role of SUMO in the dynamics of telomere maintenance in fission yeast. Proc. Natl. Acad. Sci. USA 2007, 104, 893–898. [Google Scholar]
- Xhemalce, B.; Seeler, J.S.; Thon, G.; Dejean, A.; Arcangioli, B. Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J. 2004, 23, 3844–3853. [Google Scholar] [CrossRef]
- Ferreira, H.C.; Luke, B.; Schober, H.; Kalck, V.; Lingner, J.; Gasser, S.M. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat. Cell Biol. 2011, 13, 867–874. [Google Scholar] [CrossRef]
- Lindroos, H.B.; Strom, L.; Itoh, T.; Katou, Y.; Shirahige, K.; Sjogren, C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 2006, 22, 755–767. [Google Scholar] [CrossRef]
- Pebernard, S.; Schaffer, L.; Campbell, D.; Head, S.R.; Boddy, M.N. Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 2008, 27, 3011–3023. [Google Scholar] [CrossRef]
- Torres-Rosell, J.; Machin, F.; Farmer, S.; Jarmuz, A.; Eydmann, T.; Dalgaard, J.Z.; Aragon, L. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 2005, 7, 412–419. [Google Scholar] [CrossRef]
- Chavez, A.; George, V.; Agrawal, V.; Johnson, F.B. Sumoylation and the structural maintenance of chromosomes (Smc) 5/6 complex slow senescence through recombination intermediate resolution. J. Biol. Chem. 2010, 285, 11922–11930. [Google Scholar] [CrossRef]
- Noel, J.F.; Wellinger, R.J. Abrupt telomere losses and reduced end-resection can explain accelerated senescence of Smc5/6 mutants lacking telomerase. DNA Repair (Amst) 2011, 10, 271–282. [Google Scholar]
- Blackburn, E.H. Switching and signaling at the telomere. Cell 2001, 106, 661–673. [Google Scholar] [CrossRef]
- Dunham, M.A.; Neumann, A.A.; Fasching, C.L.; Reddel, R.R. Telomere maintenance by recombination in human cells. Nat. Genet 2000, 26, 447–450. [Google Scholar] [CrossRef]
- Crabbe, L.; Verdun, R.E.; Haggblom, C.I.; Karlseder, J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 2004, 306, 1951–1953. [Google Scholar]
- Henson, J.D.; Neumann, A.A.; Yeager, T.R.; Reddel, R.R. Alternative lengthening of telomeres in mammalian cells. Oncogene 2002, 21, 598–610. [Google Scholar]
- Muntoni, A.; Reddel, R.R. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 2005, 14 Spec No. 2. R191–R196. [Google Scholar]
- Yeager, T.R.; Neumann, A.A.; Englezou, A.; Huschtscha, L.I.; Noble, J.R.; Reddel, R.R. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999, 59, 4175–4179. [Google Scholar]
- Lang, M.; Jegou, T.; Chung, I.; Richter, K.; Munch, S.; Udvarhelyi, A.; Cremer, C.; Hemmerich, P.; Engelhardt, J.; Hell, S.W.; Rippe, K. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J. Cell Sci. 2010, 123, 392–400. [Google Scholar]
- Shen, T.H.; Lin, H.K.; Scaglioni, P.P.; Yung, T.M.; Pandolfi, P.P. The mechanisms of PML-nuclear body formation. Mol. Cell 2006, 24, 331–339. [Google Scholar] [CrossRef]
- Chung, I.; Leonhardt, H.; Rippe, K. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J. Cell Sci. 2011, 124, 3603–3618. [Google Scholar] [CrossRef]
- Nabetani, A.; Ishikawa, F. Alternative lengthening of telomeres pathway: Recombination-mediated telomere maintenance mechanism in human cells. J. Biochem. 2011, 149, 5–14. [Google Scholar] [CrossRef]
- Potts, P.R.; Yu, H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 2007, 14, 581–590. [Google Scholar] [CrossRef]
- Lin, D.Y.; Huang, Y.S.; Jeng, J.C.; Kuo, H.Y.; Chang, C.C.; Chao, T.T.; Ho, C.C.; Chen, Y.C.; Lin, T.P.; Fang, H.I.; et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 2006, 24, 341–354. [Google Scholar]
- Takahashi, H.; Hatakeyama, S.; Saitoh, H.; Nakayama, K.I. Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J. Biol. Chem. 2005, 280, 5611–5621. [Google Scholar]
- Brouwer, A.K.; Schimmel, J.; Wiegant, J.C.; Vertegaal, A.C.; Tanke, H.J.; Dirks, R.W. Telomeric DNA mediates de novo PML body formation. Mol. Biol. Cell 2009, 20, 4804–4815. [Google Scholar] [CrossRef]
- Koshiyama, A.; Hamada, F.N.; Namekawa, S.H.; Iwabata, K.; Sugawara, H.; Sakamoto, A.; Ishizaki, T.; Sakaguchi, K. Sumoylation of a meiosis-specific RecA homolog, Lim15/Dmc1, via interaction with the small ubiquitin-related modifier (SUMO)-conjugating enzyme Ubc9. FEBS J. 2006, 273, 4003–4012. [Google Scholar] [CrossRef]
- Ito, T.; Tashiro, K.; Muta, S.; Ozawa, R.; Chiba, T.; Nishizawa, M.; Yamamoto, K.; Kuhara, S.; Sakaki, Y. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 1143–1147. [Google Scholar]
- Zavec, A.B.; Comino, A.; Lenassi, M.; Komel, R. Ecm11 protein of yeast Saccharomyces cerevisiae is regulated by sumoylation during meiosis. FEMS Yeast Res. 2008, 8, 64–70. [Google Scholar] [CrossRef]
- Hunter, N. Meiotic Recombination; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2007; pp. 381–441. [Google Scholar]
- Szekvolgyi, L.; Nicolas, A. From meiosis to postmeiotic events: Homologous recombination is obligatory but flexible. FEBS J. 2010, 277, 571–589. [Google Scholar] [CrossRef]
- Hooker, G.W.; Roeder, G.S. A Role for SUMO in meiotic chromosome synapsis. Curr. Biol. 2006, 16, 1238–1243. [Google Scholar] [CrossRef]
- Eichinger, C.S.; Jentsch, S. Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc. Natl. Acad. Sci. USA 2010, 107, 11370–11375. [Google Scholar] [CrossRef]
- Lin, F.M.; Lai, Y.J.; Shen, H.J.; Cheng, Y.H.; Wang, T.F. Yeast axial-element protein, Red1, binds SUMO chains to promote meiotic interhomologue recombination and chromosome synapsis. EMBO J. 2010, 29, 586–596. [Google Scholar]
- Perry, J.; Kleckner, N.; Borner, G.V. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc. Natl. Acad.Sci. USA 2005, 102, 17594–17599. [Google Scholar]
- Spirek, M.; Estreicher, A.; Csaszar, E.; Wells, J.; McFarlane, R.J.; Watts, F.Z.; Loidl, J. SUMOylation is required for normal development of linear elements and wild-type meiotic recombination in Schizosaccharomyces pombe. Chromosoma 2010, 119, 59–72. [Google Scholar] [CrossRef]
- Brown, P.W.; Hwang, K.; Schlegel, P.N.; Morris, P.L. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum. Reprod. 2008, 23, 2850–2857. [Google Scholar]
- Wilkinson, K.A.; Henley, J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010, 428, 133–145. [Google Scholar] [CrossRef]
- Hietakangas, V.; Anckar, J.; Blomster, H.A.; Fujimoto, M.; Palvimo, J.J.; Nakai, A.; Sistonen, L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA 2006, 103, 45–50. [Google Scholar]
- Nathan, D.; Ingvarsdottir, K.; Sterner, D.E.; Bylebyl, G.R.; Dokmanovic, M.; Dorsey, J.A.; Whelan, K.A.; Krsmanovic, M.; Lane, W.S.; Meluh, P.B.; et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006, 20, 966–976. [Google Scholar] [CrossRef]
- Ulrich, H.D. Mutual interactions between the SUMO and ubiquitin systems: A plea of no contest. Trends Cell Biol. 2005, 15, 525–532. [Google Scholar] [CrossRef]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Davies, A.A.; Huttner, D.; Daigaku, Y.; Chen, S.; Ulrich, H.D. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol. Cell 2008, 29, 625–636. [Google Scholar] [CrossRef]
- Bienko, M.; Green, C.M.; Crosetto, N.; Rudolf, F.; Zapart, G.; Coull, B.; Kannouche, P.; Wider, G.; Peter, M.; Lehmann, A.R.; et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 2005, 310, 1821–1824. [Google Scholar]
- Chen, J.; Bozza, W.; Zhuang, Z. Ubiquitination of PCNA and its essential role in eukaryotic translesion synthesis. Cell Biochem. Biophys. 2011, 60, 47–60. [Google Scholar] [CrossRef]
- Ulrich, H.D.; Jentsch, S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000, 19, 3388–3397. [Google Scholar] [CrossRef]
- Branzei, D.; Vanoli, F.; Foiani, M. SUMOylation regulates Rad18-mediated template switch. Nature 2008, 456, 915–920. [Google Scholar]
- Zhang, H.; Lawrence, C.W. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. USA 2005, 102, 15954–15959. [Google Scholar]
- Kats, E.S.; Enserink, J.M.; Martinez, S.; Kolodner, R.D. The Saccharomyces cerevisiae Rad6 postreplication repair and Siz1/Srs2 homologous recombination-inhibiting pathways process DNA damage that arises in asf1 mutants. Mol. Cell Biol. 2009, 29, 5226–5237. [Google Scholar] [CrossRef]
- Das-Bradoo, S.; Nguyen, H.D.; Bielinsky, A.K. Damage-specific modification of PCNA. Cell Cycle 2010, 9, 3674–3679. [Google Scholar]
- Papouli, E.; Chen, S.; Davies, A.A.; Huttner, D.; Krejci, L.; Sung, P.; Ulrich, H.D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 2005, 19, 123–133. [Google Scholar]
- Pfander, B.; Moldovan, G.L.; Sacher, M.; Hoege, C.; Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005, 436, 428–433. [Google Scholar]
- Armstrong, A.A.; Mohideen, F.; Lima, C.D. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 2012, 483, 59–63. [Google Scholar]
- Burgess, R.C.; Lisby, M.; Altmannova, V.; Krejci, L.; Sung, P.; Rothstein, R. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J. Cell Biol. 2009, 185, 969–981. [Google Scholar] [CrossRef]
- Kim, S.O.; Yoon, H.; Park, S.O.; Lee, M.; Shin, J.S.; Ryu, K.S.; Lee, J.O.; Seo, Y.S.; Jung, H.S.; Choi, B.S. Srs2 possesses a non-canonical PIP box in front of its SBM for precise recognition of SUMOylated PCNA. J. Mol. Cell Biol. 2012. [Google Scholar] [CrossRef]
- Burkovics, P.; Sebesta, M.; Sisakova, A.; Plault, N.; Szukacsov, V.; Robert, T.; Pinter, L.; Kolesar, P.; Marini, V.; Haracska, L.; Gangloff, S.; Krejci, L. Srs2 mediates PCNA-SUMO dependent Inhibition of DNA repair synthesis. 2012. Submitted. [Google Scholar]
- Lydeard, J.R.; Lipkin-Moore, Z.; Sheu, Y.J.; Stillman, B.; Burgers, P.M.; Haber, J.E. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010, 24, 1133–1144. [Google Scholar]
- Freudenthal, B.D.; Brogie, J.E.; Gakhar, L.; Kondratick, C.M.; Washington, M.T. Crystal structure of SUMO-modified proliferating cell nuclear antigen. J. Mol. Biol. 2011, 406, 9–17. [Google Scholar] [CrossRef]
- Freudenthal, B.D.; Gakhar, L.; Ramaswamy, S.; Washington, M.T. Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat. Struct. Mol. Biol. 2010, 17, 479–484. [Google Scholar]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 2006, 23, 723–732. [Google Scholar] [CrossRef]
- Arakawa, H.; Moldovan, G.L.; Saribasak, H.; Saribasak, N.N.; Jentsch, S.; Buerstedde, J.M. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006, 4. [Google Scholar] [CrossRef]
- Kannouche, P.L.; Wing, J.; Lehmann, A.R. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 2004, 14, 491–500. [Google Scholar] [CrossRef]
- Leach, C.A.; Michael, W.M. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J. Cell Biol. 2005, 171, 947–954. [Google Scholar] [CrossRef]
- Moldovan, G.L.; Dejsuphong, D.; Petalcorin, M.I.; Hofmann, K.; Takeda, S.; Boulton, S.J.; D’Andrea, A.D. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 2012, 45, 75–86. [Google Scholar] [CrossRef]
- Gali, H.; Juhasz, S.; Morocz, M.; Hajdu, I.; Fatyol, K.; Szukacsov, V.; Burkovics, P.; Haracska, L. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 2011. [Google Scholar] [CrossRef]
- Wang, S.C.; Nakajima, Y.; Yu, Y.L.; Xia, W.; Chen, C.T.; Yang, C.C.; McIntush, E.W.; Li, L.Y.; Hawke, D.H.; Kobayashi, R.; Hung, M.C. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 2006, 8, 1359–1368. [Google Scholar] [CrossRef]
- Naryzhny, S.N.; Lee, H. The post-translational modifications of proliferating cell nuclear antigen: Acetylation, not phosphorylation, plays an important role in the regulation of its function. J. Biol. Chem. 2004, 279, 20194–20199. [Google Scholar] [CrossRef]
- Yu, Y.; Cai, J.P.; Tu, B.; Wu, L.; Zhao, Y.; Liu, X.; Li, L.; McNutt, M.A.; Feng, J.; He, Q.; et al. Proliferating cell nuclear antigen is protected from degradation by forming a complex with MutT Homolog2. J. Biol. Chem. 2009, 284, 19310–19320. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Altmannová, V.; Kolesár, P.; Krejčí, L. SUMO Wrestles with Recombination. Biomolecules 2012, 2, 350-375. https://doi.org/10.3390/biom2030350
Altmannová V, Kolesár P, Krejčí L. SUMO Wrestles with Recombination. Biomolecules. 2012; 2(3):350-375. https://doi.org/10.3390/biom2030350
Chicago/Turabian StyleAltmannová, Veronika, Peter Kolesár, and Lumír Krejčí. 2012. "SUMO Wrestles with Recombination" Biomolecules 2, no. 3: 350-375. https://doi.org/10.3390/biom2030350
APA StyleAltmannová, V., Kolesár, P., & Krejčí, L. (2012). SUMO Wrestles with Recombination. Biomolecules, 2(3), 350-375. https://doi.org/10.3390/biom2030350





