Efficacy of an Optimised Bacteriophage Cocktail to Clear Clostridium difficile in a Batch Fermentation Model
Abstract
:1. Introduction
2. Results
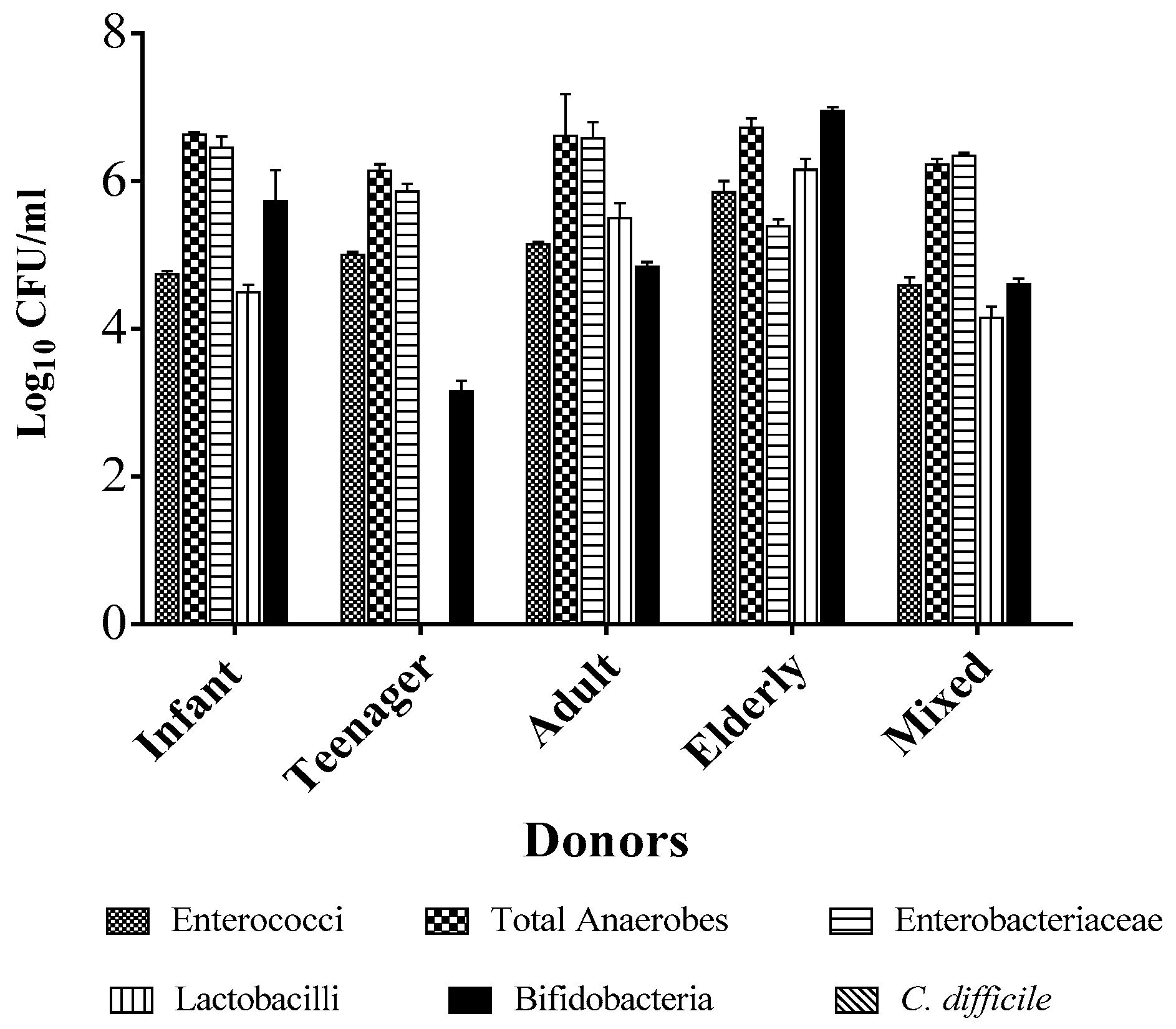
2.1. Individual Donors Have a Unique Microbiome Composition
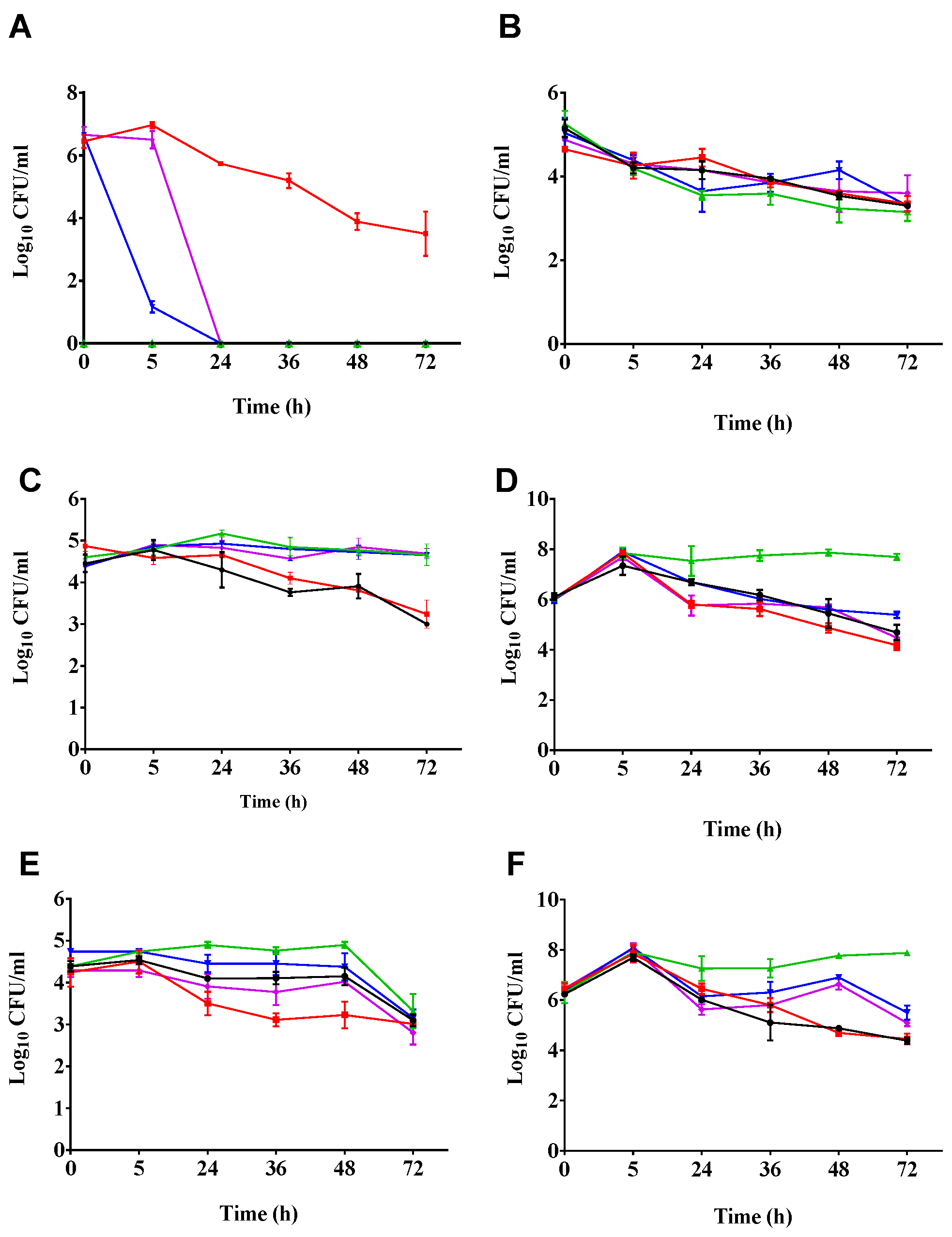
2.2. Phages Cleared C. difficile in the Batch Fermentation Model
2.3. Impact of Phage Treatmens on the Viability of other Components of the Culturable Gut Microbiota
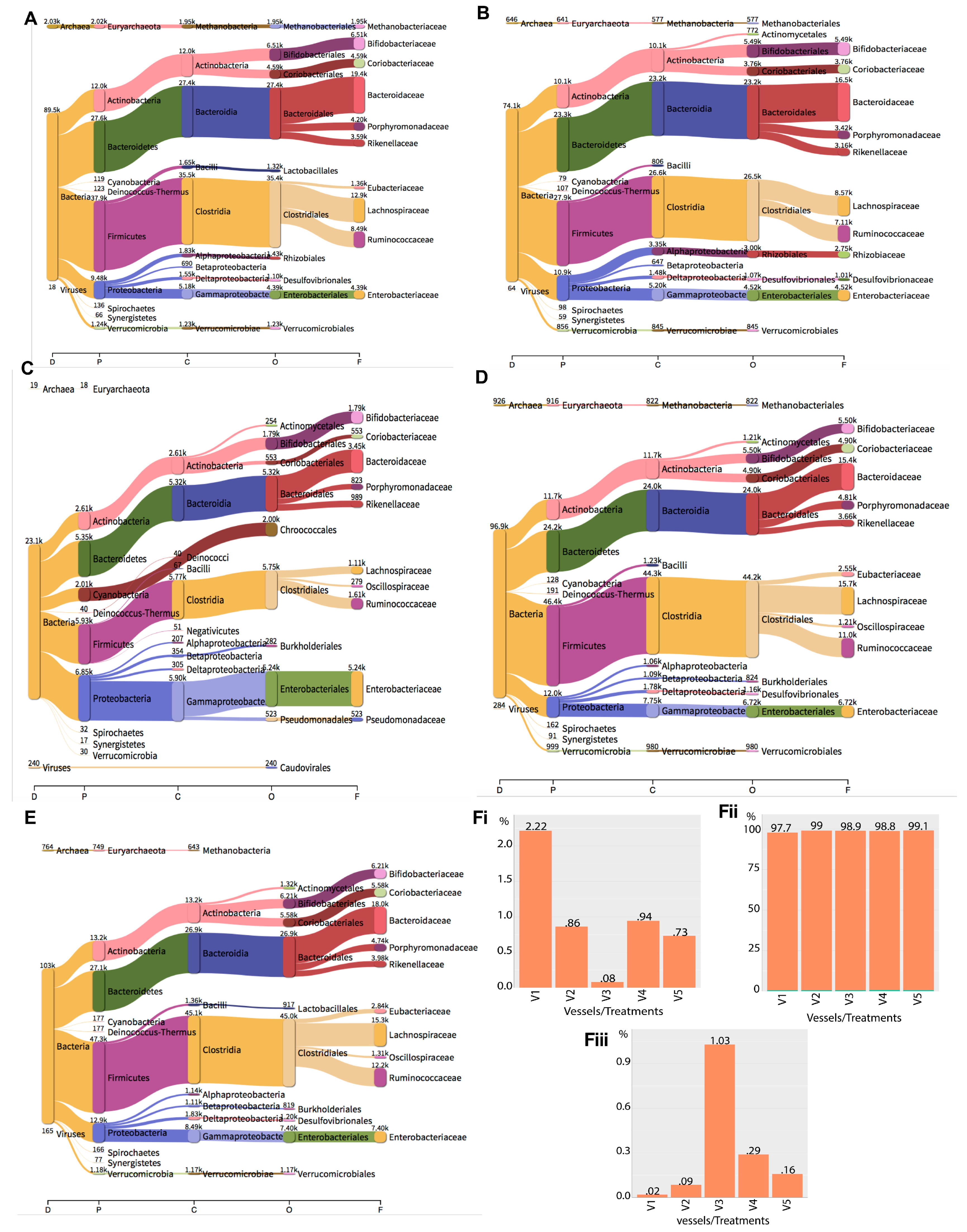
2.4. Metagenomics Analysis of the Impact of Phage Treatment on the Total Microbiome within the Gut Fermentation Vessels
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates and Phage Cocktail Used in This Study
4.2. Gut Fermentation Model Set-Up
4.3. Bacteria and Phage Treatment of the Fermenation Vessels
4.4. DNA Extraction, Metagenomics Sequencing, and Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Patents
Ethics
References
- De Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed]
- De Kraker, M.E.A.; Davey, P.G.; Grundmann, H. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: Estimating the burden of antibiotic resistance in Europe. PLoS Med. 2011, 8, e1001104. [Google Scholar] [CrossRef] [PubMed]
- Stewardson, A.J.; Allignol, A.; Beyersmann, J.; Graves, N.; Schumacher, M.; Meyer, R.; Tacconelli, E.; De Angelis, G.; Farina, C.; Pezzoli, F.; et al. The health and economic burden of bloodstream infections caused by antimicrobial-susceptible and non-susceptible Enterobacteriaceae and Staphylococcus aureus in European hospitals, 2010 and 2011: A multicentre retrospective cohort study. Euro Surveill. 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Naylor, N.R.; Pouwels, K.B.; Hope, R.; Green, N.; Henderson, K.L.; Knight, G.M.; Atun, R.; Robotham, J.V.; Deeny, S. A national estimate of the health and cost burden of Escherichia coli bacteraemia in the hospital setting: The importance of antibiotic resistance. bioRxiv 2017. [Google Scholar] [CrossRef]
- O’Neil, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. The Review on Antimicrobial Resistance 2014. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 31 December 2017).
- Zucca, M.; Scutera, S.; Savoia, D. Novel avenues for Clostridium difficile infection drug discovery. Expert Opin. Drug Discov. 2013, 8, 459–477. [Google Scholar] [CrossRef] [PubMed]
- Wise, R.; Blaser, M.J.; Carrs, O.; Cassell, G.; Fishman, N.; Guidos, R.; Levy, S.; Powers, J.; Norrby, R.; Tillotson, G.; et al. The urgent need for new antibacterial agents. J. Antimicrob. Chemother. 2011, 66, 1939–1940. [Google Scholar] [CrossRef] [PubMed]
- Rai, J.; Randhawa, G.K.; Kaur, M. Recent advances in antibacterial drugs. Int. J. Appl. Med. Res. 2013, 3, 3–10. [Google Scholar]
- Kutter, E.M.; Kuhl, S.J.; Abedon, S.T. Re-establishing a place for phage therapy in Western medicine. Future Microbiol. 2015, 10, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and bacterial plant diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Cisek, A.A.; Dabrowska, I.; Gregorczyk, K.P.; Wyzewski, Z. Phage therapy in bacterial infections treatment: One hundred years after the discovery of bacteriophages. Curr. Microbiol. 2016, 74, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Regeimbal, J.M.; Jacobs, A.C.; Corey, B.W.; Henry, M.S.; Thompson, M.G.; Pavlicek, R.L.; Quinones, J.; Hannah, R.M.; Ghebremedhin, M.; Crane, N.J.; et al. Personalized therapeutic cocktail of wild environmental phages rescues mice from Acinetobacter baumannii wound infections. Antimicrob. Agents Chemother. 2016, 60, 5806–5816. [Google Scholar] [CrossRef] [PubMed]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G.J. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Speck, P.; Smithyman, A. Safety and efficacy of phage therapy via the intravenous route. FEMS Microbiol. Lett. 2015, 363. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H. What is needed for phage therapy to become a reality in Western medicine? Virology 2012, 434, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Slopek, S.; Weber-Dabrowska, B.; Dabrowski, M.; Kucharewicz-Krukowska, A. Results of bacteriophage treatment of suppurative bacterial infections in the years 1981–1986. Arch. Immunol. Ther. Exp. 1987, 35, 569–583. [Google Scholar]
- EBI Food Safety. FDA and USDA Extend GRAS Approval for LISTEX for All Food Products. 2007. Available online: http://www.ebifoodsafety.com/en/news-2007.aspx (accessed on 31 December 2017).
- Kwon, J.H.; Olsen, M.A.; Dubberke, E.R. The morbidity, mortality, and costs associated with Clostridium difficile infection. Infect. Dis. Clin. N. Am. 2015, 29, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Olsen, M.A.; Young-Xu, Y.; Stwalley, D.; Kelly, C.P.; Gerding, D.N.; Saeed, M.J.; Mahé, C.; Dubberke, E.R. The burden of Clostridium difficile infection: Estimates of the incidence of cdi from U.S. Administrative databases. BMC Infect. Dis. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Dubberke, E.R.; Olsen, M.A. Burden of Clostridium difficile on the healthcare system. Clin. Infect. Dis. 2012, 55, S88–S92. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.P.; Notermans, D.W.; van Benthem, B.H.B.; Brazier, J.S.; Wilcox, M.H.; Rupnik, M.; Monnet, D.L.; van Dissel, J.T. Clostridium difficile infection in Europe: A hospital-based survey. Lancet 2011, 377, 63–73. [Google Scholar] [CrossRef]
- Boyle, N.M.; Magaret, A.; Stednick, Z.; Morrison, A.; Butler-Wu, S.; Zerr, D.; Rogers, K.; Podczervinski, S.; Cheng, A.; Wald, A.; et al. Evaluating risk factors for Clostridium difficile infection in adult and pediatric hematopoietic cell transplant recipients. Antimicrob. Resist. Infect. Control 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Lessa, F.C.; Mu, Y.; Bamberg, W.M.; Beldavs, Z.G.; Dumyati, G.K.; Dunn, J.R.; Farley, M.M.; Holzbauer, S.M.; Meek, J.I.; Phipps, E.C.; et al. Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 2015, 372, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Lessa, F.C.; Gould, C.V.; McDonald, L.C. Current status of Clostridium difficile infection epidemiology. Clin. Infect. Dis. 2012, 55, S65–S70. [Google Scholar] [CrossRef] [PubMed]
- Kociolek, L.K.; Gerding, D.N. Breakthroughs in the treatment and prevention of Clostridium difficile infection. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 150–160. [Google Scholar] [CrossRef] [PubMed]
- DuPont, H.L. Diagnosis and management of Clostridium difficile infection. Clin. Gastroenterol. Hepatol. 2013, 11, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Dang, M.D.; Hasbun, R.; Koo, H.L.; Jiang, Z.D.; DuPont, H.L.; Garey, K.W. Clostridium difficile infection: Update on emerging antibiotic treatment options and antibiotic resistance. Expert Rev. Anti-Infect. Ther. 2010, 8, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Debast, S.B.; Bauer, M.P.; Kuijper, E.J. European society of clinical microbiology and infectious diseases: Update of the treatment guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 2014, 20, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Moura, I.; Spigaglia, P.; Barbanti, F.; Mastrantonio, P. Analysis of metronidazole susceptibility in different Clostridium difficile PCR ribotypes. J. Antimicrob. Chemother. 2013, 68, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Koss, C.A.; Baras, D.C.; Lane, S.D.; Aubry, R.; Marcus, M.; Markowitz, L.E.; Koumans, E.H. Investigation of metronidazole use during pregnancy and adverse birth outcomes. Antimicrob. Agents Chemother. 2012, 56, 4800–4805. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.L.; Suda, K.J.; Evans, C.T. Antibiotic treatment for Clostridium difficile-associated diarrhoea in adults. Cochrane Database Syst. Rev. 2017, 3. [Google Scholar] [CrossRef] [PubMed]
- Poduval, R.D.; Kamath, R.P.; Corpuz, M.; Norkus, E.P.; Pitchumoni, C.S. Clostridium difficile and vancomycin-resistant enterococcus: The new nosocomial alliance. Am. J. Gastroenterol. 2000, 95, 3513–3515. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.P.; Kuijper, E.J.; van Dissel, J.T. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): Treatment guidance document for Clostridium difficile infection (CDI). Clin. Microbiol. Infect. 2009, 15, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Baines, S.D.; Wilcox, M.H. Antimicrobial resistance and reduced susceptibility in Clostridium difficile: Potential consequences for induction, treatment, and recurrence of C. difficile infection. Antibiotics 2015, 4, 267–298. [Google Scholar] [CrossRef] [PubMed]
- Chilton, C.H.; Crowther, G.S.; Freeman, J.; Todhunter, S.L.; Nicholson, S.; Longshaw, C.M.; Wilcox, M.H. Successful treatment of simulated Clostridium difficile infection in a human gut model by fidaxomicin first line and after vancomycin or metronidazole failure. J. Antimicrob. Chemother. 2014, 69, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; Umscheid, C.A.; Fishman, N.; Lee, B.Y. Is fidaxomicin worth the cost? An economic analysis. Clin. Infect. Dis. 2013, 57, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Stranges, P.M.; Hutton, D.W.; Collins, C.D. Cost-effectiveness analysis evaluating fidaxomicin versus oral vancomycin for the treatment of Clostridium difficile infection in the United States. Value Health 2013, 16, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.P. Fidaxomicin (Dificid), a novel oral macrocyclic antibacterial agent for the treatment of Clostridium difficile–associated diarrhea in adults. Pharmacol. Ther. 2012, 37, 278–281. [Google Scholar]
- Hargreaves, K.R.; Clokie, M.R.J. Clostridium difficile phages: Still difficult? Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Sangster, W.; Hegarty, J.P.; Stewart, D.B. Phage therapy for Clostridium difficile infection: An alternative to antibiotics? Semin. Colon Rectal Surg. 2014, 25, 167–170. [Google Scholar] [CrossRef]
- Meader, E.; Mayer, M.J.; Steverding, D.; Carding, S.R.; Narbad, A. Evaluation of bacteriophage therapy to control Clostridium difficile and toxin production in an in vitro human colon model system. Anaerobe 2013, 22, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Nale, J.Y.; Spencer, J.; Hargreaves, K.R.; Buckley, A.M.; Trzepiński, P.; Douce, G.R.; Clokie, M.R.J. Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob. Agents Chemother. 2016, 60, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Nale, J.Y.; Chutia, M.; Carr, P.; Hickenbotham, P.; Clokie, M.R.J. ‘Get in early’; biofilm and wax moth (Galleria mellonella) models reveal new insights into the therapeutic potential of Clostridium difficile bacteriophages. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, V.; Fralick, J.A.; Rolfe, R.D. Prevention of Clostridium difficile-induced ileocecitis with bacteriophage. Anaerobe 1999, 5, 69–78. [Google Scholar] [CrossRef]
- Govind, R.; Fralick, J.A.; Rolfe, R.D. In vivo lysogenization of a Clostridium difficile bacteriophage фCD119. Anaerobe 2011, 17, 125–129. [Google Scholar]
- Price, A.B.; Larson, H.E.; Crow, J. Morphology of experimental antibiotic-associated enterocolitis in the hamster: A model for human pseudomembranous colitis and antibiotic-associated diarrhoea. Gut 1979, 20, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Best, E.L.; Freeman, J.; Wilcox, M.H. Models for the study of Clostridium difficile infection. Gut Microbes 2012, 3, 145–167. [Google Scholar] [CrossRef] [PubMed]
- Meader, E.; Mayer, M.J.; Gasson, M.J.; Steverding, D.; Carding, S.R.; Narbad, A. Bacteriophage treatment significantly reduces viable Clostridium difficile and prevents toxin production in an in vitro model system. Anaerobe 2010, 16, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Chilton, C.H.; Crowther, G.S.; Todhunter, S.L.; Nicholson, S.; Freeman, J.; Chesnel, L.; Wilcox, M.H. Efficacy of surotomycin in an in vitro gut model of Clostridium difficile infection. J. Antimicrob. Chemother. 2014, 69, 2426–2433. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, K.R.; Kropinski, A.M.; Clokie, M.R.J. What does the talking?: Quorum sensing signalling genes discovered in a bacteriophage genome. PLoS ONE 2014, 9, e85131. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Baines, S.D.; Chilton, C.H.; Crowther, G.S.; Todhunter, S.L.; Freeman, J.; Wilcox, M.H. Evaluation of antimicrobial activity of ceftaroline against Clostridium difficile and propensity to induce C. difficile infection in an in vitro human gut model. J. Antimicrob. Chemother. 2013, 68, 1842–1849. [Google Scholar] [CrossRef] [PubMed]
- Gould, I.M. The epidemiology of antibiotic resistance. Int. J. Antimicrob. Agents 2008, 32, S2–S9. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; McFarland, L.V.; Mullany, P.; Mulligan, M.E. Characterization of the genetic basis of antibiotic resistance in Clostridium difficile. J. Antimicrob. Chemother. 1994, 33, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Abhilash, M.; Vidya, A.G.; Jagadevi, T. Bacteriophage therapy: A war against antibiotic resistant bacteria. Internet J. Altern. Med. 2009, 7, 1. [Google Scholar]
- Bauer, M.P.; van Dissel, J.T. Alternative strategies for Clostridium difficile infection. Int. J. Antimicrob. Agents 2009, 33, S51–S56. [Google Scholar] [CrossRef]
- Wall, S.K.; Zhang, J.; Rostagno, M.H.; Ebner, P.D. Phage therapy to reduce preprocessing salmonella infections in market-weight swine. Appl. Environ. Microbiol. 2010, 76, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Macfarlane, S.; Gibson, G.R. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 1998, 35, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Taslim, H. Clostridium difficile infection in the elderly. Acta Medica Indonesiana 2009, 41, 148–151. [Google Scholar] [PubMed]
- McGowan, K.L.; Kader, H.A. Clostridium difficile infection in children. Clin. Microbiol. Newsl. 1999, 21, 49–53. [Google Scholar] [CrossRef]
- Lagier, J.C.; Million, M.; Hugon, P.; Armougom, F.; Raoult, D. Human gut microbiota: Repertoire and variations. Front. Cell. Infect. Microbiol. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Antharam, V.C.; Li, E.C.; Ishmael, A.; Sharma, A.; Mai, V.; Rand, K.H. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013, 51, 2884–2892. [Google Scholar] [CrossRef] [PubMed]
- Miyajima, F.; Roberts, P.; Swale, A.; Price, V.; Jones, M.; Horan, M.; Beeching, N.; Brazier, J.; Parry, C.; Pendleton, N.; et al. Characterisation and carriage ratio of Clostridium difficile strains isolated from a community-dwelling elderly population in the United Kingdom. PLoS ONE 2011, 6, e22804. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Britton, R.A.; Young, V.B. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol. 2012, 20, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Wong, S.H.; Lam, K.; Lui, R.; Cheung, K.; Tang, W.; Ching, J.Y.L.; Chan, P.K.S.; Chan, M.C.W.; Wu, J.C.Y.; et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 2017. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; Waetzig, G.H.; Rehman, A.; Moltzau-Anderson, J.; Bharti, R.; Grasis, J.A.; Cassidy, L.; Tholey, A.; Fickenscher, H.; Seegert, D.; et al. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 2017, 152, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Breitwieser, F.P.; Salzberg, S.L. Pavian: Interactive analysis of metagenomics data for microbiomics and pathogen identification. bioRxiv 2016. [Google Scholar] [CrossRef]
- Heng, L. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with Bwa-Mem. Available online: https://arxiv.org/abs/1303.3997 (accessed on 31 December 2017).
| Fermentation Vessels | Treatments | Time to Dose (h) | ||||||
|---|---|---|---|---|---|---|---|---|
| −2 | 0 | 5 | 24 | 36 | 48 | 72 | ||
| 1 | Control untreated | - | M | M | M | M | M | - |
| 2 | C. difficile control | - | B | M | M | M | M | - |
| 3 | Control phage | - | P | P | P | P | M | - |
| 4 | Prophylaxis | P | P+B | P | P | M | M | - |
| 5 | Remedial | - | B | P | P | P | P | - |
| Domain | Clade Reads in Each Vessel (%) | ||||
|---|---|---|---|---|---|
| V1 | V2 | V3 | V4 | V5 | |
| Bacteria | 89,526 (97.76) | 74,116 (99.05) | 23,061 (98.89) | 96,888 (98.77) | 103,406 (99.11) |
| Archaea | 2030 (2.217) | 646 (0.8633) | 19 (0.08148) | 926 (0.944) | 764 (0.7323) |
| Viruses | 18 (0.01966) | 64 (0.08553) | 240 (1.029) | 284 (0.2895) | 165 (0.1581) |
| Total | 91,574 | 74,826 | 23,320 | 98,098 | 104,335 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nale, J.Y.; Redgwell, T.A.; Millard, A.; Clokie, M.R.J. Efficacy of an Optimised Bacteriophage Cocktail to Clear Clostridium difficile in a Batch Fermentation Model. Antibiotics 2018, 7, 13. https://doi.org/10.3390/antibiotics7010013
Nale JY, Redgwell TA, Millard A, Clokie MRJ. Efficacy of an Optimised Bacteriophage Cocktail to Clear Clostridium difficile in a Batch Fermentation Model. Antibiotics. 2018; 7(1):13. https://doi.org/10.3390/antibiotics7010013
Chicago/Turabian StyleNale, Janet Y., Tamsin A. Redgwell, Andrew Millard, and Martha R. J. Clokie. 2018. "Efficacy of an Optimised Bacteriophage Cocktail to Clear Clostridium difficile in a Batch Fermentation Model" Antibiotics 7, no. 1: 13. https://doi.org/10.3390/antibiotics7010013
APA StyleNale, J. Y., Redgwell, T. A., Millard, A., & Clokie, M. R. J. (2018). Efficacy of an Optimised Bacteriophage Cocktail to Clear Clostridium difficile in a Batch Fermentation Model. Antibiotics, 7(1), 13. https://doi.org/10.3390/antibiotics7010013






