The Staphylococcus aureus Membrane Protein SA2056 Interacts with Peptidoglycan Synthesis Enzymes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Expression of sa2056 During Growth
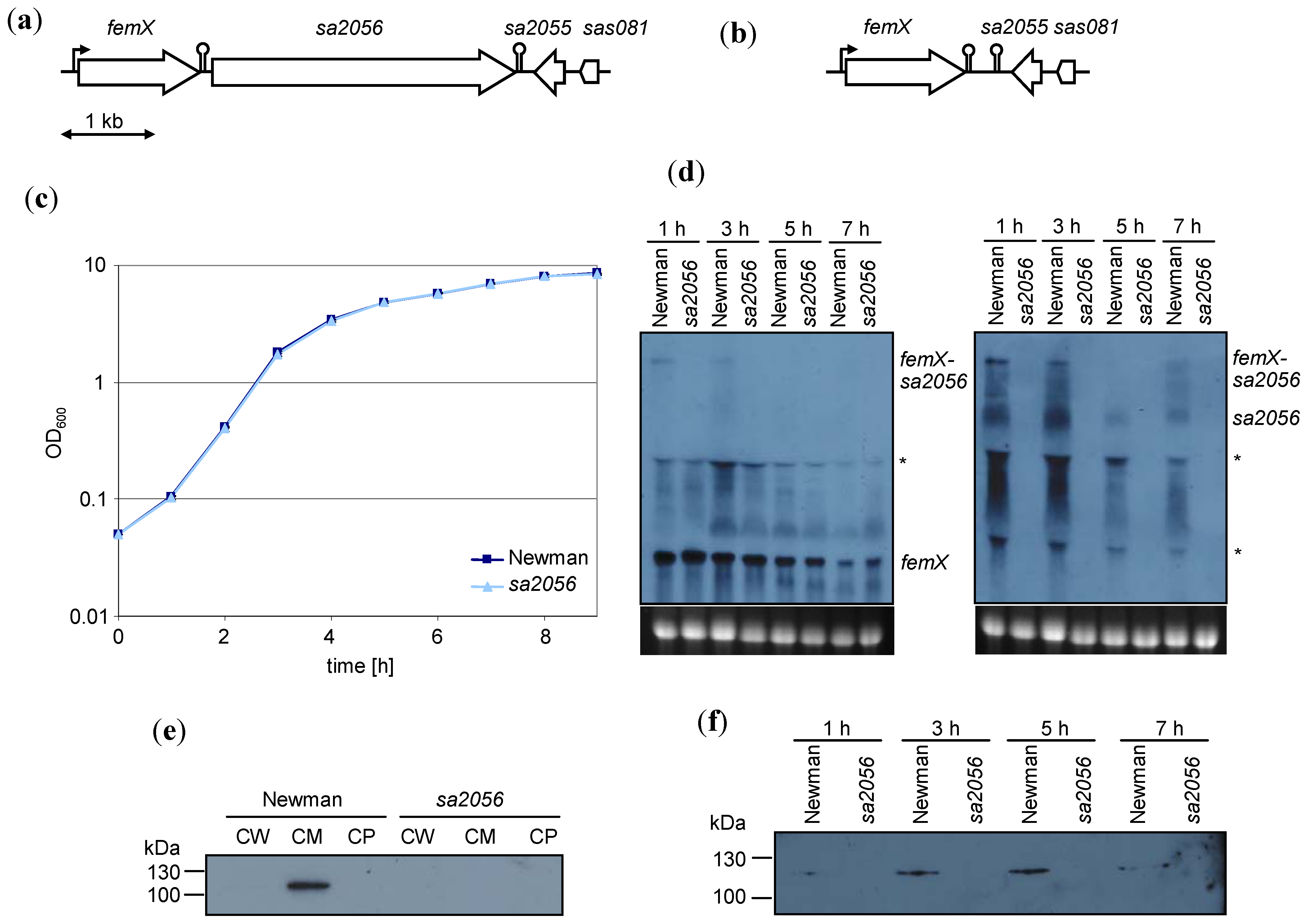
2.2. Analyses of Mutant Phenotype
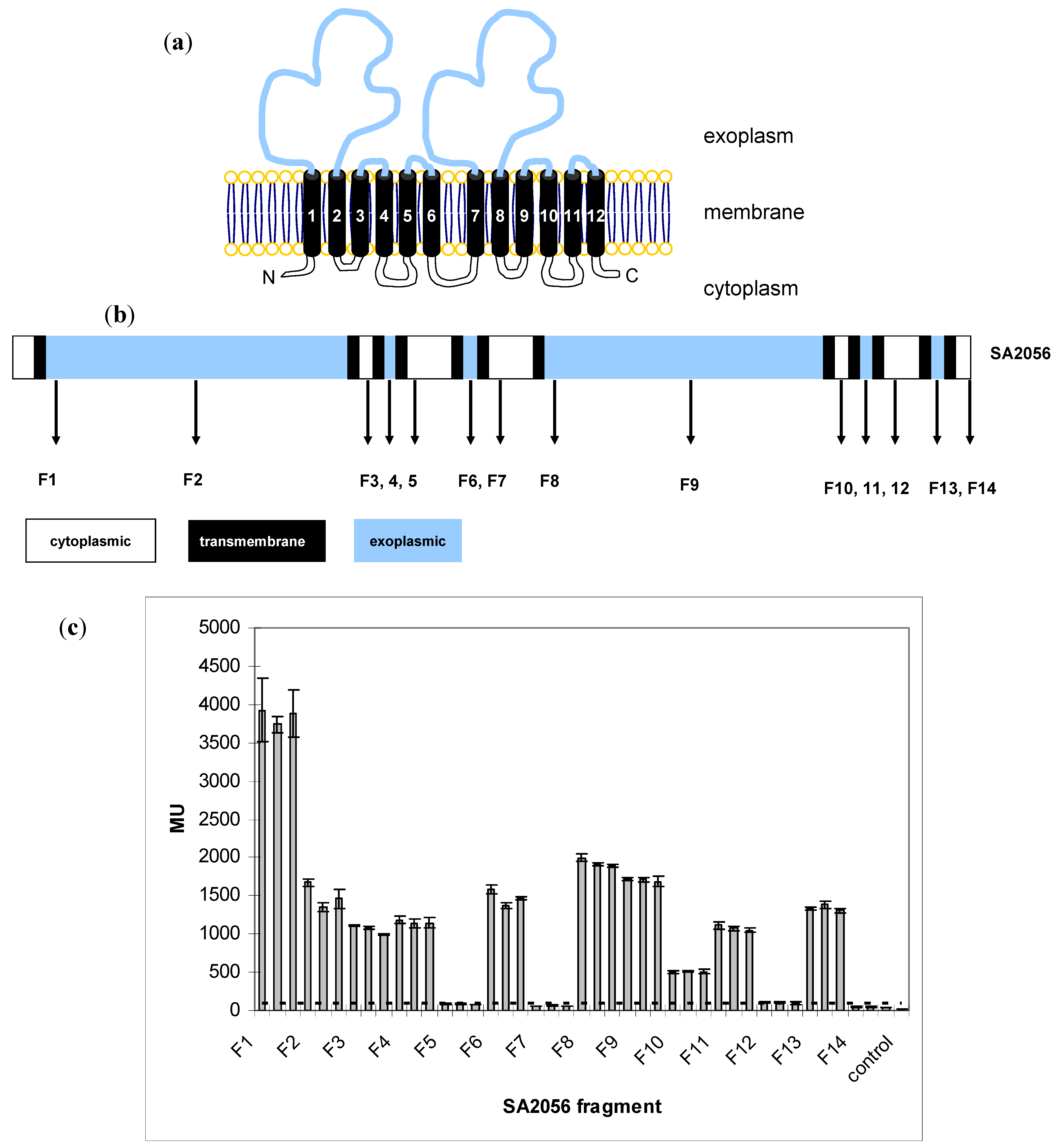
2.3. Topology of SA2056
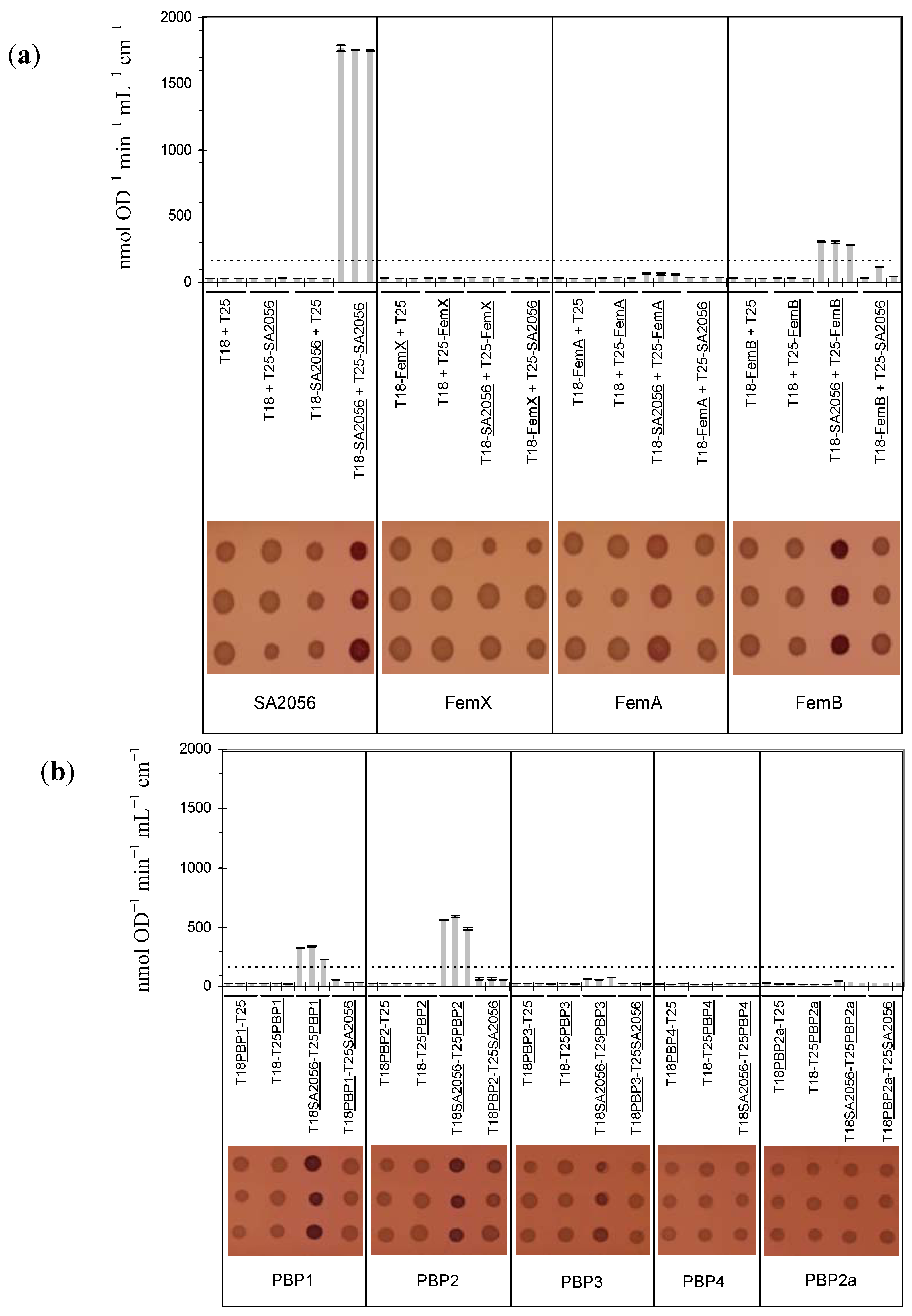
2.4. Interactions of SA2056 Identified in a Bacterial Two-Hybrid System
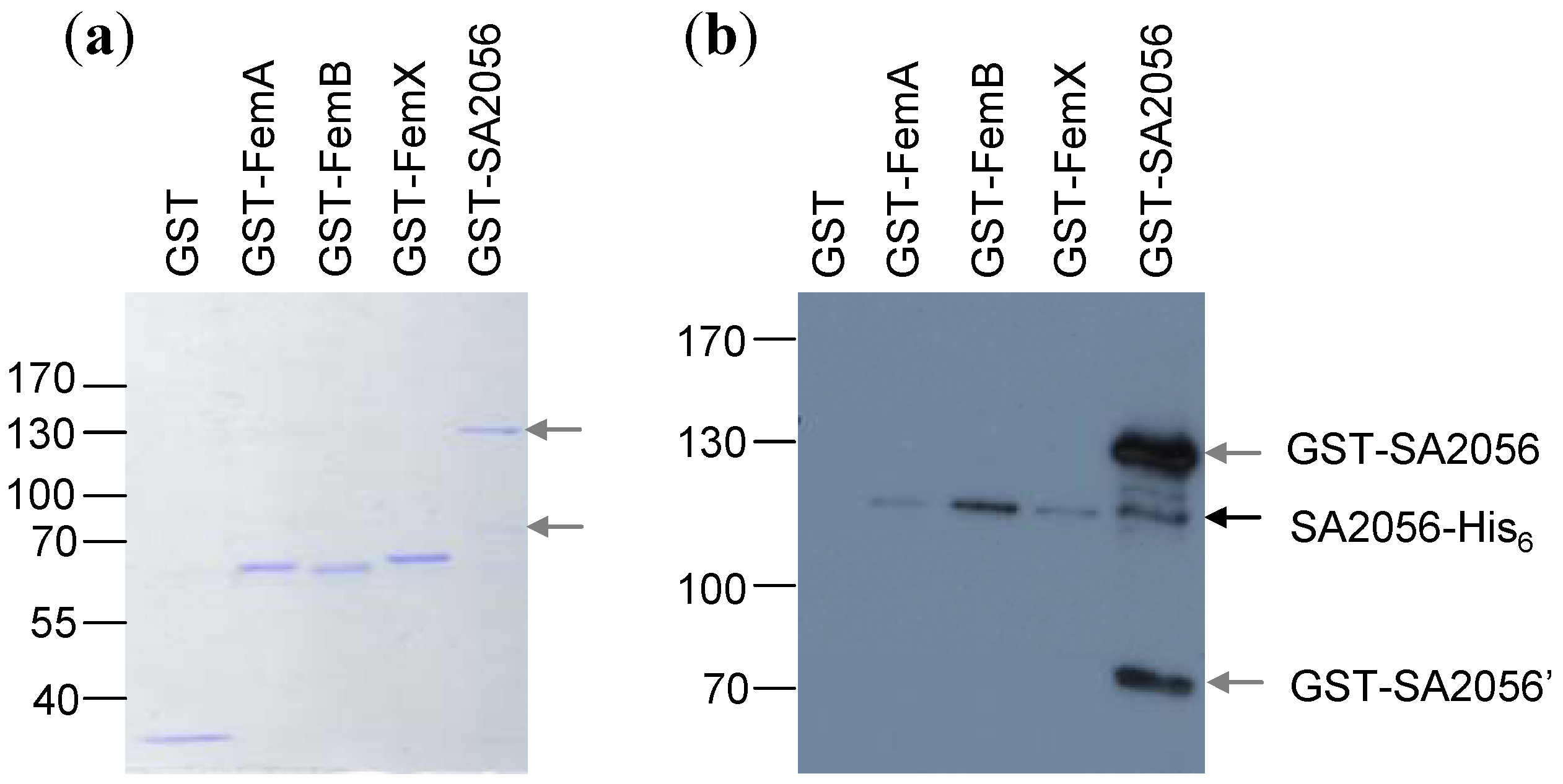
2.5. Resistance Phenotype of a femB sa2056 Double Mutant
3. Experimental
3.1. Bacterial Strains and Growth Conditions
3.2. Construction of MRSA Strains
3.3. Northern Blot Analyses
3.4. Expression of Recombinant Proteins
3.5. S. aureus Cell Fractionation
3.6. Western Blot Analyses
3.7. Construction of phoA Fusions and PhoA Activity Assay

3.8. Topology Prediction Programs
3.9. Bacterial Two-Hybrid System
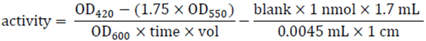
3.10. Pull-Down Experiments
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Conflict of Interest
References and Notes
- Maidhof, H.; Reinicke, B.; Blümel, P.; Berger-Bächi, B.; Labischinski, H. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Bacteriol. 1991, 173, 3507–3513. [Google Scholar]
- Rohrer, S.; Ehlert, K.; Tschierske, M.; Labischinski, H.; Berger-Bächi, B. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. USA. 1999, 96, 9351–9356. [Google Scholar]
- Stranden, A.M.; Ehlert, K.; Labischinski, H.; Berger-Bächi, B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 1997, 179, 9–16. [Google Scholar]
- Schneider, T.; Senn, M.M.; Berger-Bächi, B.; Tossi, A.; Sahl, H.-G.; Wiedemann, I. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol. Microbiol. 2004, 53, 675–685. [Google Scholar] [CrossRef]
- Ton-That, H.; Labischinski, H.; Berger-Bächi, B.; Schneewind, O. Anchor structure of staphylococcal surface proteins. III. Role of the FemA, FemB and FemX factors in anchoring surface proteins to the bacterial cell wall. J. Biol. Chem. 1998, 273, 29143–29149. [Google Scholar]
- Ling, B.; Berger-Bächi, B. Increased overall antibiotic susceptibility in Staphylococcus aureus femAB null mutants. Antimicrob. Agents Chemother. 1998, 42, 936–938. [Google Scholar]
- Hübscher, J.; Jansen, A.; Kotte, O.; Schafer, J.; Majcherczyk, P.; Harris, L.; Bierbaum, G.; Heinemann, M.; Berger-Bächi, B. Living with an imperfect cell wall: Compensation of femAB inactivation in Staphylococcus aureus. BMC Genomics 2007, 8. [Google Scholar] [CrossRef] [Green Version]
- Berger-Bächi, B. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J. Bacteriol. 1983, 154, 479–487. [Google Scholar]
- Kornblum, J.; Hartman, B.J.; Novick, R.P.; Tomasz, A. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur. J. Clin. Microbiol. 1986, 5, 714–718. [Google Scholar] [CrossRef]
- Murakami, K.; Tomasz, A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J. Bacteriol. 1989, 171, 874–879. [Google Scholar]
- Tseng, T.T.; Gratwick, K.S.; Kollman, J.; Park, D.; Nies, D.H.; Goffeau, A.; Saier, M.H.J. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1999, 1, 107–25. [Google Scholar]
- Transporter Classification Database, TCDB. Available online: http://www.tcdb.org (accessed on 24 December 2012).
- Kyoto Encyclopedia of Genes and Genomes Database, KEGG. Available online: http://www.genome.jp/kegg/kegg2.html (accessed on 24 December 2012).
- Rohrer, S. Studies on members of the FemABX protein family in Staphylococcus aureus. Ph.D. Thesis, Swiss Federal Institute of Technology, Zürich, Switzerland, 2002; p. 152. [Google Scholar]
- TransTermHP. Available online: http://transterm.cbcb.umd.edu/ (accessed on 24 December 2012).
- Softberry. Available online: http://linux1.softberry.com/berry.phtml (accessed on 24 December 2012).
- Muthaiyan, A.; Silverman, J.A.; Jayaswal, R.K.; Wilkinson, B.J. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 2008, 52, 980–990. [Google Scholar] [CrossRef]
- Chang, W.; Small, D.A.; Toghrol, F.; Bentley, W.E. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 2006, 188, 1648–1659. [Google Scholar] [CrossRef]
- Chang, M.W.; Toghrol, F.; Bentley, W.E. Toxicogenomic response to chlorination includes induction of major virulence genes in Staphylococcus aureus. Environ. Sci. Technol. 2007, 41, 7570–7575. [Google Scholar] [CrossRef]
- Anderson, K.L.; Roberts, C.; Disz, T.; Vonstein, V.; Hwang, K.; Overbeek, R.; Olson, P.D.; Projan, S.J.; Dunman, P.M. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 2006, 188, 6739–6756. [Google Scholar]
- Herbert, S.; Bera, A.; Nerz, C.; Kraus, D.; Peschel, A.; Goerke, C.; Meehl, M.; Cheung, A.; Götz, F. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 2007, 3, e102. [Google Scholar] [CrossRef]
- Michel, A.; Agerer, F.; Hauck, C.R.; Herrmann, M.; Ullrich, J.; Hacker, J.; Ohlsen, K. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 2006, 188, 5783–5796. [Google Scholar] [CrossRef]
- Banerjee, R.; Gretes, M.; Harlem, C.; Basuino, L.; Chambers, H.F. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level β-lactam resistance contains mutations in three genes. Antimicrob. Agents Chemother. 2010, 54, 4900–4902. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Abbott, J.C.; Burhenne, H.; Kaever, V.; Gründling, A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011, 7, e1002217. [Google Scholar] [CrossRef]
- Henze, U.U.; Roos, M.; Berger-Bächi, B. Effects of penicillin-binding protein 4 overproduction in Staphylococcus aureus. Microb. Drug Resist. 1996, 2, 193–199. [Google Scholar] [CrossRef]
- Sieradzki, K.; Pinho, M.G.; Tomasz, A. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 1999, 274, 18942–18946. [Google Scholar]
- Leski, T.A.; Tomasz, A. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: Evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J. Bacteriol. 2005, 187, 1815–1824. [Google Scholar] [CrossRef]
- Memmi, G.; Filipe, S.R.; Pinho, M.G.; Fu, Z.; Cheung, A. Staphylococcus aureus PBP4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 2008, 52, 3955–3966. [Google Scholar] [CrossRef]
- Griffiths, J.M.; O'Neill, A.J. Loss of function of the GdpP protein leads to joint β-lactam/ glycopeptide tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 56, 579–581. [Google Scholar] [CrossRef]
- Pozzi, C.; Waters, E.M.; Rudkin, J.K.; Schaeffer, C.R.; Lohan, A.J.; Tong, P.; Loftus, B.J.; Pier, G.B.; Fey, P.D.; Massey, R.C.; et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012, 8, e1002626. [Google Scholar] [CrossRef]
- Quiblier, C.; Zinkernagel, A.; Schuepbach, R.; Berger-Bächi, B.; Senn, M. Contribution of SecDF to Staphylococcus aureus resistance and expression of virulence factors. BMC Microbiol. 2011, 11. [Google Scholar] [CrossRef] [Green Version]
- Ender, M.; McCallum, N.; Berger-Bächi, B. Impact of mecA promoter mutations on mecA expression and β-lactam resistance levels. Int. J. Med. Microbiol. 2008, 298, 607–617. [Google Scholar] [CrossRef]
- Garen, A.; Levinthal, C. A fine-structure genetic and chemical study of the enzyme alkaline phosphatase of E. coli I. Purification and characterization of alkaline phosphatase. Biochim. Biophys. Acta 1960, 38, 470–483. [Google Scholar] [CrossRef]
- Derman, A.I.; Beckwith, J. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J. Bacteriol. 1991, 173, 7719–7722. [Google Scholar]
- Akiyama, Y.; Ito, K. Folding and assembly of bacterial alkaline phosphatase in vitro and in vivo. J. Biol. Chem. 1993, 268, 8146–8150. [Google Scholar]
- Drew, D.; Sjöstrand, D.; Nilsson, J.; Urbig, T.; Chin, C.N.; de Gier, J.W.; von Heijne, G. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. USA 2002, 99, 2690–2695. [Google Scholar]
- Karimova, G.; Pidoux, J.; Ullmann, A.; Ladant, D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5752–5756. [Google Scholar]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 2002, 419, 587–593. [Google Scholar] [CrossRef]
- Henze, U.; Sidow, T.; Wecke, J.; Labischinski, H.; Berger-Bächi, B. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J. Bacteriol. 1993, 175, 1612–1620. [Google Scholar]
- Berger-Bächi, B.; Tschierske, M. Role of fem factors in methicillin resistance. Drug Resist. Updat. 1998, 1, 325–335. [Google Scholar] [CrossRef]
- Over, B.; Heusser, R.; McCallum, N.; Schulthess, B.; Kupferschmied, P.; Gaiani, J.M.; Sifri, C.D.; Berger-Bächi, B.; Stutzmann Meier, P. LytR-CpsA-Psr proteins in Staphylococcus aureus display partial functional redundancy and the deletion of all three severely impairs septum placement and cell separation. FEMS Microbiol. Lett. 2011, 320, 142–151. [Google Scholar] [CrossRef]
- Cheung, A.; Eberhardt, K.; Fischetti, V. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 1994, 222, 511–514. [Google Scholar] [CrossRef]
- Rohrer, S.; Berger-Bächi, B. Application of a bacterial two-hybrid system for the analysis of protein-protein interactions between FemABX family proteins. Microbiology 2003, 149, 2733–2738. [Google Scholar] [CrossRef] [Green Version]
- Miroux, B.; Walker, J.E. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996, 260, 289–298. [Google Scholar] [CrossRef]
- Schneewind, O.; Mihaylova-Petkov, D.; Model, P. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 1993, 12, 4803–4811. [Google Scholar]
- TMHMM. Available online: http://cbs.dtu.dk/services/TMHMM-2.0/ (accessed on 24 December 2012).
- DAS. Available online: http://www.sbc.su.se/~miklos/DAS/ (accessed on 24 December 2012).
- HMMTOP. Available online: http://www.enzim.hu/hmmtop/ (accessed on 24 December 2012).
- MEMSAT. Available online: http://bioinf.cs.ucl.ac.uk/software_downloads/memsat/ (accessed on 24 December 2012).
- SOSUI. Available online: http://bp.nuap.nagoya-u.ac.jp/sosui/ (accessed on 24 December 2012).
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Quiblier, C.; Luczak-Kadlubowska, A.; Holdener, E.; Alborn, D.; Schneider, T.; Wiedemann, I.; Pinho, M.G.; Sahl, H.-G.; Rohrer, S.; Berger-Bächi, B.; et al. The Staphylococcus aureus Membrane Protein SA2056 Interacts with Peptidoglycan Synthesis Enzymes. Antibiotics 2013, 2, 11-27. https://doi.org/10.3390/antibiotics2010011
Quiblier C, Luczak-Kadlubowska A, Holdener E, Alborn D, Schneider T, Wiedemann I, Pinho MG, Sahl H-G, Rohrer S, Berger-Bächi B, et al. The Staphylococcus aureus Membrane Protein SA2056 Interacts with Peptidoglycan Synthesis Enzymes. Antibiotics. 2013; 2(1):11-27. https://doi.org/10.3390/antibiotics2010011
Chicago/Turabian StyleQuiblier, Chantal, Agnieszka Luczak-Kadlubowska, Esther Holdener, Daniela Alborn, Tanja Schneider, Imke Wiedemann, Mariana G. Pinho, Hans-Georg Sahl, Susanne Rohrer, Brigitte Berger-Bächi, and et al. 2013. " The Staphylococcus aureus Membrane Protein SA2056 Interacts with Peptidoglycan Synthesis Enzymes" Antibiotics 2, no. 1: 11-27. https://doi.org/10.3390/antibiotics2010011



