Reduction in Toxicity of Nano-Ag-Polyvinyl-pyrrolidone Using Hydra Proteins and Peptides during Zebrafish Embryogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of N-Ag-PVP, HP, and Hym176
2.2. Physicochemical Properties of N-Ag-PVP and N-Ag-PVP+HP
2.3. Experimental Animals
2.4. Exposure Conditions
2.5. Microarray Analysis
2.6. qRT-PCR Analysis
2.7. Statistical Analysis
3. Results and Discussion
3.1. Properties of N-Ag-PVP and HP
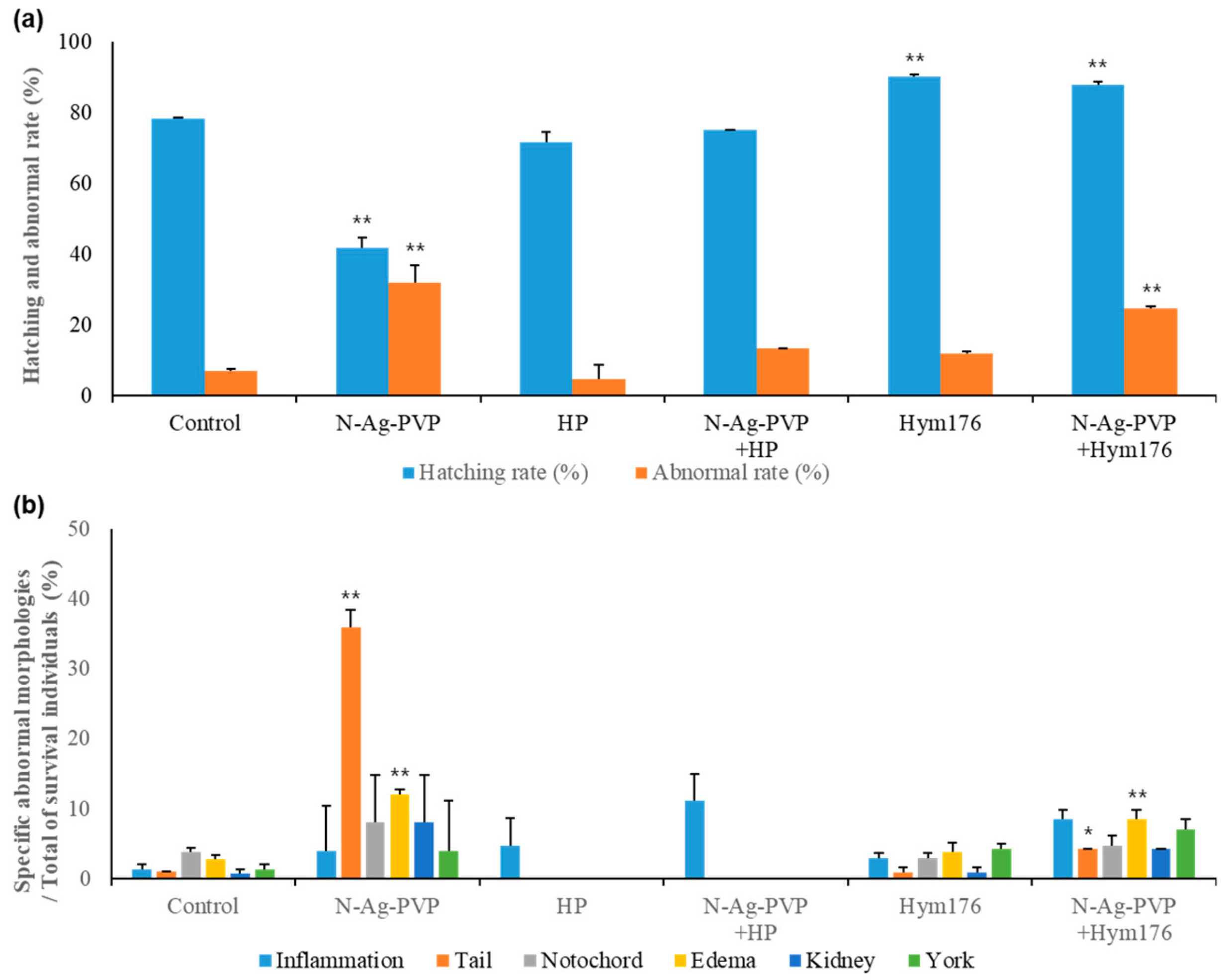
3.2. Effects of HP and Hym176 on N-Ag-PVP-Exposed Zebrafish Embryos
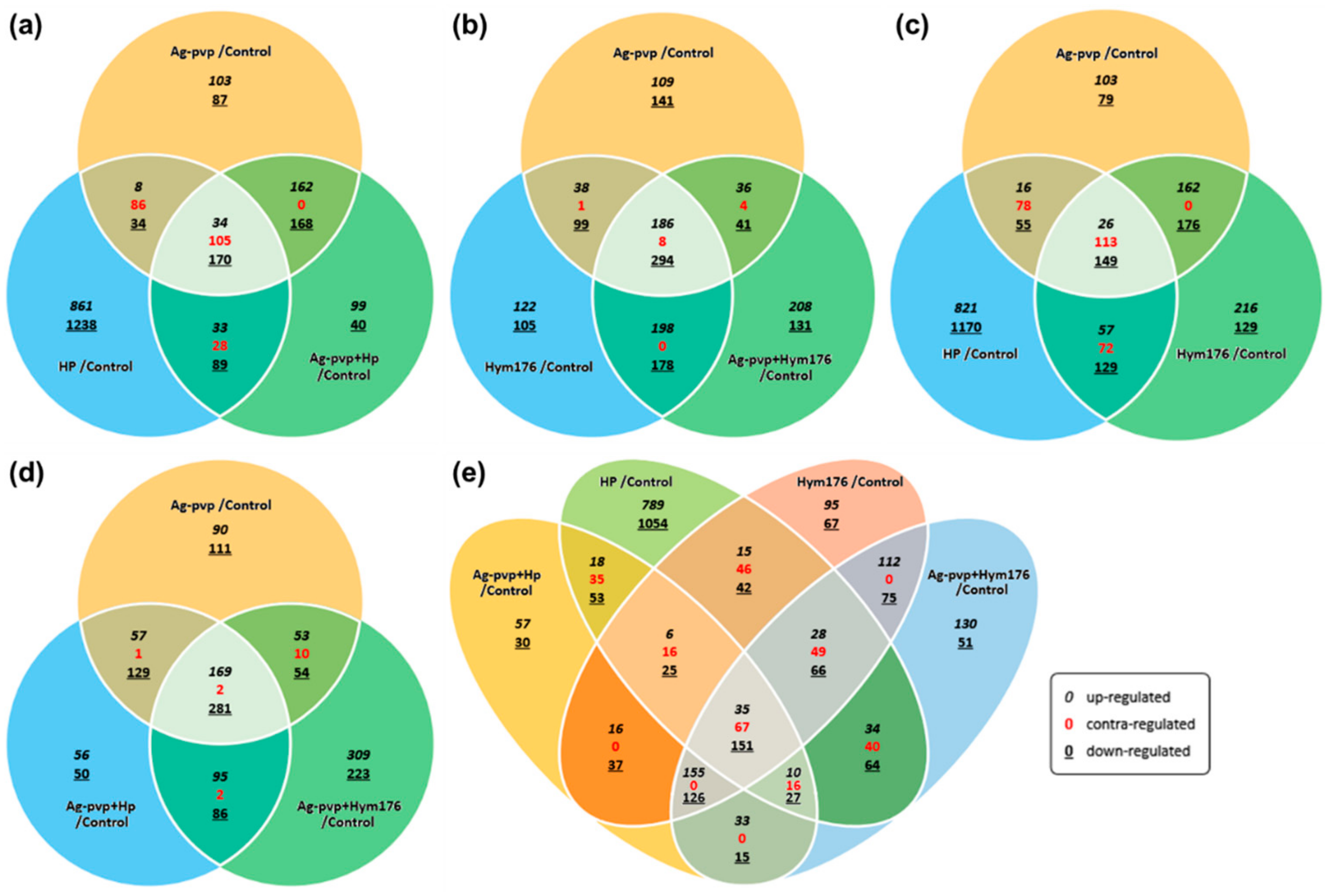
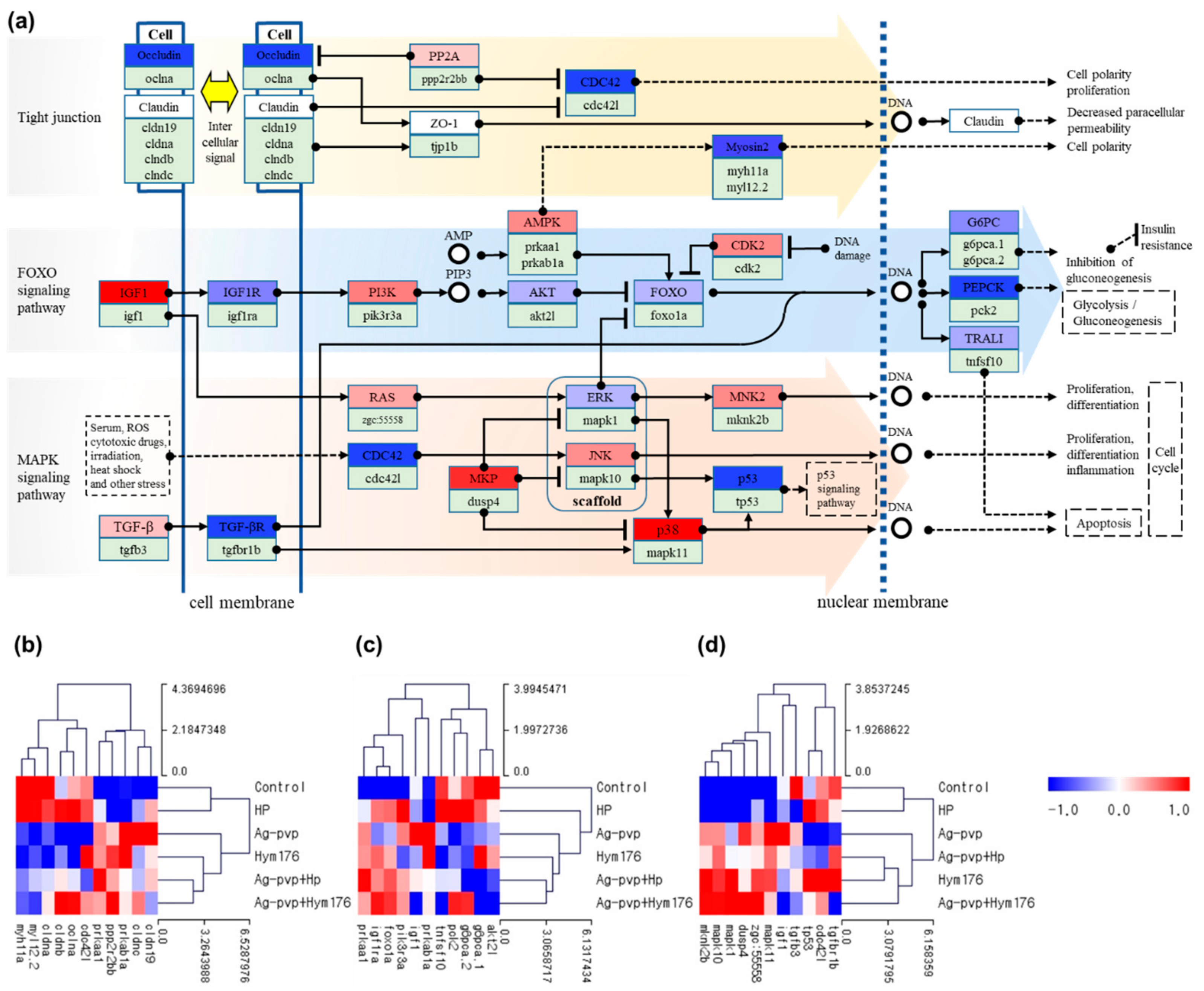
3.3. Microarray and qRT-PCR Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Auffan, M.; Rose, J.; Bottero, J.Y.; Lowry, G.V.; Jolivet, J.P.; Wiesner, M.R. Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat. Nanotechnol. 2009, 4, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Durán, N.; Durán, M.; de Jesus, M.B.; Seabra, A.B.; Fávaro, W.J.; Nakazato, G. Silver nanoparticles: A new view on mechanistic aspects on antimicrobial activity. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 789–799. [Google Scholar]
- Blaser, S.A.; Scheringer, M.; MacLeod, M.; Hungerbühler, K. Estimation of cumulative aquatic exposure and risk due to silver: Contribution of nano-functionalized plastics and textiles. Sci. Total Environ. 2008, 390, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Benn, T.M.; Westerhoff, P. Nanoparticle silver released into water from commercially available sock fabrics. Environ. Sci. Technol. 2008, 42, 4133–4139. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, Q.; Scotter, M.; Blackburn, J.; Ross, B.; Boxall, A.; Castle, L.; Aitken, R.; Watkins, R. Applications and implications of nanotechnologies for the food sector. Food Addit. Contam. 2008, 25, 241–258. [Google Scholar] [CrossRef]
- Chen, X.; Schluesener, H.J. Nanosilver: A nanoproduct in medical application. Toxicol. Lett. 2008, 176, 1–12. [Google Scholar] [CrossRef]
- Yeo, M.K.; Kang, M.S. Effects of nanometer sized silver materials on biological toxicity during zebrafish embryogenesis. Bull. Korean Chem. Soc. 2008, 29, 1179–1184. [Google Scholar]
- Park, H.G.; Yeo, M.K. Toxic effects against bacteria of silver nanocolloids and silver nanotubes in the presence of hydra cells. Mol. Cell. Toxicol. 2017, 13, 37–47. [Google Scholar]
- Wijnhoven, S.W.P.; Peijnenburg, W.J.G.M.; Herberts, C.A.; Hagens, W.I.; Oomen, A.G.; Heugens, E.H.W.; Roszek, B.; Bisschops, J.; Gosens, I.; van de Meent, D.; et al. Nano-silver–a review of available data and knowledge gaps in human and environmental risk assessment. Nanotoxicology 2009, 3, 109–138. [Google Scholar] [CrossRef]
- Gottschalk, F.; Sun, T.; Nowack, B. Environmental concentrations of engineered nanomaterials: Review of modeling and analytical studies. Environ. Pollut. 2013, 181, 287–300. [Google Scholar] [CrossRef]
- Klaine, S.J.; Koelmans, A.A.; Horne, N.; Carley, S.; Handy, R.D.; Kapustka, L.; Nowack, B.; von der Kammer, F. Paradigms to assess the environmental impact of manufactured nanomaterials. Environ. Toxicol. Chem. 2012, 31, 3–14. [Google Scholar] [CrossRef]
- Westerhoff, P.K.; Kiser, M.A.; Hristovski, K. Nanomaterial removal and transformation during biological wastewater treatment. Environ. Eng. Sci. 2013, 30, 109–117. [Google Scholar]
- Meyer, D.E.; Curran, M.A.; Gonzalez, M.A. An examination of existing data for the industrial manufacture and use of nanocomponents and their role in the life cycle impact of nanoproducts. Environ. Sci Technol. 2009, 43, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Lombi, E.; Donner, E.; Taheri, S.; Tavakkoli, E.; Jämting, Å.K.; McClure, S.; Naidu, R.; Miller, B.W.; Scheckel, K.G.; Vasilev, K. Transformation of four silver/silver chloride nanoparticles during anaerobic treatment of wastewater and post-processing of sewage sludge. Environ. Pollut. 2013, 176, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.S.; Wei, D.B.; Wei, J.; Hu, H.Y. Screening and estimating of toxicity formation with photobacterium bioassay during chlorine disinfection of wastewater. J. Hazard. Mater. 2007, 141, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Park, H.G.; Yeo, M.K. Comparison of gene expression patterns from zebrafish embryos between pure silver nanomaterial and mixed silver nanomaterial containing cells of Hydra magnipapillata. Mol. Cell. Toxicol. 2015, 11, 307–314. [Google Scholar] [CrossRef]
- Yeo, M.K.; Yoon, J.W. Comparison of the effects of nano-silver antibacterial coatings and silver ions on zebrafish embryogenesis. Mol. Cell. Toxicol. 2009, 5, 23–31. [Google Scholar]
- Bosch, T.C. Understanding complex host-microbe interactions in Hydra. Gut Microbes 2012, 3, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Sarras, M.P., Jr.; Deutzmann, R. Hydra and Niccolo Paganini (1782–1840)—two peas in a pod? The molecular basis of extracellular matrix structure in the invertebrate, Hydra. BioEssays 2001, 23, 716–724. [Google Scholar] [CrossRef]
- Fujisawa, T. Hydra regeneration and epitheliopeptides. Dev. Dyn. 2003, 226, 182–189. [Google Scholar] [CrossRef]
- Mergaert, P. Role of Antimicrobial Peptides in Controlling Symbiotic Bacterial Populations. Nat. Prod. Rep. 2018, 35, 336–356. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T. Hydra peptide project 1993–2007. Dev. Growth Differ. 2008, 50, S257–S268. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Paulson, H.L.; Perez, M.K.; Trottier, Y.; Trojanowski, J.Q.; Subramony, S.H.; Das, S.S.; Vig, P.; Mandel, J.L.; Fischbeck, K.H.; Pittman, R.N. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 1997, 19, 333–344. [Google Scholar] [CrossRef]
- Yum, S.; Woo, S.; Lee, A.; Won, H.; Kim, J. Hydra, a candidate for an alternative model in environmental genomics. Mol. Cell. Toxicol. 2014, 10, 339–346. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar]
- Li, X.; Song, G.; Zhao, Y.; Zhao, F.; Liu, C.; Liu, D.; Li, Q.; Cui, Z. Claudin7b is required for the formation and function of inner ear in zebrafish. J. Cell. Physiol. 2018, 233, 3195–3206. [Google Scholar]
- Wakayama, Y.; Fukuhara, S.; Ando, K.; Matsuda, M.; Mochizuki, N. Cdc42 mediates Bmp-induced sprouting angiogenesis through Fmnl3-driven assembly of endothelial filopodia in zebrafish. Dev. Cell 2015, 32, 109–122. [Google Scholar] [CrossRef]
- Eivers, E.; Mccarthy, K.; Glynn, C.; Nolan, C.M.; Byrnes, L. Insulin-like growth factor (IGF) signalling is required for early dorso-anterior development of the zebrafish embryo. Int. J. Dev. Biol. 2004, 48, 1131–1140. [Google Scholar] [CrossRef]
- Park, H.; Kim, J.; Chang, K.; Lee, C.; Eom, I.; Kim, P.; Nam, D.; Yeo, M. Trophic transfer of citrate, PVP coated silver nanomaterials, and silver ions in a paddy microcosm. Environ. Pollut. 2018, 235, 435–445. [Google Scholar] [CrossRef]
- Meloche, S.; Pouysségur, J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1-to S-phase transition. Oncogene 2007, 26, 3227. [Google Scholar] [CrossRef]
- Gharibshahi, L.; Saion, E.; Gharibshahi, E.; Shaari, A.H.; Matori, K.A. Influence of Poly (vinylpyrrolidone) concentration on properties of silver nanoparticles manufactured by modified thermal treatment method. PLoS ONE 2017, 12, e0186094. [Google Scholar] [CrossRef] [PubMed]
- Naja, G.; Bouvrette, P.; Hrapovic, S.; Luong, J.H. Raman-based detection of bacteria using silver nanoparticles conjugated with antibodies. Analyst 2007, 132, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Feng, J.; Ma, X.; Wu, C.; Zhao, X. One-dimensional silver nanowires synthesized by self-seeding polyol process. J. Nanopart. Res. 2012, 14, 887. [Google Scholar] [CrossRef]
- Lowry, G.V.; Gregory, K.B.; Apte, S.C.; Lead, J.R. Transformations of nanomaterials in the environment. Environ. Sci. Technol. 2012, 46, 6893–6899. [Google Scholar] [CrossRef]
- Levard, C.; Hotze, E.M.; Lowry, G.V.; Brown, G.E., Jr. Environmental transformations of silver nanoparticles: Impact on stability and toxicity. Environ. Sci. Technol. 2012, 46, 6900–6914. [Google Scholar] [PubMed]
- McShan, D.; Ray, P.C.; Yu, H. Molecular toxicity mechanism of nanosilver. J. Food Drug Anal. 2014, 22, 116–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramirez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346. [Google Scholar] [CrossRef]
- Liu, J.; Hurt, R.H. Ion release kinetics and particle persistence in aqueous nano-silver colloids. Environ. Sci. Technol. 2010, 44, 2169–2175. [Google Scholar] [CrossRef]
- Berghmans, S.; Butler, P.; Goldsmith, P.; Waldron, G.; Gardner, I.; Golder, Z.; Richards, F.M.; Kimber, G.; Roach, A.; Alderton, W.; et al. Zebrafish based assays for the assessment of cardiac, visual and gut function-potential safety screens for early drug discovery. J. Pharmacol. Toxicol. Methods 2008, 58, 59–68. [Google Scholar] [CrossRef]
- Lee, K.J.; Nallathamby, P.D.; Browning, L.M.; Osgood, C.J.; Nancy, X.H. In vivo imaging of transport and biocompatibility of single silver nanoparticles in early development of zebrafish embryos. ACS Nano 2007, 1, 133–143. [Google Scholar] [CrossRef]
- Anselmo, A.C.; Zhang, M.; Kumar, S.; Vogus, D.R.; Menegatti, S.; Helgeson, M.E.; Mitragotri, S. Elasticity of nanoparticles influences their blood circulation, phagocytosis, endocytosis, and targeting. ACS Nano 2015, 9, 3169–3177. [Google Scholar] [CrossRef] [PubMed]
- Metreveli, G.; Frombold, B.; Seitz, F.; Grun, A.; Philippe, A.; Rosenfeldt, R.R.; Bundschuh, M.; Schulz, R.; Manz, W.; Schaumann, G.E. Impact of chemical composition of ecotoxicological test media on the stability and aggregation status of silver nanoparticles. Environ. Sci. Nano 2016, 3, 418–433. [Google Scholar] [CrossRef] [Green Version]
- Ott, E.B.; van den Akker, N.M.; Sakalis, P.A.; Gittenberger-de Groot, A.C.; Te Velthuis, A.J.; Bagowski, C.P. The lim domain only protein 7 is important in zebrafish heart development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2008, 237, 3940–3952. [Google Scholar] [CrossRef] [PubMed]
- Yeh, L.K.; Liu, C.Y.; Kao, W.W.W.Y.; Huang, C.J.; Hu, F.R.; Chien, C.L.; Wang, I.J. Knockdown of zebrafish lumican gene (zlum) causes scleral thinning and increased size of scleral coats. J. Biol. Chem. 2010, 285, 28141–28155. [Google Scholar] [CrossRef] [PubMed]
- Boyadjiev, S.A.; Fromme, J.C.; Ben, J.; Chong, S.S.; Nauta, C.; Hur, D.J.; Zhang, G.; Hamamoto, S.; Schekman, R.; Ravazzola, M.; et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat. Genet. 2006, 38, 1192–1197. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.J.; Unlu, G.; Bisnett, B.J.; Meister, T.R.; Condon, B.M.; Luo, P.M.; Smith, T.J.; Hanna, M.; Chhetri, A.; Soderblom, E.J.; et al. Dynamic glycosylation governs the vertebrate COPII protein trafficking pathway. Biochemistry 2017, 57, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zou, Z.; Collodi, P.; Xu, F.; Xu, X.; Zhao, Q. Identification and characterization of a second fibronectin gene in zebrafish. Matrix Biol. 2005, 24, 69–77. [Google Scholar] [CrossRef]
- Shibata, E.; Ando, K.; Murase, E.; Kawakami, A. Heterogeneous fates and dynamic rearrangement of regenerative epidermis-derived cells during zebrafish fin regeneration. Development 2018, 145, 162016. [Google Scholar] [CrossRef]
- Ghoumid, J.; Drevillon, L.; Alavi-Naini, S.M.; Bondurand, N.; Rio, M.; Briand-Suleau, A.; Nasser, M.; Goodwin, L.; Raymond, P.; Yanicostas, C.; et al. ZEB2 zinc-finger missense mutations lead to hypomorphic alleles and a mild Mowat-Wilson syndrome. Hum. Mol. Genet. 2013, 22, 2652–2661. [Google Scholar] [CrossRef]
- Taibi, A.; Mandavawala, K.P.; Noel, J.; Okoye, E.V.; Milano, C.R.; Martin, B.L.; Sirotkin, H.I. Zebrafish churchill regulates developmental gene expression and cell migration. Dev. Dyn. 2013, 242, 614–621. [Google Scholar] [CrossRef]
- Say, E.; Tay, H.G.; Zhao, Z.S.; Baskaran, Y.; Li, R.; Lim, L.; Manser, E. A functional requirement for PAK1 binding to the KH (2) domain of the fragile X protein-related FXR1. Mol. Cell. 2010, 38, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Van’t Padje, S.; Chaudhry, B.; Severijnen, L.A.; van der Linde, H.C.; Mientjes, E.J.; Oostra, B.A.; Willemsen, R. Reduction in fragile X related 1 protein causes cardiomyopathy and muscular dystrophy in zebrafish. J. Exp. Biol. 2009, 212, 2564–2570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, I.; Dawson, K.A. Protein-nanoparticle interactions. Nano Today 2008, 3, 40–47. [Google Scholar] [CrossRef]
- Fruh, V.; IJzerman, A.P.; Siegal, G. How to catch a membrane protein in action: A review of functional membrane protein immobilization strategies and their applications. Chem. Rev. 2010, 111, 640–656. [Google Scholar] [CrossRef] [PubMed]
- Chatzopoulou, A.; Heijmans, J.P.; Burgerhout, E.; Oskam, N.; Spaink, H.P.; Meijer, A.H.; Schaaf, M.J. Glucocorticoid-induced attenuation of the inflammatory response in zebrafish. Endocrinology 2016, 157, 2772–2784. [Google Scholar] [CrossRef] [PubMed]
- AshaRani, P.V.; Sethu, S.; Lim, H.K.; Balaji, G.; Valiyaveettil, S.; Hande, M.P. Differential regulation of intracellular factors mediating cell cycle, DNA repair and inflammation following exposure to silver nanoparticles in human cells. Genome Integr. 2012, 3, 2. [Google Scholar] [CrossRef]
- Gonzalez-Mariscal, L.; Tapia, R.; Chamorro, D. Crosstalk of tight junction components with signaling pathways. Biochim. Et Biophys. Acta (Bba)-Biomembr. 2008, 1778, 729–756. [Google Scholar] [CrossRef] [Green Version]
- Simon, D.B.; Lu, Y.; Choate, K.A.; Velazquez, H.; Al-Sabban, E.; Praga, M.; Casari, G.; Bettinelli, A.; Colussi, G.; Rodriguez-Soriano, J.; et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 1999, 285, 103–106. [Google Scholar]
- Calnan, D.R.; Brunet, A. The foxo code. Oncogene 2008, 27, 2276. [Google Scholar] [CrossRef]
- Eijkelenboom, A.; Burgering, B.M.T. FOXOs: Signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013, 14, 83. [Google Scholar] [CrossRef]
- Raman, M.; Chen, W.; Cobb, M.H. Differential regulation and properties of MAPKs. Oncogene 2007, 26, 3100. [Google Scholar] [CrossRef] [PubMed]
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Stoy, H.; Gurevich, V.V. How genetic errors in GPCRs affect their function: Possible therapeutic strategies. Genes Dis. 2015, 2, 108–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene Name | Gene Symbol | Function | Accession No. | Primer Sequences (5′ → 3′) |
|---|---|---|---|---|
| claudin a | cldna | structural molecule activity | NM _131762 | (F) TGTGGCAAGTCACTGCTTTT (R) CACGCAACTCATCCAAATTC |
| cell division cycle 42, like | cdc42l | GTP binding, GTPase activity, nucleotide binding, protein kinase binding | NM _199865 | (F) GGCAGGAAGACTACGACAGA (R) GGAGGAAGGAGAAACAACTGA |
| insulin-like growth factor 1 | igf1 | Growth factor activity, hormone activity, insulin-like growth factor receptor binding | NM _131825 | (F) GTGGACGAATGCTGCTTTCA (R) CTGTCTTCACAGGCGCACAA |
| zgc:55558 | zgc:55558 | GTP binding, GTPase activity, nucleotide binding, | NM _200258 | (F) TTTTTACACCCCCATCCTTT (R) GGTCTCGTGTGCACAGACAT |
| mitogen-activated protein kinase 10 | mapk10 | ATP binding, JUN kinase activity, MAP kinase activity, kinase activity, nucleotide binding, protein kinase activity, protein serine/threonine kinase activity, transferase activity | NM _001037701 | (F) TCGAGGAGAGAACAAAGAATGG (R) AGGCTCTCGCTGCTGTTCAC |
| mitogen-activated protein kinase 11 | mapk11 | ATP binding, MAP kinase activity, kinase activity, nucleotide binding, protein kinase activity, protein serine/threonine kinase activity, transferase activity | NM _001002095 | (F) CAGTACTGCCCTCTCCTTCTT (R) ATCGTCTCGTCTGGCTGAAC |
| tumor protein p53 | tp53 | DNA binding, DNA-binding transcription factor activity, metal ion binding, promoter-specific chromatin binding, protein binding, sequence-specific DNA binding, transcription regulatory region DNA binding, | NM _001271820 | (F) TGCGATACATGTGATCCATT (R) CAGTGTCCAGCAACAAAGGT |
| actin, beta 2 | actb2 | ATP binding, nucleotide binding | NM _181601 | (F) GACTCAAACTGCGCAGAGAA (R) AGTCAAGCGCCAAAAATAAC |
| N-Ag-PVP | N-Ag-PVP+HP | |
|---|---|---|
| Zeta potential (mV) | −22.84 ± 0.16 | −5.54 ± 0.80 |
| Diameter (nm) | 285.50 ± 16.40 | 534.40 ± 122.05 |
| Total Genes | Up-Regulated | Down-Regulated | ||
|---|---|---|---|---|
| N-Ag-PVP | vs. Control | 957 (6.61%) | 377 | 580 |
| HP | vs. Control | 2686 (18.56%) | 1066 | 1620 |
| N-Ag-PVP+HP | vs. Control | 928 (6.41%) | 379 | 549 |
| Hym176 | vs. Control | 1229 (8.49%) | 545 | 684 |
| N-Ag-PVP+Hym176 | vs. Control | 1284 (8.87%) | 632 | 652 |
| N-Ag-PVP+HP | vs. HP | 2879 (19.89%) | 1652 | 1227 |
| N-Ag-PVP+Hym176 | vs. Hym176 | 164 (1.13%) | 112 | 52 |
| N-Ag-PVP+Hym176 | vs. N-Ag-PVP+HP | 353 (2.44%) | 202 | 151 |
| N-Ag-PVP+HP | vs. N-Ag-PVP | 110 (0.76%) | 49 | 61 |
| N-Ag-PVP+Hym176 | vs. N-Ag-PVP | 552 (3.81%) | 332 | 220 |
| Gene Description | Gene Symbol | Regulation Profile and Ratio(Fold change) | ||||
|---|---|---|---|---|---|---|
| N-Ag-PVP /Control | HP /Control | N-Ag-PVP+HP /Control | Hym176 /Control | N-Ag-PVP +Hym176 /Control | ||
| Heart morphogenesis (GO:0003007) | ||||||
| LIM domain 7b | lmo7b | 2.708 | 0.743 | 1.415 | 2.264 | 1.425 |
| fibronectin 1b | fn1b | 2.179 | 1.643 | 1.723 | 3.874 | 5.859 |
| myosin binding protein C, cardiac | mybpc3 | 0.357 | 1.706 | 0.569 | 0.493 | 0.959 |
| Fin morphogenesis (GO:0033334) | ||||||
| Sec23 homolog A, COPII coat complex component | sec23a | 0.448 | 1.369 | 0.709 | 0.588 | 0.832 |
| procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1a | plod1a | 0.441 | 0.869 | 0.611 | 0.462 | 0.665 |
| Embryonic organ morphogenesis (GO:0048562) | ||||||
| LIM domain 7b | lmo7b | 2.708 | 0.743 | 1.415 | 2.264 | 1.425 |
| lumican | lum | 0.491 | 1.359 | 0.620 | 0.701 | 0.625 |
| Sec23 homolog A, COPII coat complex component | sec23a | 0.448 | 1.369 | 0.709 | 0.588 | 0.832 |
| Eye morphogenesis (GO:0048592) | ||||||
| intraflagellar transport 80 homolog (Chlamydomonas) | ift80 | 2.179 | 0.722 | 1.356 | 1.416 | 1.203 |
| tubulin, gamma complex associated protein 4 | tubgcp4 | 2.017 | 0.698 | 1.563 | 1.177 | 0.896 |
| lumican | lum | 0.491 | 1.359 | 0.620 | 0.701 | 0.625 |
| adiponectin receptor 1b | adipor1b | 0.444 | 0.924 | 0.620 | 0.271 | 0.371 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.S.; Lee, J.A.; Yeo, M.-K. Reduction in Toxicity of Nano-Ag-Polyvinyl-pyrrolidone Using Hydra Proteins and Peptides during Zebrafish Embryogenesis. Nanomaterials 2019, 9, 1210. https://doi.org/10.3390/nano9091210
Kim SS, Lee JA, Yeo M-K. Reduction in Toxicity of Nano-Ag-Polyvinyl-pyrrolidone Using Hydra Proteins and Peptides during Zebrafish Embryogenesis. Nanomaterials. 2019; 9(9):1210. https://doi.org/10.3390/nano9091210
Chicago/Turabian StyleKim, Soon Seok, Jin Ah Lee, and Min-Kyeong Yeo. 2019. "Reduction in Toxicity of Nano-Ag-Polyvinyl-pyrrolidone Using Hydra Proteins and Peptides during Zebrafish Embryogenesis" Nanomaterials 9, no. 9: 1210. https://doi.org/10.3390/nano9091210




