Effect of Different Feed Structures and Bedding on the Horizontal Spread of Campylobacter jejuni within Broiler Flocks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Challenge Strains
2.3. Sample Preparation
2.4. DNA Isolation
2.5. C. jejuni Detection
2.6. Typing of Colonizing C. jejuni Strain
2.7. Microbiota Analyses in Cecum
2.7.1. Pyrosequencing
2.7.2. Real-Time Quantification of Specific Bacterial Groups
3. Results
3.1. Horizontal Spread of C. jejuni within Chicken Flocks
| Treatment | Cage a | Day 2 pi | Day 3 pi | Day 4 pi | Day 5 pi | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Feed | Bedding | Positive | Total | Positive | Total | Positive | Total | Positive | Total | |
| Control | Wood | 2 | 0 | 7 | 2 | 7 | 3 | 7 | 4 | 7 |
| 4 | 0 | 7 | 2 | 7 | 4 | 7 | 6 | 7 | ||
| Sum | 0 | 14 | 4 | 14 | 7 | 14 | 10 | 14 | ||
| Oat | Wood | 1 | 0 | 7 | 2 | 7 | 5 | 7 | 7 | 7 |
| 3 | 0 | 7 | 1 | 7 | 2 | 7 | 4 | 7 | ||
| 4 | 0 | 7 | 0 | 7 | 1 | 7 | 3 | 7 | ||
| Sum | 0 | 21 a | 3 | 21 | 8 | 21 | 14 | 21 | ||
| Wheat | Wood | 1 | 0 | 7 | 0 | 7 | 0 | 7 | 2 | 7 |
| 2 | 0 | 7 | 0 | 7 | 2 | 7 | 5 | 6 | ||
| 3 | 0 | 7 | 0 | 7 | 4 | 7 | 6 | 7 | ||
| Sum | 0 | 21 a | 0 | 21 | 6 | 21 | 13 | 20 b | ||
| Control | Rubber | 1 | 0 | 7 | 1 | 7 | 6 | 7 | 7 | 7 |
| 2 | 2 | 7 | 3 | 7 | 6 | 7 | 6 | 7 | ||
| 3 | 0 | 7 | 1 | 7 | 2 | 6 | 2 | 6 | ||
| 4 | 0 | 7 | 2 | 7 | 7 | 7 | 7 | 7 | ||
| Sum | 2 | 28 | 7 | 28 | 21 | 27 b | 22 | 27 | ||
| Oat | Rubber | 1 | 0 | 7 | 0 | 7 | 0 | 7 | 2 | 7 |
| 3 | 0 | 7 | 0 | 7 | 0 | 7 | 0 | 7 | ||
| 4 | 0 | 7 | 0 | 7 | 0 | 7 | 1 | 7 | ||
| Sum | 0 | 21 a | 0 | 21 | 0 c | 21 | 3 | 21 | ||
| Wheat | Rubber | 1 | 0 | 7 | 0 | 7 | 3 | 7 | 4 | 7 |
| 2 | 0 | 7 | 0 | 7 | 0 | 7 | 0 | 7 | ||
| 3 | 0 | 7 | 0 | 7 | 4 | 7 | 4 | 7 | ||
| Sum | 0 | 21 a | 0 | 21 | 7 c | 21 | 8 | 21 | ||
| Total | 2 | 126 | 14 | 126 | 49 | 125 | 70 | 124 | ||
| Days Post-Inoculation | P-values | ||
|---|---|---|---|
| Feed × Bedding a | Feed b | Bedding c | |
| Day 2 pi | 0.212 | 0.131 | 0.202 |
| Day 3 pi | 0.007 | 0.001 | 0.896 |
| Day 4 pi | <0.001 | <0.001 | 0.578 |
| Day 5 pi | <0.001 | 0.002 | 0.030 |
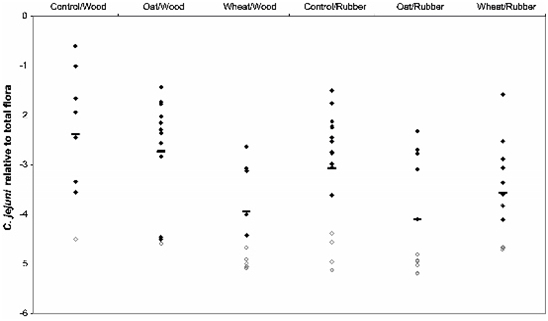
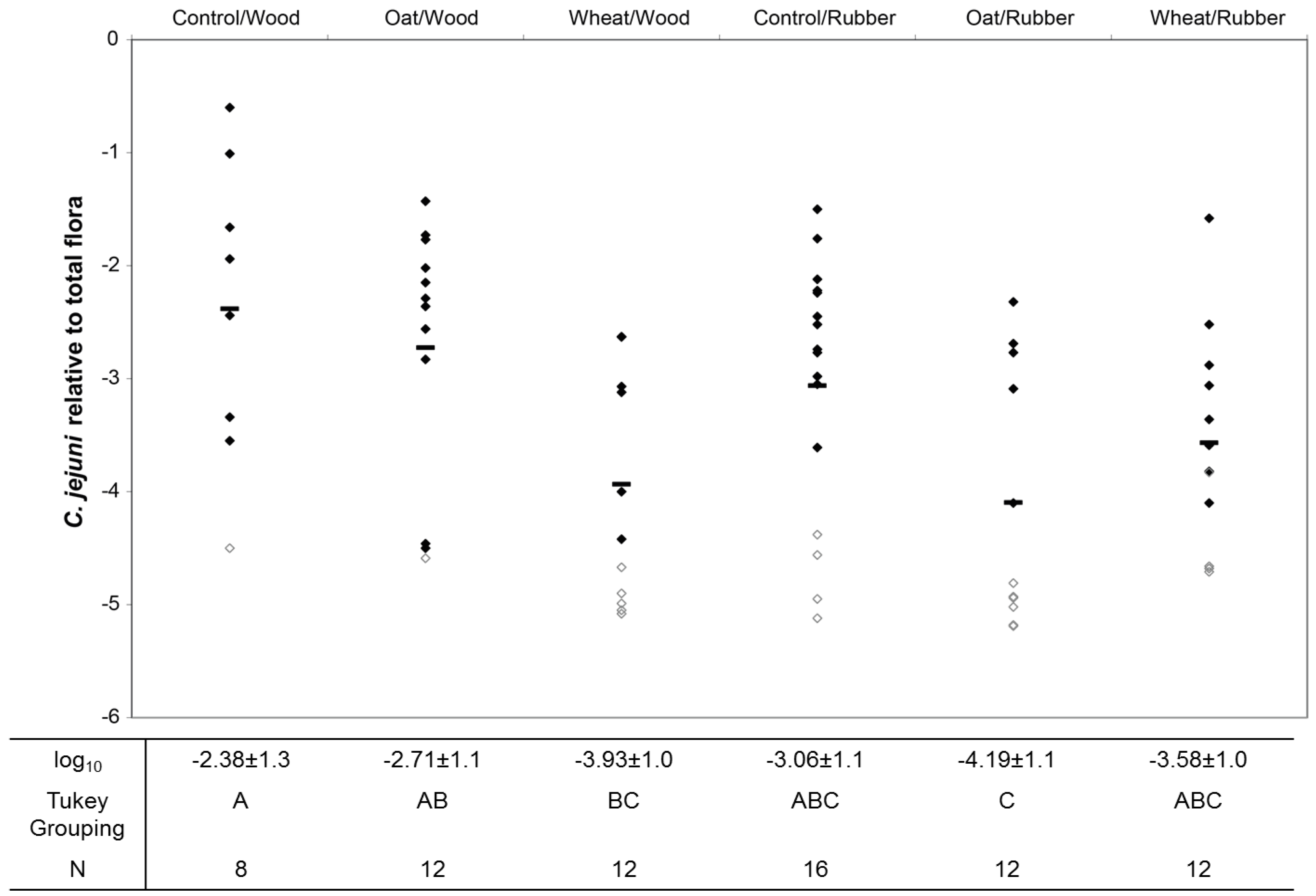
3.2. Colonization Levels of C. jejuni in Cecum
3.3. Effect of Treatment on the Microbiota in Cecum

3.4. Slaughter Weight and pH in Gizzard
4. Discussion
| Strains | Control/Wood (n =16) | Oat/Wood (n =16) | Wheat/Wood (n =16) | Control/Rubber (n =16) | Oat/Rubber (n =15) | Wheat/Rubber (n =15) | P-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Feed | Bedding | F × B | |||||||
| Lactobacillusspp. | −2.73 ± 0.6 | −2.21 ± 0.6 | −2.10 ± 0.5 | −2.40 ± 0.3 | −2.25 ± 0.4 | −2.18 ± 0.4 | 0.001 | 0.470 | 0.160 |
| Enterococcusspp. | −3.63 ± 0.8 | −4.11 ± 0.8 | −3.98 ± 0.7 | −4.28 ± 0.9 | −4.19 ± 1.0 | −4.11 ± 0.5 | 0.629 | 0.084 | 0.304 |
| E. coli | −4.45 ± 0.9 | −4.98 ± 0.9 | −4.93 ± 0.7 | −4.63 ± 0.8 | −4.48 ± 0.5 | −5.10 ± 0.7 | 0.052 | 0.759 | 0.138 |
| Cl. Perfringens | −4.65 ± 0.6 | −4.44 ± 0.7 | −4.19 ± 0.7 | −4.49 ± 0.8 | −4.40 ± 0.9 | −4.06 ± 0.9 | 0.068 | 0.480 | 0.946 |
| Gizzard Weight/pH | Control/Wood | Oat/Wood | Wheat/Wood | Control/Rubber | Oat/Rubber | Wheat/Rubber | P-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Feed | Bedding | F × B | |||||||
| Weight (g) | 2981 ± 172 | 2853 ± 197 | 3009 ± 202 | 2921 ± 221 | 2745 ± 191 | 2920 ± 234 | 0.002 | 0.043 | 0.893 |
| pH in gizzard | 3.89 ± 0.4 | 2.98 ± 0.7 | 3.63 ± 0.4 | 3.79 ± 0.3 | 2.98 ± 0.6 | 3.37 ± 0.6 | <0.001 | 0.258 | 0.563 |
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Blaser, M.J. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 1997, 176, S103–S105. [Google Scholar]
- Silva, J.; Leite, D.; Fernandes, M.; Mena, C.; Gibbs, P.A.; Teixeira, P. Campylobacter spp. as a foodborne pathogen: A review. Front. Microbiol. 2011, 2, 200. [Google Scholar]
- Gillespie, I.A.; O’Brien, S.J.; Frost, J.A.; Adak, G.K.; Horby, P.; Swan, A.V.; Painter, M.J.; Neal, K.R. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: A tool for generating hypotheses. Emerg. Infect. Dis. 2002, 8, 937–942. [Google Scholar] [CrossRef]
- Wilson, D.J.; Gabriel, E.; Leatherbarrow, A.J.; Cheesbrough, J.; Gee, S.; Bolton, E.; Fox, A.; Fearnhead, P.; Hart, C.A.; Diggle, P.J. Tracing the source of campylobacteriosis. PLoS Genet. 2008, 4, e1000203. [Google Scholar] [CrossRef]
- Hermans, D.; van Deun, K.; Martel, A.; van Immerseel, F.; Messens, W.; Heyndrickx, M.; Haesebrouck, F.; Pasmans, F. Colonization factors of Campylobacter jejuni in the chicken gut. Vet. Res. 2011, 42, 82. [Google Scholar] [CrossRef]
- Janssen, R.; Krogfelt, K.A.; Cawthraw, S.A.; van Pelt, W.; Wagenaar, J.A.; Owen, R.J. Host-pathogen interactions in Campylobacter infections: The host perspective. Clin. Microbiol. Rev. 2008, 21, 505–518. [Google Scholar] [CrossRef]
- Solomon, E.B.; Hoover, D.G. Campylobacter jejuni: A bacterial paradox. J. Food Saf. 1999, 19, 121–136. [Google Scholar] [CrossRef]
- Humphrey, T.; O’Brien, S.; Madsen, M. Campylobacters as zoonotic pathogens: A food production perspective. Int. J. Food Microbiol. 2007, 117, 237–257. [Google Scholar] [CrossRef]
- Nauta, M.; Hill, A.; Rosenquist, H.; Brynestad, S.; Fetsch, A.; van der Logt, P.; Fazil, A.; Christensen, B.; Katsma, E.; Borck, B.; et al. A comparison of risk assessments on Campylobacter in broiler meat. Int. J. Food Microbiol. 2009, 129, 107–123. [Google Scholar] [CrossRef]
- Van Gerwe, T.; Miflin, J.K.; Templeton, J.M.; Bouma, A.; Wagenaar, J.A.; Jacobs-Reitsma, W.F.; Stegeman, A.; Klinkenberg, D. Quantifying transmission of Campylobacter jejuni in commercial broiler flocks. Appl. Environ. Microbiol. 2009, 75, 625–628. [Google Scholar] [CrossRef]
- Beery, J.T.; Hugdahl, M.B.; Doyle, M.P. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 1988, 54, 2365–2370. [Google Scholar]
- Shane, S.M. The significance of Campylobacter jejuni infection in poultry—A review. Avian Pathol. 1992, 21, 189–213. [Google Scholar] [CrossRef]
- Stern, N.J.; Bailey, J.S.; Blankenship, L.C.; Cox, N.A.; McHan, F. Colonization characteristics of Campylobacter jejuni in chick ceca. Avian Dis. 1988, 32, 330–334. [Google Scholar] [CrossRef]
- Rosenquist, H.; Sommer, H.M.; Nielsen, N.L.; Christensen, B.B. The effect of slaughter operations on the contamination of chicken carcasses with thermotolerant Campylobacter. Int. J. Food Microbiol. 2006, 108, 226–232. [Google Scholar] [CrossRef]
- Solis de los Santos, F.; Donoghue, A.M.; Venkitanarayanan, K.; Reyes-Herrera, I.; Metcalf, J.H.; Dirain, M.L.; Aguiar, V.F.; Blore, P.J.; Donoghue, D.J. Therapeutic supplementation of caprylic acid in feed reduces Campylobacter jejuni colonization in broiler chicks. Appl. Environ. Microbiol. 2008, 74, 4564–4566. [Google Scholar] [CrossRef]
- Hariharan, H.; Murphy, G.A.; Kempf, I. Campylobacter jejuni: Public health hazards and potential control methods in poultry: A review. Vet. Med. 2004, 49, 441–446. [Google Scholar]
- Saris, P.E.J.; Hilmi, H.T.A.; Surakka, A.; Apajalahti, J. Identification of the most abundant Lactobacillus species in the crop of 1- and 5-week-old broiler chickens. Appl. Environ. Microb. 2007, 73, 7867–7873. [Google Scholar] [CrossRef]
- Engberg, R.M.; Hedemann, M.S.; Jensen, B.B. The influence of grinding and pelleting of feed on the microbial composition and activity in the digestive tract of broiler chickens. Br. Poult. Sci. 2002, 43, 569–579. [Google Scholar] [CrossRef]
- Huang, D.S.; Li, D.F.; Xing, J.J.; Ma, Y.X.; Li, Z.J.; Lv, S.Q. Effects of feed particle size and feed form on survival of Salmonella typhimurium in the alimentary tract and cecal S. typhimurium reduction in growing broilers. Poult. Sci. 2006, 85, 831–836. [Google Scholar]
- Moen, B.; Rudi, K.; Svihus, B.; Skånseng, B. Reduced spread of Campylobacter jejuni in broiler chickens by stimulating the bird’s natural barriers. J. Appl. Microbiol. 2012, 113, 1176–1183. [Google Scholar] [CrossRef]
- Svihus, B. The gizzard: Function, influence of diet structure and effects on nutrient availability. World Poult. Sci. J. 2011, 67, 207–224. [Google Scholar] [CrossRef]
- Hetland, H.; Svihus, B. Inclusion of dust bathing materials affects nutrient digestion and gut physiology of layers. J. Appl. Poult. Res. 2007, 16, 22–26. [Google Scholar]
- Hetland, H.; Svihus, B.; Choct, M. Role of insoluble fiber on gizzard activity in layers. J. Appl. Poult. Res. 2005, 14, 38–46. [Google Scholar]
- Norwegian Food Safety Authority. Available online: http://www.mattilsynet.no/fdu/ (accessed on 6 June 2013).
- SAS; Version 9.2; SAS Institute Inc.: Cary, NC, USA, 2008.
- Rudi, K.; Moen, B.; Drømtorp, S.M.; Holck, A.L. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 2005, 71, 1018–1024. [Google Scholar] [CrossRef]
- Skånseng, B.; Trosvik, P.; Zimonja, M.; Johnsen, G.; Bjerrum, L.; Pedersen, K.; Wallin, N.; Rudi, K. Co-infection dynamics of a major food-borne zoonotic pathogen in chicken. PLoS Pathog. 2007, 3, e175. [Google Scholar] [CrossRef]
- Skånseng, B.; Kaldhusdal, M.; Moen, B.; Gjevre, A.G.; Johannessen, G.S.; Sekelja, M.; Trosvik, P.; Rudi, K. Prevention of intestinal Campylobacter jejuni colonization in broilers by combinations of in-feed organic acids. J. Appl. Microbiol. 2010, 109, 1265–1273. [Google Scholar] [CrossRef]
- Skånseng, B.; Kaldhusdal, M.; Rudi, K. Comparison of chicken gut colonisation by the pathogens Campylobacter jejuni and Clostridium perfringens by real-time quantitative PCR. Mol. Cell. Probes 2006, 20, 269–279. [Google Scholar] [CrossRef]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148, 257–266. [Google Scholar]
- Nogva, H.K.; Bergh, A.; Holck, A.; Rudi, K. Application of the 5′-nuclease PCR assay in evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 2000, 66, 4029–4036. [Google Scholar] [CrossRef]
- SDS Plate Utility Software; Version 2.2; Applied Biosystems: Foster City, CA, USA, 2004.
- SYSTAT 12; Version 9.2; Systat Software Inc.: Chicago, IL, USA, 2007.
- Berget, I.; Heir, E.; Petcovic, J.; Rudi, K. Discriminatory power, typability, and accuracy of single nucleotide extension microarrays. J. AOAC Int. 2007, 90, 802–809. [Google Scholar]
- Trosvik, P.; Skånseng, B.; Jakobsen, K.S.; Stenseth, N.C.; Næs, T.; Rudi, K. Multivariate analysis of complex DNA-sequence electropherograms for high-throughput quantitative analysis of mixed microbial populations. Appl. Environ. Microbiol. 2007, 73, 4975–4983. [Google Scholar] [CrossRef]
- Michigan State University. RDP’s Pyrosequencing Pipeline. Available online: http://pyro.cme.msu.edu/pyro/help.jsp (accessed on 6 June 2013).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Krzanowski, W.J. Principles of Multivariate Analysis. A User’s Perspective; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Haarman, M.; Knol, J. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 2005, 71, 2318–2324. [Google Scholar] [CrossRef]
- Frahm, E.; Obst, U. Application of the fluorogenic probe technique (TaqMan PCR) to the detection of Enterococcus spp. and Escherichia coli in water samples. J. Microbiol. Methods 2003, 52, 123–131. [Google Scholar] [CrossRef]
- Walter, J.; Hertel, C.; Tannock, G.W.; Lis, C.M.; Munro, K.; Hammes, W.P. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2001, 67, 2578–2585. [Google Scholar]
- Heilig, H.G.; Zoetendal, E.G.; Vaughan, E.E.; Marteau, P.; Akkermans, A.D.; de Vos, W.M. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 2002, 68, 114–123. [Google Scholar] [CrossRef]
- Bjerrum, L.; Pedersen, A.B.; Engberg, R.M. The influence of whole wheat feeding on Salmonella infection and gut flora composition in broilers. Avian Dis. 2005, 49, 9–15. [Google Scholar] [CrossRef]
- Svihus, B.; Hetland, H.; Choct, M.; Sundby, F. Passage rate through the anterior digestive tract of broiler chickens fed on diets with ground and whole wheat. Br. Poult. Sci. 2002, 43, 662–668. [Google Scholar] [CrossRef]
- Jackson, D.N.; Davis, B.; Tirado, S.M.; Duggal, M.; van Frankenhuyzen, J.K.; Deaville, D.; Wijesinghe, M.A.; Tessaro, M.; Trevors, J.T. Survival mechanisms and culturability of Campylobacter jejuni under stress conditions. Antonie Van Leeuwenhoek 2009, 96, 377–394. [Google Scholar] [CrossRef]
- Amerah, A.M.; Ravindran, V. Influence of method of whole-wheat feeding on the performance, digestive tract development and carcass traits of broiler chickens. Anim. Feed Sci. Tech. 2008, 147, 326–339. [Google Scholar] [CrossRef]
- Hetland, H.; Svihus, B.; Krogdahl, A. Effects of oat hulls and wood shavings on digestion in broilers and layers fed diets based on whole or ground wheat. Br. Poult. Sci. 2003, 44, 275–282. [Google Scholar] [CrossRef]
- Santos, F.B.; Sheldon, B.W.; Santos, A.A., Jr.; Ferket, P.R. Influence of housing system, grain type, and particle size on Salmonella colonization and shedding of broilers fed triticale or corn-soybean meal diets. Poult. Sci. 2008, 87, 405–420. [Google Scholar] [CrossRef]
- Rosenquist, H.; Nielsen, N.L.; Sommer, H.M.; Nørrung, B.; Christensen, B.B. Quantitative risk assessment of human campylobacteriosis associated with thermophilic Campylobacter species in chickens. Int. J. Food Microbiol. 2003, 83, 87–103. [Google Scholar] [CrossRef]
- Hermans, D.; Pasmans, F.; Heyndrickx, M.; van Immerseel, F.; Martel, A.; van Deun, K.; Haesebrouck, F. A tolerogenic mucosal immune response leads to persistent Campylobacter jejuni colonization in the chicken gut. Crit. Rev. Microbiol. 2012, 38, 17–29. [Google Scholar] [CrossRef]
- Gabriel, I.; Mallet, S.; Leconte, M.; Travel, A.; Lalles, J.P. Effects of whole wheat feeding on the development of the digestive tract of broiler chickens. Anim. Feed Sci. Tech. 2008, 142, 144–162. [Google Scholar] [CrossRef]
- Rehman, H.U.; Vahjen, W.; Awad, W.A.; Zentek, J. Indigenous bacteria and bacterial metabolic products in the gastrointestinal tract of broiler chickens. Arch. Tierernahr. 2007, 61, 319–335. [Google Scholar]
- Yegani, M.; Korver, D.R. Factors affecting intestinal health in poultry. Poult. Sci. 2008, 87, 2052–2063. [Google Scholar] [CrossRef]
- Van Immerseel, F.; de Buck, J.; Pasmans, F.; Huyghebaert, G.; Haesebrouck, F.; Ducatelle, R. Clostridium perfringens in poultry: An emerging threat for animal and public health. Avian Pathol. 2004, 33, 537–549. [Google Scholar] [CrossRef]
- Sekelja, M.; Rud, I.; Knutsen, S.H.; Denstadli, V.; Westereng, B.; Naes, T.; Rudi, K. Abrupt temporal fluctuations in the chicken fecal microbiota are explained by its gastrointestinal origin. Appl. Environ. Microbiol. 2012, 78, 2941–2948. [Google Scholar]
- Choct, M. Managing gut health through nutrition. Br. Poult. Sci. 2009, 50, 9–15. [Google Scholar] [CrossRef]
- Jozefiak, D.; Rutkowski, A.; Kaczmarek, S.; Jensen, B.B.; Engberg, R.M.; Højberg, O. Effect of beta-glucanase and xylanase supplementation of barley- and rye-based diets on caecal microbiota of broiler chickens. Br. Poult. Sci. 2010, 51, 546–557. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Zhong, T.Y.; Pandya, Y.; Joerger, R.D. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 2002, 68, 124–137. [Google Scholar] [CrossRef]
Supplementary Information
| Control/Whole Wheat Diet | Oat Hulls Diet | |
|---|---|---|
| Corn, % | 25.34 | 22.30 |
| Ground/whole wheat, % | 42.00 | 36.96 |
| Soybean meal, % | 17.34 | 15.26 |
| Fish meal, % | 7.00 | 6.16 |
| Coarse oat hulls, % | 0.00 | 12.00 |
| Soybean oil, % | 4.00 | 3.52 |
| Limestone, % | 1.30 | 1.14 |
| Monocalcium phosphate, % | 1.00 | 0.88 |
| l-lysine-HCl, % | 0.50 | 0.44 |
| dl-methionine, % | 0.30 | 0.26 |
| l-threonine, % | 0.10 | 0.09 |
| Salt, % | 0.20 | 0.18 |
| Sodium bicarbonate, % | 0.30 | 0.26 |
| Choline chlorate, % | 0.15 | 0.13 |
| Microminerals/vitamins, % | 0.44 | 0.39 |
| Species | Strain a | Medium b | TaqMan PCR c | Relative to total DNA d | Std e |
|---|---|---|---|---|---|
| Positive Controls | |||||
| Lb. acidophilus | ATCC 4356 | MRS | ++ | −0.65 | 0.03 |
| Lb. brevis | DSM 20556 | MRS | ++ | −0.61 | 0.09 |
| Lb. casei | ATCC 393 | MRS | ++ | −0.72 | 0.04 |
| Lb. curvatus | DSM 20019 | MRS | ++ | −0.67 | 0.11 |
| Lb. delbrueckii subsp. lactis | ATCC 12315 | MRS | ++ | −0.62 | 0.01 |
| Lb. gasseri | ATCC 33323 | MRS | ++ | −0.66 | 0.02 |
| Lb. helveticus | ATCC 15009 | MRS | ++ | −0.61 | 0.08 |
| Lb. pentosus | ATCC 8041 | MRS | ++ | −0.64 | 0.04 |
| Lb. plantarum | NCIMB 8826 | MRS | ++ | −0.73 | 0.07 |
| Lb. reuteri | DSM 17938 | MRS | ++ | −0.59 | 0.04 |
| Lb. rhamnosus (GG) | ATCC 53103 | MRS | ++ | −0.66 | 0.10 |
| Lb. sakei | DSM 20017 | MRS | ++ | −0.51 | 0.30 |
| Lb. salivarius | DSM 20555 | MRS | ++ | −0.55 | 0.14 |
| Other LAB | |||||
| Lactococcus cremoris | ATCC 19257 | MRS | - | −5.64 | 0.06 |
| Lc. Lactis | ATCC 15346 | MRS | - | −5.41 | 0.07 |
| Leuconostoc mesenteroides f | ATCC 19255 | MRS | ++ | −0.61 | 0.05 |
| Carnobacterium divergens f | NCDO 2306 | MRS | + | −3.85 | 0.09 |
| Enterococcus faecalis | DSM 12956 | MRS | - | −6.04 | 0.02 |
| Streptococcus thermophilus | MF2403 | MRS | - | −5.47 | 0.07 |
| Negative controls | |||||
| Bifidobacterium longum | DSM 20219 | W-C | - | −5.08 | 0.09 |
| Staphylococcus aureus f | ATCC 12600 | BHI | + | −3.96 | 0.02 |
| Bacillus cereus | ATCC 4516 | BHI | - | −6.61 | 0.08 |
| Clostridium perfringens | ATCC 13124 | BHI | - | −6.25 | 0.14 |
| Escherichia coli | ATCC 47076 | BHI | - | −6.86 | 0.13 |
| Campylobacter jejuni | DSM 4688 | M-H | - | −6.29 | 0.19 |

© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Skånseng, B.; Svihus, B.; Rudi, K.; Trosvik, P.; Moen, B. Effect of Different Feed Structures and Bedding on the Horizontal Spread of Campylobacter jejuni within Broiler Flocks. Agriculture 2013, 3, 741-760. https://doi.org/10.3390/agriculture3040741
Skånseng B, Svihus B, Rudi K, Trosvik P, Moen B. Effect of Different Feed Structures and Bedding on the Horizontal Spread of Campylobacter jejuni within Broiler Flocks. Agriculture. 2013; 3(4):741-760. https://doi.org/10.3390/agriculture3040741
Chicago/Turabian StyleSkånseng, Beate, Birger Svihus, Knut Rudi, Pål Trosvik, and Birgitte Moen. 2013. "Effect of Different Feed Structures and Bedding on the Horizontal Spread of Campylobacter jejuni within Broiler Flocks" Agriculture 3, no. 4: 741-760. https://doi.org/10.3390/agriculture3040741