Nanobodies that Neutralize HIV
Abstract
1. Introduction
2. HIV-1 Broadly Neutralizing Nanobodies
2.1. Isolation and Characeterization of HIV-1 Neutralizing Nanobodies IUV-1, for Instance
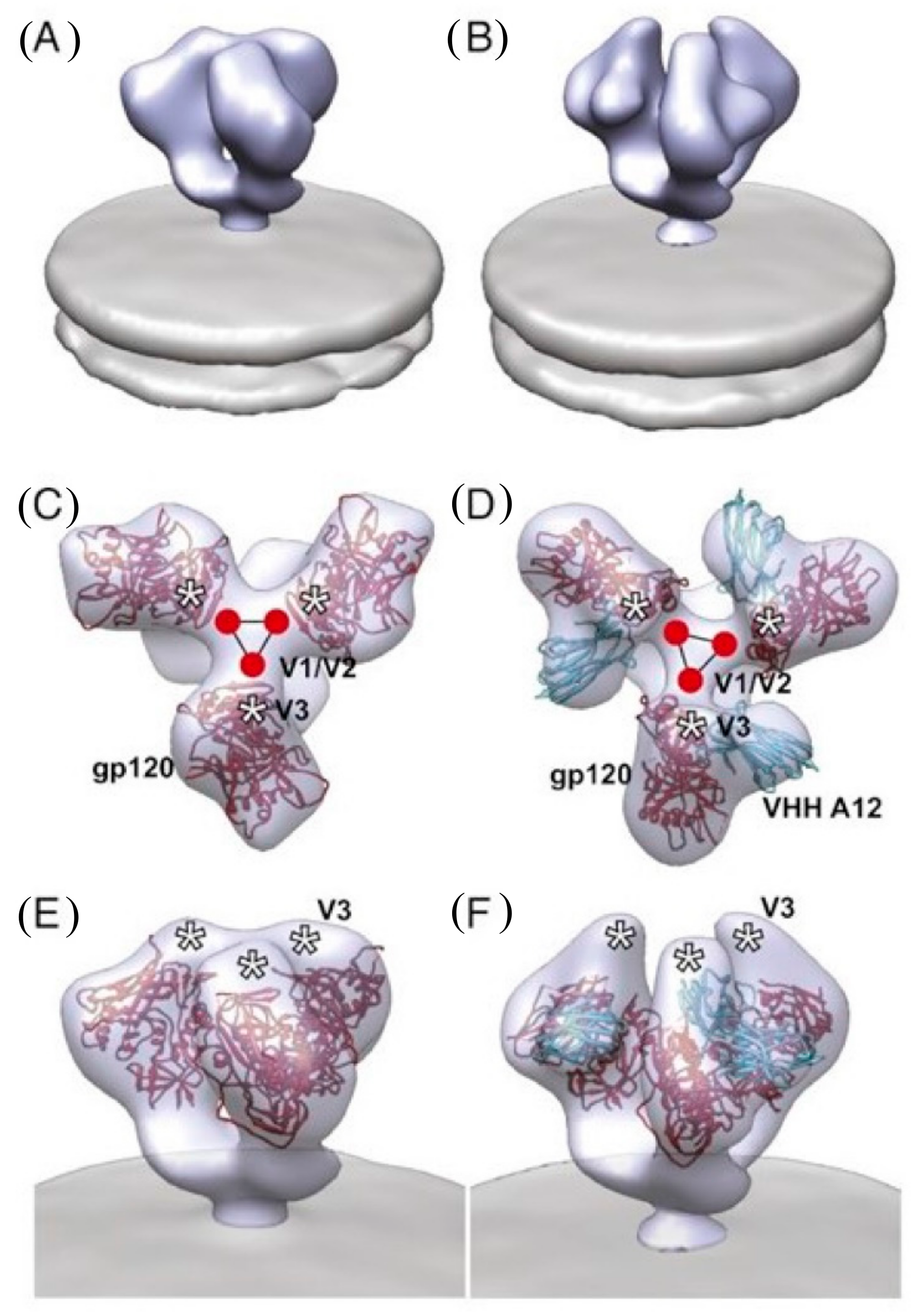
2.2. Structural Studies of HIV-1 Nanobodies
2.3. Molecular Manipulation and Modification of Anti-HIV-1 Nanobodies
Selection and Induction of Nanobody Variants
2.4. ‘Humanized’ Reconstituted Antibodies with Nanobody Heads
2.5. Mono- and Bispecific Bi-Head and Tri-Head Nanobodies
2.6. Intracellular Expression of Nanobodies to Combat HIV
3. Application of Nanobodies for Clinical Use Including Prevention and Treatment of HIV Infection
3.1. Production of Nanobodies
3.2. Nanobody Expression in Commensal Lactobacillus Strains
3.3. Nanobodies as Potential HIV Microbicides
3.4. Nanobodies as Labeling Agents to Detect Latent Reservoirs of HIV Infected Cells
4. Conclusions and Prospect
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hammers, C.; Songa, E.B.; Bendahman, N.; Hammers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.S.; Avila, D.; Hughes, M.; Hughes, A.; McKinney, E.C.; Flajnik, M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995, 374, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F.; Deschacht, N.; Muyldermans, S. A case of convergence: Why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol. 2011, 9, e1001120. [Google Scholar] [CrossRef] [PubMed]
- Desmyter, A.; Spinelli, S.; Roussel, A.; Cambillau, C. Camelid nanobodies: Killing two birds with one stone. Curr. Opin. Struct. Biol. 2015, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Helma, J.; Cardoso, M.C.; Muyldermans, S.; Leonhardt, H. Nanobodies and recombinant binders in cell biology. J. Cell Biol. 2015, 209, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Prole, D.L.; Taylor, C.W. A genetically encoded toolkit of functionalized nanobodies against fluorescent proteins for visualizing and manipulating intracellular signalling. BMC Biol. 2019, 17, 41. [Google Scholar] [CrossRef] [PubMed]
- Jähnichen, S.; Blanchetot, C.; Maussang, D.; Gonzalez-Pajuelo, M.; Chow, K.Y.; Bosch, L.; De Vrieze, S.; Serruys, B.; Ulrichts, H.; Vandevelde, W.; et al. CXCR4 nanobodies (VHH-1 based single variable domains) potently inhibit chemotaxis and HIV replication in mobile stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 20565–20570. [Google Scholar] [CrossRef]
- Vercruysse, T.; Pardon, E.; Vanstreels, E.; Steyaert, J.; Daelemans, D. An intrabody based on a llama single-domain antibody targeting the N-terminal alpha-helical multimerization domain of HIV-1 Rev prevents viral production. J. Biol. Chem. 2010, 285, 21768–21780. [Google Scholar]
- Gray, E.R.; Brookes, J.C.; Caillat, C.; Turbé, V.; Webb, B.L.J.; Granger, L.A.; Miller, B.S.; McCoy, L.E.; El Khattabi, M.; Verrips, C.T.; et al. Unravelling the molecular basis of high affinity nanobodies against HIV p24: In vitro functional, structural, and in silico insights. ACS Infect. Dis. 2017, 3, 479–491. [Google Scholar] [CrossRef]
- Turbé, V.; Gray, E.R.; Lawson, V.E.; Nastouli, E.; Brookes, J.C.; Weiss, R.A.; Pillay, D.; Emery, V.C.; Verrips, C.T.; Yatsuda, H.; et al. Towards an ultra-rapid smartphone connected test for infectious diseases. Sci. Rep. 2017, 7, 11971. [Google Scholar] [CrossRef] [PubMed]
- Van der Vaart, J.M.; Pant, N.; Wolvers, D.; Bezemer, S.; Hermans, P.W.; Bellamy, K.; Sarker, S.A.; Van der Logt, C.P.; Svensson, L.; Verrips, C.T.; et al. Reduction in morbidity of rotavirus induced diarrhoea in mice by yeast-produced monovalent llama-derived antibody fragments. Vaccine 2006, 24, 4130–4137. [Google Scholar] [CrossRef] [PubMed]
- Hultberg, A.; Temperton, N.J.; Rosseels, V.; Koenders, M.; Gonzalez-Pajuelo, M.; Schepens, B.; Ibañez, L.I.; Vanlandschoot, P.; Schillemans, J.; Saunders, M.; et al. Llama-derived single chain antibodies to build multivalent, superpotent, broadened neutralizing anti-viral molecules. PLoS ONE 2011, 6, e17665. [Google Scholar] [CrossRef] [PubMed]
- Hufton, S.E.; Risley, P.; Ball, C.R.; Major, D.; Engelhardt, O.G.; Poole, S. The breadth of cross sub-type neutralisation activity of a single domain antibody to influenza hemagglutinin can be increased by antibody valency. PLoS ONE 2014, 9, e103294. [Google Scholar] [CrossRef] [PubMed]
- Koromyslova, A.D.; Hansman, G.S. Nanobodies targeting norovirus capsid reveal functional epitopes and potential mechanisms of neutralization. PLoS Pathog. 2017, 13, e1006636. [Google Scholar] [CrossRef] [PubMed]
- Ibañez, L.I.; De Filette, M.; Hultberg, A.; Verrips, C.T.; Temperton, N.; Weiss, R.A.; Vandevelde, W.; Schepens, B.; Vanlandschoot, P.; Saelens, X. Nanobodies with in vitro neutralizing activity protect mice against H5N1 influenza virus infection. J. Infect. Dis. 2011, 203, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Günaydın, G.; Yu, S.; Gräslund, T.; Hammarström, L.; Marcotte, H. Fusion of the mouse IgG1 Fc domain to the VHH fragment (ARP1) enhances protection in a mouse model of rotavirus. Sci. Rep. 2016, 6, 30171. [Google Scholar] [CrossRef] [PubMed]
- Haim, H.; Si, Z.; Madani, N.; Wang, L.; Courter, J.R.; Princiotto, A.; Kassa, A.; DeGrace, M.; McGee-Estrada, K.; Mefford, M.; et al. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009, 5, e1000360. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.R.; Kattenhorn, L.M.; Kondur, H.R.; Von Schaewen, M.; Dorfman, T.; Chiang, J.J.; Haworth, K.G.; Decker, J.M.; Alpert, M.D.; Bailey, C.C.; et al. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 2015, 519, 87–91. [Google Scholar] [CrossRef]
- Schweizer, A.; Rusert, P.; Berlinger, L.; Ruprecht, C.R.; Mann, A.; Corthésy, S.; Turville, S.G.; Aravantinou, M.; Fischer, M.; Robbiani, M.; et al. CD4-specific designed ankyrin repeat proteins are novel potent HIV entry inhibitors with unique characteristics. PLoS Pathog. 2008, 4, e1000109. [Google Scholar] [CrossRef]
- Mann, A.; Friedrich, N.; Krarup, N.; Weber, J.; Stiegeler, E.; Dreier, B.; Pugach, P.; Robbiani, M.; Riedel, T.; Moehle, K.; et al. Conformation-dependent recognition of HIV gp120 by designed ankyrin repeat proteins provides access to novel HIV entry inhibitors. J. Virol. 2013, 87, 5868–5881. [Google Scholar] [CrossRef] [PubMed]
- Burton, D.R.; Hangartner, L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 2016, 34, 635–659. [Google Scholar] [CrossRef] [PubMed]
- McCoy, L.E.; Burton, D.R. Identification and specificity of broadly neutralizing antibodies against HIV. Immunol. Rev. 2017, 275, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Van Schooten, J.; Van Gils, M.J. HIV-1 immunogens and strategies to drive antibody responses towards neutralization breadth. Retrovirology 2018, 15, 74. [Google Scholar] [CrossRef] [PubMed]
- Forsman, A.; Beirnaert, E.; Aasa-Chapman, M.M.I.; Hoorelbeke, B.; Hijazi, K.; Koh, W.; Tack, V.; Szynol, A.; Kelly, C.; McKnight, A.; et al. Llama antibody fragments with cross-subtype human immunodeficiency virus type 1 (HIV-1)-neutralizing properties and high affinity for HIV-1 gp120. J. Virol. 2008, 82, 12069–12081. [Google Scholar] [CrossRef]
- Strokappe, N.; Szynol, A.; Aasa-Chapman, M.; Gorlani, A.; Forsman Quigley, A.; Lutje Hulsik, D.; Chen, L.; Weiss, R.A.; De Haard, H.; Verrips, C.T.; et al. Llama antibody fragments recognizing various epitopes of the CD4bs neutralize a broad range of HIV-1 subtypes A, B and C. PLoS ONE 2012, 7, e33298. [Google Scholar] [CrossRef]
- McCoy, L.E.; Forsman Quigley, A.; Strokappe, N.M.; Bulmer-Thomas, B.; Seaman, M.S.; Mortier, D.; Rutten, L.; Chander, N.; Edwards, C.J.; Kettler, R.; et al. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J. Exp. Med. 2012, 209, 1091–1103. [Google Scholar] [CrossRef]
- Lutje Hulsik, D.; Liu, Y.Y.; Strokappe, N.M.; Battella, S.; El-Khattabi, M.; McCoy, L.E.; Sabin, C.; Hinz, A.; Hock, M.; Macheboeuf, P.; et al. A gp41 MPER-specific llama VHH requires a hydrophobic CDR3 for neutralization but not for antigen recognition. PLoS Pathog. 2013, 9, e1003202. [Google Scholar] [CrossRef]
- Matz, J.; Kessler, P.; Bouchet, J.; Combes, O.; Pereira Ramos, O.H.; Barin, F.; Daniel Baty, D.; Martin, L.; Benichou, S.; Chames, P. Straightforward selection of broadly neutralizing single-domain antibodies targeting the conserved CD4 and coreceptor binding sites of HIV-1 gp120. J. Virol. 2013, 87, 1137–1149. [Google Scholar] [CrossRef]
- Acharya, P.; Luongo, T.S.; Georgiev, I.S.; Matz, J.; Schmidt, S.D.; Louder, M.K.; Kessler, P.; Yang, Y.; McKee, K.; O’Dell, S.; et al. Heavy chain-only IgG2b llama antibody effects near-pan HIV-1 neutralization by recognizing a CD4-induced epitope that includes elements of coreceptor- and CD4-binding sites. J. Virol. 2013, 87, 10173–10181. [Google Scholar] [CrossRef]
- McCoy, L.E.; Rutten, L.; Frampton, D.; Anderson, I.; Granger, L.; Bashford-Rogers, R.; Dekkers, G.; Strokappe, N.M.; Seaman, M.S.; Koh, W.; et al. Molecular evolution of broadly neutralizing llama antibodies to the CD4-binding site of HIV-1. PLoS Pathog. 2014, 10, e1004552. [Google Scholar] [CrossRef]
- Koch, K.; Kalusche, S.; Torres, J.L.; Stanfield, R.L.; Danquah, W.; Khazanehdari, K.; Von Briesen, H.; Geertsma, E.R.; Wilson, I.A.; Wernery, U.; et al. Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers. Sci. Rep. 2017, 7, 8390. [Google Scholar] [CrossRef]
- Strokappe, N.M.; Hock, M.; Rutten, L.; McCoy, L.E.; Back, J.W.; Caillat, C.; Haffke, M.; Weiss, R.A.; Weissenhorn, W.; Verrips, C.T. Super potent bispecific llama VHH antibodies neutralize HIV via a combination of gp41 and gp120 epitopes. Antibodies 2019, 8, 38. [Google Scholar] [CrossRef]
- Sabin, C.; Corti, D.; Buzon, V.; Seaman, M.S.; Lutje Hulsik, D.; Hinz, A.; Vanzetta, F.; Agatic, G.; Silacci, C.; Mainetti, L.; et al. Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41. PLoS Pathog. 2010, 6, e1001195. [Google Scholar] [CrossRef]
- Seaman, M.S.; Janes, H.; Hawkins, N.; Grandpre, L.E.; Devoy, C.; Giri, A.; Coffey, R.T.; Harris, L.; Wood, B.; Daniels, M.G.; et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 2010, 84, 1439–1452. [Google Scholar] [CrossRef]
- Barbian, H.J.; Decker, J.M.; Bibollet-Ruche, F.; Galimidi, R.P.; West, A.P.; Learn, G.H.; Parrish, N.F.; Iyer, S.S.; Li, Y.; Pace, C.S.; et al. Neutralization properties of simian immunodeficiency viruses infecting chimpanzees and gorillas. MBio 2015, 6, e00296-15. [Google Scholar] [CrossRef]
- Zhou, T.; Lynch, R.M.; Chen, L.; Acharya, P.; Wu, X.; Doria-Rose, N.A.; Joyce, M.G.; Lingwood, D.; Soto, C.; Bailer, R.T.; et al. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 2015, 161, 1280–1292. [Google Scholar] [CrossRef]
- Pardon, E.; Laeremans, T.; Triest, S.; Rasmussen, S.G.F.; Wohlkönig, A.; Ruf, A.; Muyldermans, S.; Hol, W.G.J.; Kobilka, B.K.; Steyaert, J. A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 2014, 9, 674–693. [Google Scholar] [CrossRef]
- Hinz, A.; Lutje Hulsik, D.; Forsman, A.; Koh, W.W.; Belrhali, H.; Gorlani, A.; De Haard, H.; Weiss, R.A.; Verrips, T.; Weissenhorn, W. Crystal structure of the neutralizing llama V(HH) D7 and its mode of HIV-1 gp120 interaction. PLoS ONE 2010, 5, e10482. [Google Scholar] [CrossRef]
- Sok, D.; Le, K.M.; Vadnais, M.; Saye-Francisco, K.L.; Jardine, J.G.; Torres, J.L.; Berndsen, Z.T.; Kong, L.; Stanfield, R.; Ruiz, J.; et al. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature 2017, 548, 108–111. [Google Scholar] [CrossRef]
- Meyerson, J.R.; Trana, E.E.H.; Kuybedac, O.; Chend, W.; Dimitrov, D.S.; Gorlani, A.; Verrips, C.T.; Lifson, J.D.; Subramaniam, S. Molecular structures of trimeric HIV-1 Env in complex with small antibody derivatives. Proc. Natl. Acad. Sci. USA 2013, 110, 513–518. [Google Scholar] [CrossRef]
- Koh, W.L.; Steffensen, S.; Gonzalez-Pajuelo, M.; Hoorelbeke, B.; Gorlani, A.; Szynol, A.; Forsman, A.; Aasa-Chapman, M.M.I.; De Haard, H.; Verrips, C.T.; et al. Generation of a family-specific phage library of llama single chain antibody fragments that neutralize HIV-1. J. Biol. Chem. 2010, 285, 19116–19124. [Google Scholar] [CrossRef]
- McCoy, L.E.; Groppelli, E.; Blanchetot, C.; De Haard, H.; Verrips, C.T.; Rutten, L.; Weiss, R.A.; Jolly, C. Neutralisation of HIV-1 cell-cell spread by human and llama antibodies. Retrovirology 2014, 11, 83. [Google Scholar] [CrossRef]
- Liu, L.; Wang, W.; Matz, J.; Ye, C.; Bracq, L.; Delon, J.; Kimata, J.T.; Chen, Z.; Benichou, S.; Zhou, P. The glycosylphosphatidylinositol-anchored variable region of llama heavy chain-only antibody JM4 efficiently blocks both cell-free and T cell-T cell transmission of human immunodeficiency virus type 1. J. Virol. 2016, 90, 10642–10659. [Google Scholar] [CrossRef]
- Vincke, C.; Loris, R.; Saerens, D.; Martinez-Rodriguez, S.; Muyldermans, S.; Conrath, K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J. Biol. Chem. 2009, 284, 3273–3284. [Google Scholar] [CrossRef]
- Klarenbeek, A.; El Mazouari, K.; Desmyter, A.; Blanchetot, C.; Hultberg, A.; De Jonge, N.; Roovers, R.C.; Cambillau, C.; Spinelli, S.; Del-Favero, J.; et al. Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform. MAbs 2015, 7, 693–706. [Google Scholar] [CrossRef]
- Deschacht, N.; De Groeve, K.; Vincke, C.; Raes, G.; De Baetselier, P.; Muyldermans, S. A novel promiscuous class of camelid single-domain antibody contributes to the antigen-binding repertoire. J. Immunol. 2010, 184, 5696–5704. [Google Scholar] [CrossRef]
- Padte, N.N.; Yu, J.; Huang, Y.; Ho, D.D. Engineering multi-specific antibodies against HIV-1. Retrovirology 2018, 15, 60. [Google Scholar] [CrossRef]
- Mouquet, H.; Warncke, M.; Scheid, J.F.; Seaman, M.S.; Nussenzweig, M.C. Enhanced HIV-1 neutralization by antibody heteroligation. Proc. Natl. Acad. Sci. USA 2012, 109, 875–880. [Google Scholar] [CrossRef]
- Asokan, M.; Rudicell, R.S.; Louder, M.; McKee, K.; O’Dell, S.; Stewart-Jones, G.; Wang, K.; Xu, L.; Chen, X.; Choe, M.; et al. Bispecific antibodies targeting different epitopes on the HIV-1 envelope exhibit broad and potent neutralization. J. Virol. 2015, 89, 12501–12512. [Google Scholar] [CrossRef]
- Dolk, E.; Van Vliet, C.; Perez, J.M.; Vriend, G.; Darbon, H.; Ferrat, G.; Cambillau, C.; Frenken, L.G.; Verrips, C.T. Induced refolding of a temperature denatured llama heavy-chain antibody fragment by its antigen. Proteins 2005, 59, 555–564. [Google Scholar] [CrossRef]
- Bouchet, J.; Basmaciogullari, S.E.; Chrobak, P.; Stolp, B.; Bouchard, N.; Fackler, O.T.; Chames, P.; Jolicoeur, P.; Benichou, S.; Baty, D. Inhibition of the Nef regulatory protein of HIV-1 by a single-domain antibody. Blood 2011, 117, 3559–3568. [Google Scholar] [CrossRef]
- Matz, J.; Hérate, C.; Bouchet, J.; Dusetti, N.; Gayet, O.; Baty, D.; Benichou, S.; Chames, P. Selection of intracellular single domain antibodies targeting the HIV-1 Vpr protein by cytoplasmic yeast two-hybrid system. PLoS ONE 2014, 9, e113729. [Google Scholar] [CrossRef]
- D’Astolfo, D.S.; Pagliero, R.J.; Pras, A.; Karthaus, W.R.; Clevers, H.; Prasad, V.; Lebbink, R.J.; Rehmann, H.; Geijsen, N. Efficient intracellular delivery of native proteins. Cell 2015, 160, 674–690. [Google Scholar] [CrossRef]
- Thomassen, Y.E.; Verkleij, A.J.; Boonstra, J.; Verrips, C.T. Specific production rate of VHH antibody fragments by Saccharomyces cerevisiae is correlated with growth rate, independent of nutrient limitation. J. Biotechnol. 2005, 118, 270–277. [Google Scholar] [CrossRef]
- Del Rio, B.; Redruello, B.; Fernandez, M.; Martin, M.C.; Ladero, V.; Alvarez, M.A. Lactic acid bacteria as a live delivery system for the in situ production of nanobodies in the human gastrointestinal tract. Front. Microbiol. 2019, 9, 3179. [Google Scholar] [CrossRef]
- Andersen, K.K.; Strokappe, N.M.; Hultberg, A.; Truusalu, K.; Smidt, I.; Mikelsaar, R.H.; Verrips, C.T.; Hammarström, L.; Marcotte, H. Neutralization of Clostridium difficile toxin B mediated by engineered lactobacilli producing single domain antibodies. Infect. Immun. 2015, 84, 395–406. [Google Scholar] [CrossRef]
- Pant, N.; Marcotte, H.; Hermans, P.; Bezemer, S.; Frenken, L.; Johansen, K.; Hammarström, L. Lactobacilli producing bispecific llama-derived anti-rotavirus proteins in vivo for rotavirus-induced diarrhea. Future Microbiol. 2011, 6, 583–593. [Google Scholar] [CrossRef]
- Pendharkar, S.; Brandsborg, E.; Hammarström, L.; Marcotte, H.; Larsson, P.-G. Vaginal colonisation by probiotic lactobacilli and clinical outcome in women conventionally treated for bacterial vaginosis and yeast infection. BMC Infect. Dis. 2015, 15, 255. [Google Scholar] [CrossRef]
- Pendharkar, S.; Magopane, T.; Larsson, P.G.; De Bruyn, G.; Gray, G.E.; Hammarström, L.; Marcotte, H. Identification and characterisation of vaginal lactobacilli from South African women. BMC Infect. Dis. 2013, 13, 43. [Google Scholar] [CrossRef]
- Lagenaur, L.A.; Sanders-Beer, B.E.; Brichacek, B.; Pal, R.; Liu, X.; Liu, Y.; Yu, R.; Venzon, D.; Lee, P.P.; Hamer, D.H. Prevention of vaginal SHIV transmission in macaques by a live recombinant Lactobacillus. Mucosal Immunol. 2011, 4, 648–657. [Google Scholar] [CrossRef]
- Marcobal, A.; Liu, X.; Zhang, W.; Dimitrov, A.S.; Jia, L.; Lee, P.P.; Fouts, T.R.; Parks, T.P.; Lagenaur, L.A. Expression of human immunodeficiency virus type 1 neutralizing antibody fragments using human vaginal Lactobacillus. AIDS Res. Hum. Retroviruses 2016, 32, 964–971. [Google Scholar] [CrossRef]
- Gorlani, A.; Brouwers, J.; McConville, C.; Van der Bijl, P.; Malcolm, K.; Augustijns, P.; Forsman Quigley, A.; Weiss, R.A.; De Haard, H.; Verrips, C.T. Llama antibody fragments have good potential for application as HIV type 1 topical microbicides. AIDS Res. Hum. Retroviruses 2012, 28, 198–205. [Google Scholar] [CrossRef]
- Kijanka, M.; Dorresteijn, B.; Oliveira, S.; Van Bergen en Henegouwen, P.M.P. Nanobody-based cancer therapy of solid tumors. Nanomedicine 2015, 10, 161–174. [Google Scholar] [CrossRef]
- European Commission. European Commission 7th framework programme (Health): Combined Highly Active Anti-Retroviral Microbicides (CHAARM); Final Report Summary; European Commission: Brussels, Belgium, 2016. [Google Scholar]
- Kijanka, M.M.; Van Brussel, A.S.A.; Van der Wall, E.; Mali, W.P.T.M.; Van Diest, P.J.; Van Bergen en Henegouwen, P.M.P.; Oliveira, S. Optical imaging of pre-invasive breast cancer with a combination of VHHs targeting CAIX and HER2 increases contrast and facilitates tumour characterization. EJNMMI Res. 2016, 6, 14. [Google Scholar] [CrossRef]
- Pleiner, T.; Bates, M.; Trakhanov, S.; Chung-Tien Lee, C.-T.; Schliep, J.E.; Chug, H.; Böhning, M.; Stark, H.; Urlaub, H.; Görlich, D. Nanobodies: Site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. Elife 2015, 4, e11349. [Google Scholar]
- Togtmea, M.; Hussack, G.; Dayer, G.; Teghtmeyer, M.R.; Raphael, S.; Tanha, J.; Zehbe, I. Single-domain antibodies represent novel alternatives to monoclonal antibodies as targeting agents against the human papillomavirus 16 E6 protein. Int. J. Mol. Sci. 2019, 20, 2088. [Google Scholar] [CrossRef]
- Sifniotis, V.; Cruz, E.; Eroglu, B.; Kayser, V. Current advancements in addressing key challenges of therapeutic antibody design, manufacture, and formulation. Antibodies 2019, 8, 36. [Google Scholar] [CrossRef]
- Chowdhury, S.; Castro, S.; Coker, C.; Hinchcliffe, T.E.; Apraia, N.; Danino, T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 2019, 25, 1057–1063. [Google Scholar] [CrossRef]





| Name of VHH | Domain Recognized b | Breadth of Strains Tested c | Immunogen | Reference |
|---|---|---|---|---|
| A12 | CD4bs | 45% | gp120 | [25] |
| C8 | CD4bs | 45% | gp120 | [25] |
| D7 | CD4bs | 40% | gp120 | [25] |
| 1B5 | CoRbs | 75% | gp140 | [26,33] |
| 1F10 | V3 loop | 73% | gp140 | [26,33] |
| 2E7 | gp41 heptad | 80% | gp140 | [26,33] |
| J3 | CD4bs | 96% | gp140 | [27,31] |
| 2H10 | MPER | 45% | gp41-liposomes | [28] |
| JM2 | CD4bs | 60% | gp140 | [29,30] |
| JM3 | CoRbs | 73% | gp140/CD4 mimic | [29,30] |
| JM4 | CD4bs/CoRBs | 70% | gp140/CD4 mimic | [29,30] |
| 3E3 | CD4bs | 95% | gp140 | [31,33] |
| A14 | CD4bs | 74% | gp140 | [31] |
| B9 | CD4bs | 77% | gp140 | [31] |
| B21 | CD4bs | 72% | gp140 | [31] |
| VHH-9 | CD4bs | 53% | gp140 SOSIP | [32] |
| VHH-28 | CD4bs | 65% | gp140 SOSIP | [32] |
| VHH-A6 | CD4bs | 77% | gp140 SOSIP | [32] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiss, R.A.; Verrips, C.T. Nanobodies that Neutralize HIV. Vaccines 2019, 7, 77. https://doi.org/10.3390/vaccines7030077
Weiss RA, Verrips CT. Nanobodies that Neutralize HIV. Vaccines. 2019; 7(3):77. https://doi.org/10.3390/vaccines7030077
Chicago/Turabian StyleWeiss, Robin A., and C. Theo Verrips. 2019. "Nanobodies that Neutralize HIV" Vaccines 7, no. 3: 77. https://doi.org/10.3390/vaccines7030077
APA StyleWeiss, R. A., & Verrips, C. T. (2019). Nanobodies that Neutralize HIV. Vaccines, 7(3), 77. https://doi.org/10.3390/vaccines7030077





