Gene Dosage Analysis in a Clinical Environment: Gene-Targeted Microarrays as the Platform-of-Choice
Abstract
:1. Introduction
2. Experimental Section
2.1. Patient Samples
| Gene(s) of interest | Number of patients | Clinical indication | Mode of inheritance | Sample type |
|---|---|---|---|---|
| APC | 7 | Familial adenomatous polyposis (FAP) | Autosomal dominant | Peripheral blood |
| Dystrophin ( DMD) | 7 | Becker muscular dystrophy (BMD) | X-linked | Peripheral blood |
| 17 | Duchenne muscular dystrophy (DMD) | Peripheral blood; Guthrie spot (1) | ||
| 17 | Carrier testing for BMD/DMD | Peripheral blood | ||
| Calcium-sensing receptor (CaSR) | 1 | Familial hypocalciuric hypercalcemia | Autosomal dominant | Peripheral blood |
| E-cadherin (CDH1) | 5 | Familial gastric cancer | Autosomal dominant | Peripheral blood |
| EPCAM | 3 | Familial colon cancer | Autosomal dominant | Peripheral blood |
| HNF4α (MODY1), GCK (MODY2), HNF1α (MODY3), HNF1β (MODY5) | 3 | Maturity-onset diabetes of the young (MODY); 1 individual also with hepatic multiple adenomatosis | Autosomal dominant | Peripheral blood |
| PMP22 | 19 | Possible diagnosis of Charcot Marie Tooth Type 1A (CMT1A) | Autosomal dominant | Peripheral blood |
| 7 | Possible diagnosis of Hereditary Neuropathy with liability to Pressure Palsies (HNPP) | Autosomal dominant | Peripheral blood | |
| MSH2 | 2 | Hereditary Non-Polyposis Colorectal Cancer (HNPCC) | Autosomal dominant | Peripheral blood |
| PTEN | 3 | Cowden syndrome | Autosomal dominant | Peripheral blood |
| RET proto-oncogene, SDHAF2, SDHB, SDHC, SDHD, TMEM127, VHL | 6 | Familial phaeochromocytoma/paraganglioma | Autosomal dominant | Peripheral blood |
| 1 | Predictive testing for familial paraganglioma | |||
| VHL | 1 | Possible diagnosis of Von-Hippel-Lindau syndrome | Autosomal dominant | Peripheral blood |
2.2. DNA Extraction
2.3. Dosage Analysis
3. Results and Discussion
| Patient | Gene(s) analysed | Genotype | Significance of result |
|---|---|---|---|
| 1,2 | DMD | Hemizygous deletion of exons 45–47 (inclusive) | In-frame deletion; consistent with BMD phenotype |
| 3 | Hemizygous deletion of exons 45–48 (inclusive) | In-frame deletion; consistent with BMD phenotype | |
| 4 | c.5199_5209del (p.Thr1734SerfsX10) | Premature truncation of protein; consistent with DMD phenotype | |
| 5 | Hemizygous deletion of exons 46–50 (inclusive) | Out-of-frame deletion; consistent with DMD phenotype | |
| 6 | Hemizygous duplication of exon 12 | Out-of-frame duplication; consistent with DMD phenotype | |
| 7 | Hemizygous duplication of exons 10–11 (inclusive) | Out-of-frame duplication; consistent with DMD phenotype | |
| 8 | Hemizygous deletion of exons 53–59 (inclusive) | Out-of-frame deletion; consistent with DMD phenotype | |
| 9,10 | Hemizygous duplication of exons 8–9 (inclusive) | Out-of-frame duplication; consistent with DMD phenotype | |
| 11–19 | Various (heterozygous deletion/duplication) | Carrier of familial deletion/duplication | |
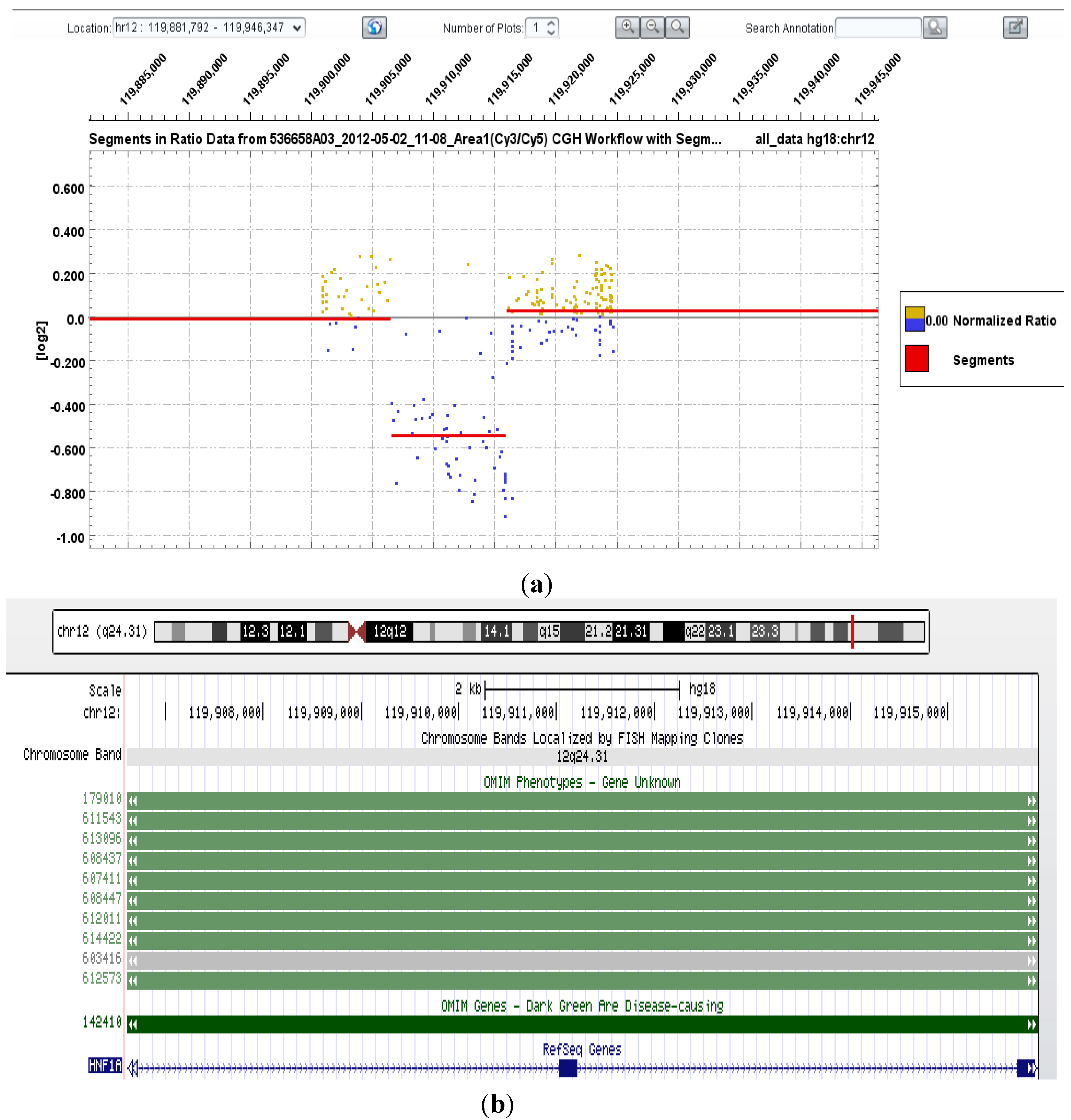
| 20 | HNF1α | Heterozygous deletion of exons 2–3 (inclusive) | Consistent with clinical phenotype—adenomatosis and MODY3 |
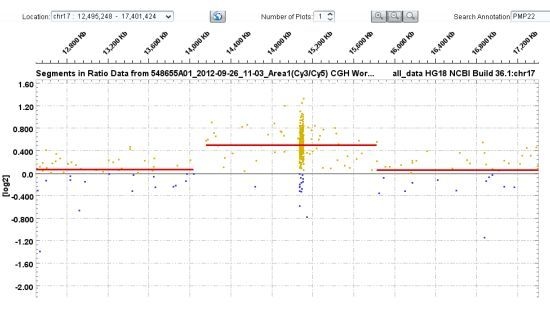
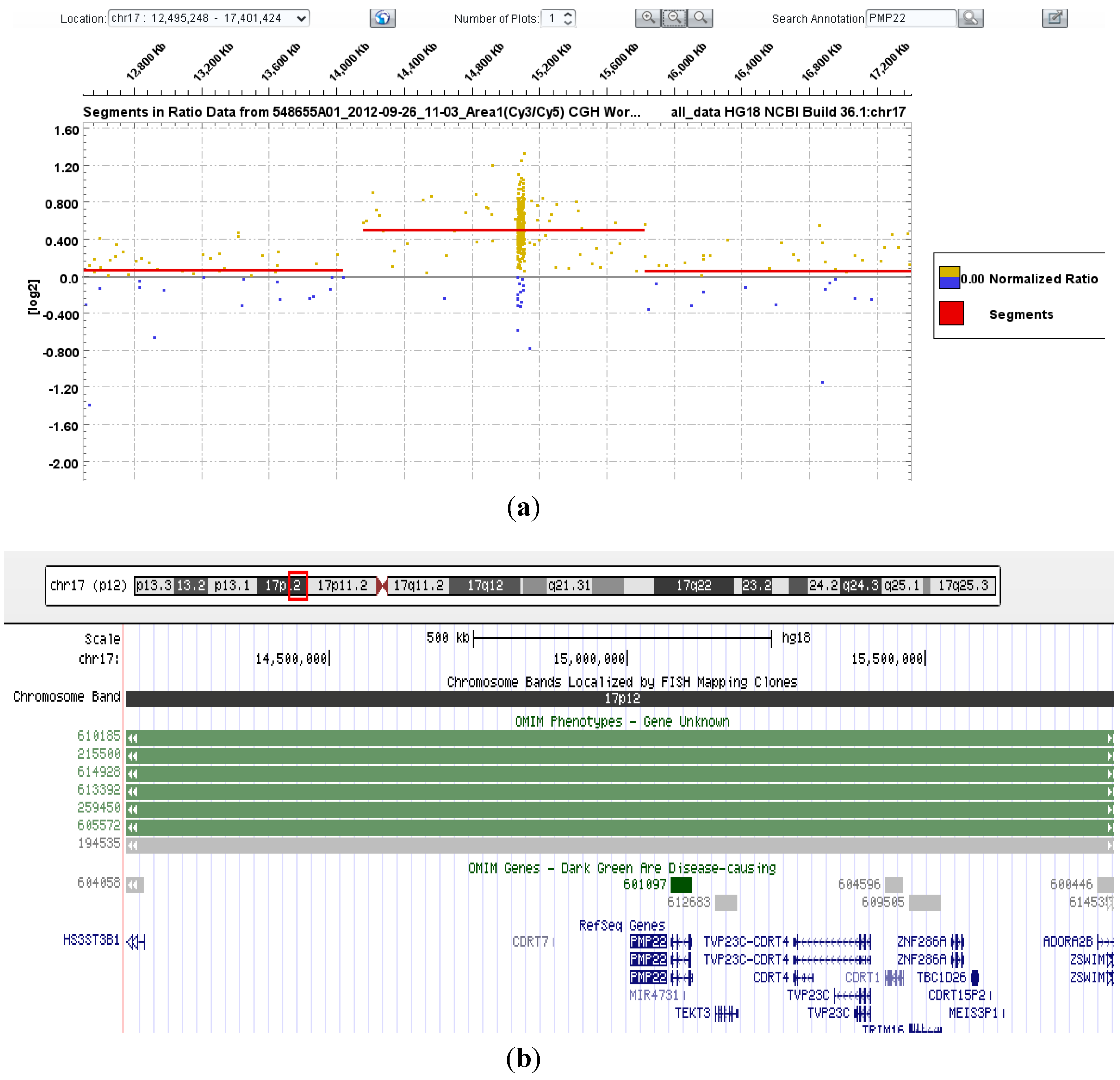
| 21–29 | PMP22 | ~1.5 Mb heterozygous duplication encompassing PMP22 gene | Consistent with CMT1A phenotype |
| 30,31 | Reciprocal deletion | Consistent with HNPP phenotype | |
| 32,33 | PTEN | Heterozygous deletion of exon 2 | Consistent with Cowden syndrome phenotype |
| 34,35 | SDHB | Heterozygous deletion of exon 1 | Consistent with clinical diagnosis of familial phaeo syndrome |
| 36 | Presence of familial deletion— appropriate surveillance/operative management required |
3.1. PMP22 Gene Analysis—Charcot-Marie-Tooth Neuropathy Type 1A (CMT1A) and Hereditary Neuropathy with Liability to Pressure Palsies (HNPP)
3.2. DMD Gene Analysis—Duchenne and Becker Muscular Dystrophy (DMD/BMD)
3.3. PTEN Gene Analysis
3.4. Familial Paraganglioma/Phaeochromocytoma Syndrome Mutation Screening—SDHAF2, SHDB, SDHC, SDHD, VHL, RET Proto-Oncogene, and TMEM127 Gene Analysis
3.5. Maturity-Onset Diabetes of the Young (MODY) Mutation Screening—HNF4α, GCK, HNF1α, HNF1β
4. Conclusions
Acknowledgments
References
- Kunkel, L.M.; Hejtmancik, J.F.; Caskey, C.T.; Speer, A.; Monaco, A.P.; Middlesworth, W.; Colletti, C.A.; Bertelson, C.; Muller, U.; Bresnan, M.; et al. Analysis of deletions in DNA from patients with Becker and Duchenne muscular dystrophy. Nature 1986, 322, 73–77. [Google Scholar]
- Roa, B.B.; Garcia, C.A.; Lupski, J.R. Charcot-Marie-Tooth disease type 1A: Molecular mechanisms of gene dosage and point mutation underlying a common inherited peripheral neuropathy. Int. J. Neurol. 1991–1992, 25–26, 97–107. [Google Scholar]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Howells, K.; Phillips, A.D.; Thomas, N.S.; Cooper, D.N. The human gene mutation database: 2008 update. Genome Med. 2009, 1. [Google Scholar] [CrossRef]
- Stenson, P.D.; Ball, E.V.; Mort, M.; Phillips, A.D.; Shiel, J.A.; Thomas, N.S.; Abeysinghe, S.; Krawczak, M.; Cooper, D.N. Human gene mutation database (hgmd): 2003 update. Hum. Mutat. 2003, 21, 577–581. [Google Scholar] [CrossRef]
- Eijk-Van Os, P.G.; Schouten, J.P. Multiplex ligation-dependent probe amplification (MLPA) for the detection of copy number variation in genomic sequences. Meth. Mol. Biol. 2011, 688, 97–126. [Google Scholar] [CrossRef]
- Sieber, O.M.; Lamlum, H.; Crabtree, M.D.; Rowan, A.J.; Barclay, E.; Lipton, L.; Hodgson, S.; Thomas, H.J.; Neale, K.; Phillips, R.K.; et al. Whole-gene APC deletions cause classical familial adenomatous polyposis, but not attenuated polyposis or “multiple” colorectal adenomas. Proc. Natl. Acad. Sci. USA 2002, 99, 2954–2958. [Google Scholar] [CrossRef]
- Bendavid, C.; Kleta, R.; Long, R. FISH diagnosis of the common 57-kb deletion in CTNS causing cystinosis. Hum. Genet. 2004, 115, 510–514. [Google Scholar] [CrossRef]
- Gouas, L.; Goumy, C.; Veronese, L.; Tchirkov, A.; Vago, P. Gene dosage methods as diagnostic tools for the identification of chromosome abnormalities. Pathol. Biol. 2008, 56, 345–353. [Google Scholar] [CrossRef]
- Armour, J.A.L.; Barton, D.E.; Cockbuen, D.J.; Taylor, G.R. The detection of large deletions or duplications in genomic DNA. Hum. Mutat. 2002, 20, 325–337. [Google Scholar] [CrossRef]
- Marquis-Nicholson, R.; Doherty, E.; Thrush, A.; Love, J.M.; Lan, C.-C.; George, A.M.; Love, D.R. Array-based identification of copy number changes: Simultaneous gene-focused and low resolution whole human genome analysis. Sultan Qaboos Univ. Med. J. 2013, 13, 69–79. [Google Scholar]
- Bird, T.D. Charcot-Marie-Tooth Neuropathy Type 1; Pagon, R.A., Bird, T.D., Dolan, C.R., Eds.; GeneReviews™: Seattle, WA, USA, 1993; Available online: http://www.ncbi.nlm.nih.gov/books/ NBK1205/ (accessed on 1 February 2013).
- Bird, T.D. Hereditary Neuropathy with Liability to pressure Palsies; Pagon, R.A., Bird, T.D., Dolan, C.R., Eds.; GeneReviews™: Seattle, WA, USA, 1993; Available online: http://www.ncbi.nlm.nih.gov/books/NBK1392/ (accessed on 1 February 2013).
- Flanigan, K.M.; Dunn, D.M.; von Niederhausern, A.; Soltanzadeh, P.; Gappmaier, E.; Howard, M.T.; Sampson, J.B.; Mendell, J.R.; Wall, C.; King, W.M.; et al. Mutational spectrum of DMD mutations in dystrophinopathy patients: Application of modern diagnostic techniques to a large cohort. Hum. Mutat. 2009, 30, 1657–1666. [Google Scholar] [CrossRef]
- Roche NimbleGen. Available online: http://www.nimblegen.com (accessed on 25 March 2013).
- UCSC Genome Browser. Available online: http://genome.ucsc.edu (accessed on 25 March 2013).
- Aartsma-Rus, A.; van Deutekom, J.C.; Fokkema, I.F.; van Ommen, G.J.; den Dunnen, J.T. Duchenne muscular dystrophy mutation database: An overview of mutation types and paradoxical cases confirm the reading-frame rule. Muscle Nerve 2006, 34, 135–144. [Google Scholar] [CrossRef]
- Beggs, A.H.; Koenig, M.; Boyce, F.M.; Kunkel, L.M. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum. Genetic. 1990, 86, 45–48. [Google Scholar]
- Monaco, A.P.; Bertelson, C.J.; Liechti-Gallati, S.; Moser, H.; Kunkel, L.M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988, 2, 90–95. [Google Scholar]
- DMD Exonic Deletions/Duplications Reading-Frame Checker 1.9. Available online: http://www.humgen.nl/scripts/DMD_frame.php (accessed on 25 March 2013).
- Abbs, S.; Tuffery-Giraud, S.; Bakker, E.; Ferlini, A.; Sejersen, T.; Mueller, C.R. Best practice guidelines on molecular diagnostics in Duchenne/Becker muscular dystrophies. Neuromuscul. Disord. 2010, 20, 422–427. [Google Scholar] [CrossRef]
- Eng, C. PTEN Hamartoma Tumor Syndrome (PHTS); Pagon, R.A., Bird, T.D., Dolan, C.R., Eds.; GeneReviews™: Seattle, WA, USA, 1993; Available online: http://www.ncbi.nlm.nih.gov/ books/NBK1488/ (accessed on 1 February 2013).
- Chibon, F.; Primois, C.; Bressieux, J.M.; Lacombe, D.; Lok, C.; Mauriac, L.; Taieb, A.; Longy, M. Contribution of PTEN large rearrangements in Cowden disease: A multiplex amplifiable probe hybridisation (MAPH) screening approach. J. Med. Genet. 2008, 45, 657–665. [Google Scholar] [CrossRef]
- McWhinney, S.R.; Pilarski, R.T.; Forrester, S.R.; Schneider, M.C.; Sarquis, M.M.; Dias, E.P.; Eng, C. Large germline deletions of mitochondrial complex II subunits SDHB and SDHD in hereditary paraganglioma. J. Clin. Endocrinol. Metab. 2004, 89, 5694–5699. [Google Scholar] [CrossRef]
- Cascon, A.; Landa, I.; Lopez-Jimenez, E.; Dieaz-Hernandez, A.; Buchta, M.; Montero-Conde, C.; Leskela, S.; Leandro-Garcia, L.J.; Leton, R.; Rodriquez-Antona, C.; et al. Molecular characterisation of a common SDHB deletion in paraganglioma patients. J. Med. Genet. 2008, 45, 233–238. [Google Scholar]
- Bluteau, O.; Jeannot, E.; Bioulac-Sage, P.; Marques, J.M.; Blanc, J.F.; Bui, H.; Beaudoin, J.C.; Franco, D.; Balabaud, C.; Laurent-Puig, P.; Zucman-Rossi, J. Bi-allelic inactivation of TCF1 in hepatic adenomas. Nat. Genet. 2002, 32, 312–315. [Google Scholar] [CrossRef]
- Ellard, S.; Thomas, K.; Edghill, E.L.; Owens, M.; Ambye, L.; Cropper, J.; Little, J.; Strachan, M.; Stride, A.; Ersoy, B. Partial and whole gene deletion mutations of the GCK and HNF1A genes in maturity-onset diabetes of the young. Diabetologia 2007, 50, 2313–2317. [Google Scholar] [CrossRef]
- Bach, I.; Pontoglio, M.; Yaniv, M. Structure of the gene encoding hepatocytes nuclear factor 1 (HNF1). NAR 1992, 20, 4199–4204. [Google Scholar] [CrossRef]
- Del Gaudio, D.; Yang, Y.; Boggs, B.A.; Schmitt, E.S.; Lee, J.A.; Sahoo, T.; Pham, H.T.; Wiszniewska, J.; Chinault, A.C.; Beaudet, A.L.; Eng, C.M. Molecular diagnosis of Duchenne/Becker muscular dystrophy: Enhanced detection of dystrophin gene rearrangements by oligonucleotide array-comparative genomic hybridization. Hum. Mutat. 2008, 29, 1100–1107. [Google Scholar] [CrossRef]
- Bovolenta, M.; Neri, M.; Fini, S.; Fabris, M.; Trabanelli, C.; Venturoli, A.; Martoni, E.; Bassi, E.; Spitali, P.; Brioschi, S.; et al. A novel custom high density-comparative genomic hybridization array detects common rearrangements as well as deep intronic mutations in dystrophinopathies. BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Marquis-Nicholson, R.; Prosser, D.; Love, J.M.; Love, D.R. Gene Dosage Analysis in a Clinical Environment: Gene-Targeted Microarrays as the Platform-of-Choice. Microarrays 2013, 2, 51-62. https://doi.org/10.3390/microarrays2020051
Marquis-Nicholson R, Prosser D, Love JM, Love DR. Gene Dosage Analysis in a Clinical Environment: Gene-Targeted Microarrays as the Platform-of-Choice. Microarrays. 2013; 2(2):51-62. https://doi.org/10.3390/microarrays2020051
Chicago/Turabian StyleMarquis-Nicholson, Renate, Debra Prosser, Jennifer M. Love, and Donald R. Love. 2013. "Gene Dosage Analysis in a Clinical Environment: Gene-Targeted Microarrays as the Platform-of-Choice" Microarrays 2, no. 2: 51-62. https://doi.org/10.3390/microarrays2020051