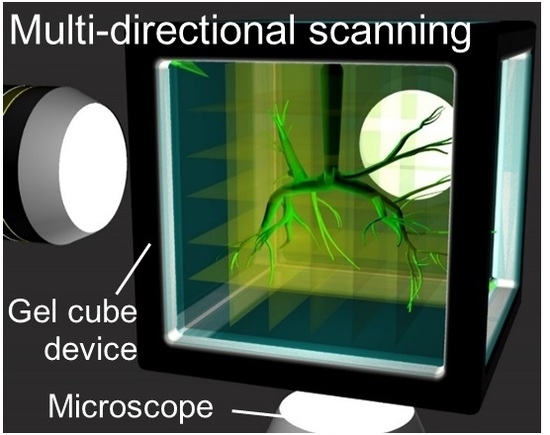
Large Scale Imaging by Fine Spatial Alignment of Multi-Scanning Data with Gel Cube Device
Abstract
:1. Introduction
2. Materials and Methods
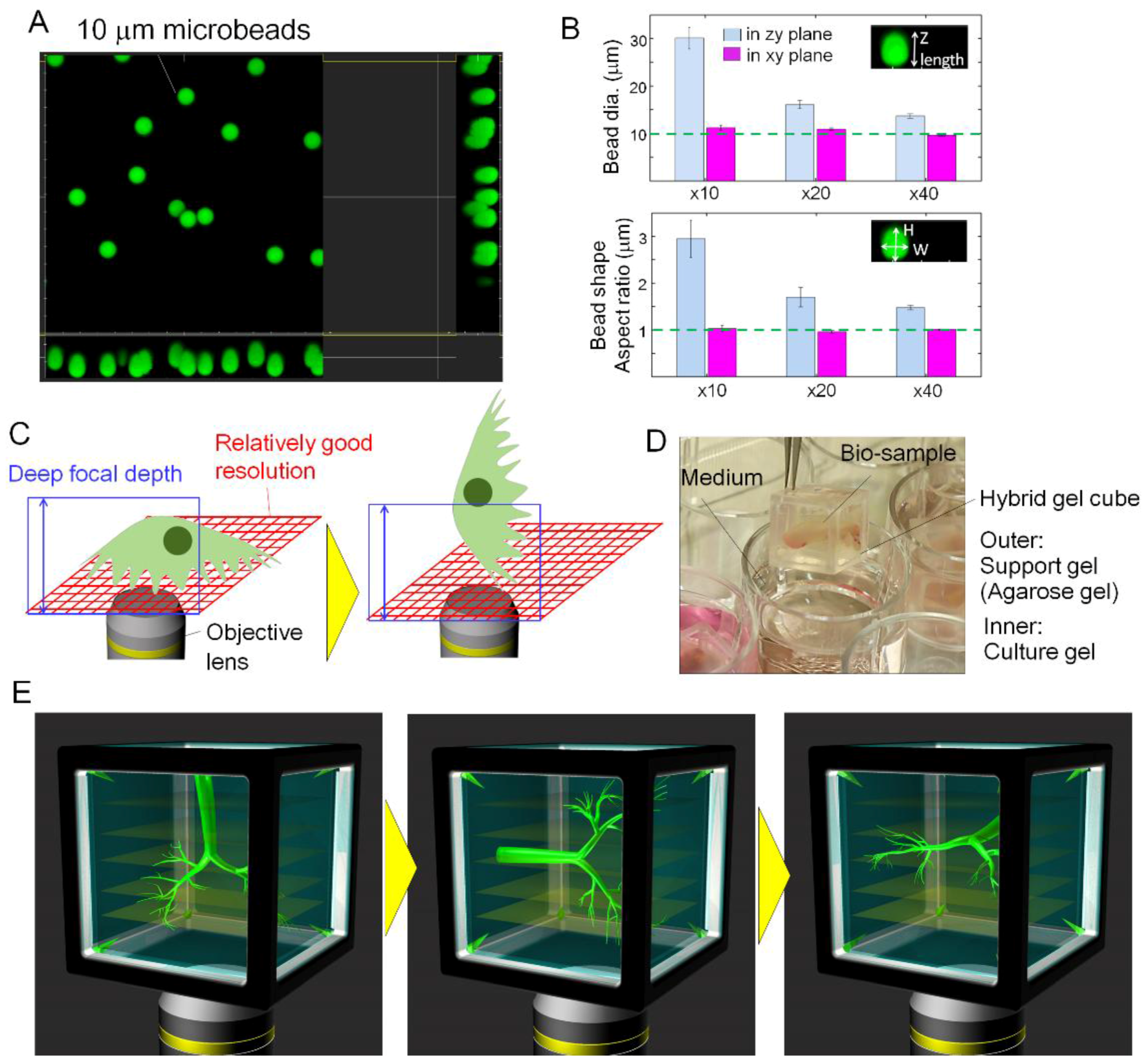
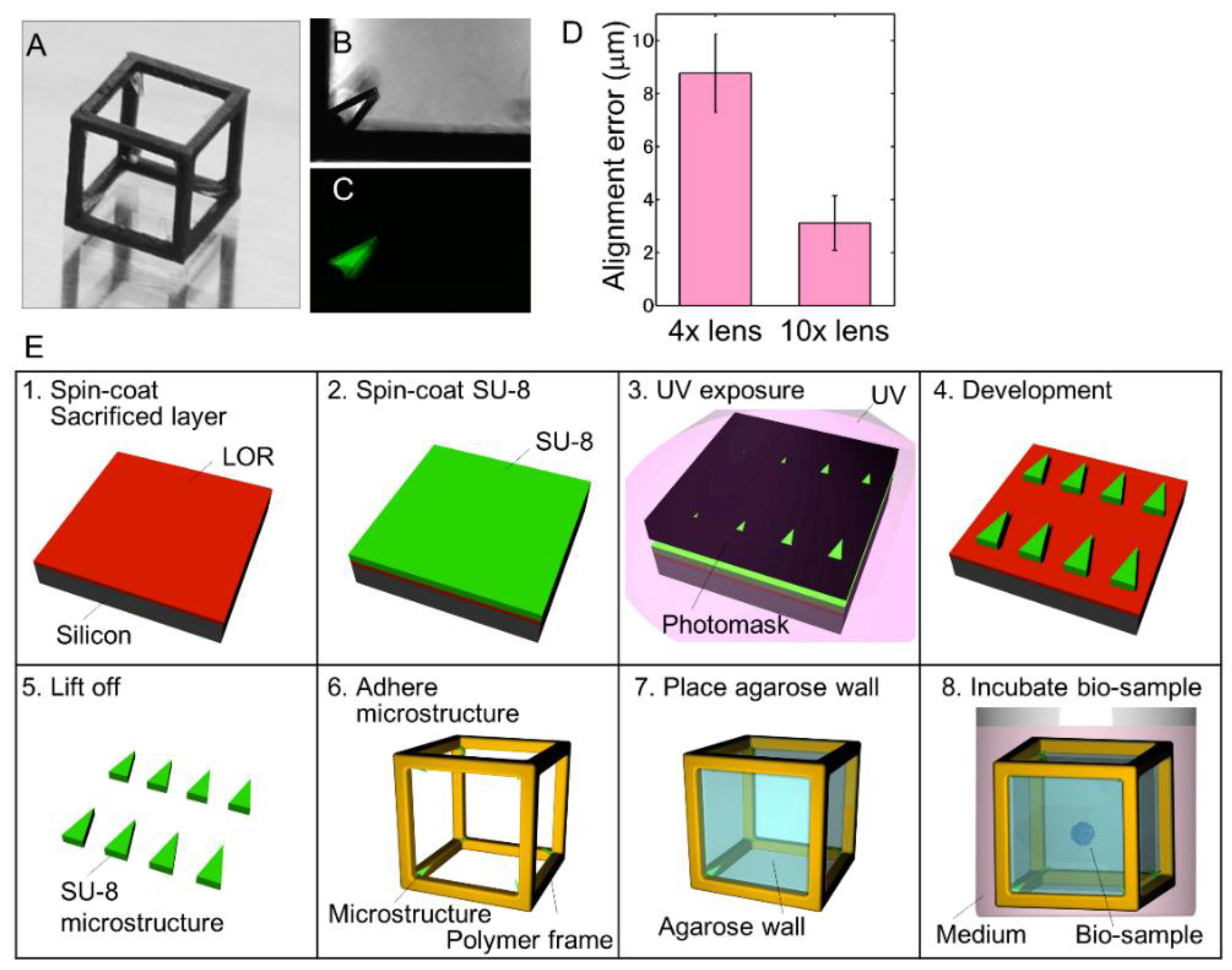
2.1. Fabrication of the Gel Cube Device with Alignment Mark
2.2. Three Dimensional Culture of NHBE in the Gel Cube Device
2.3. Immunostaining
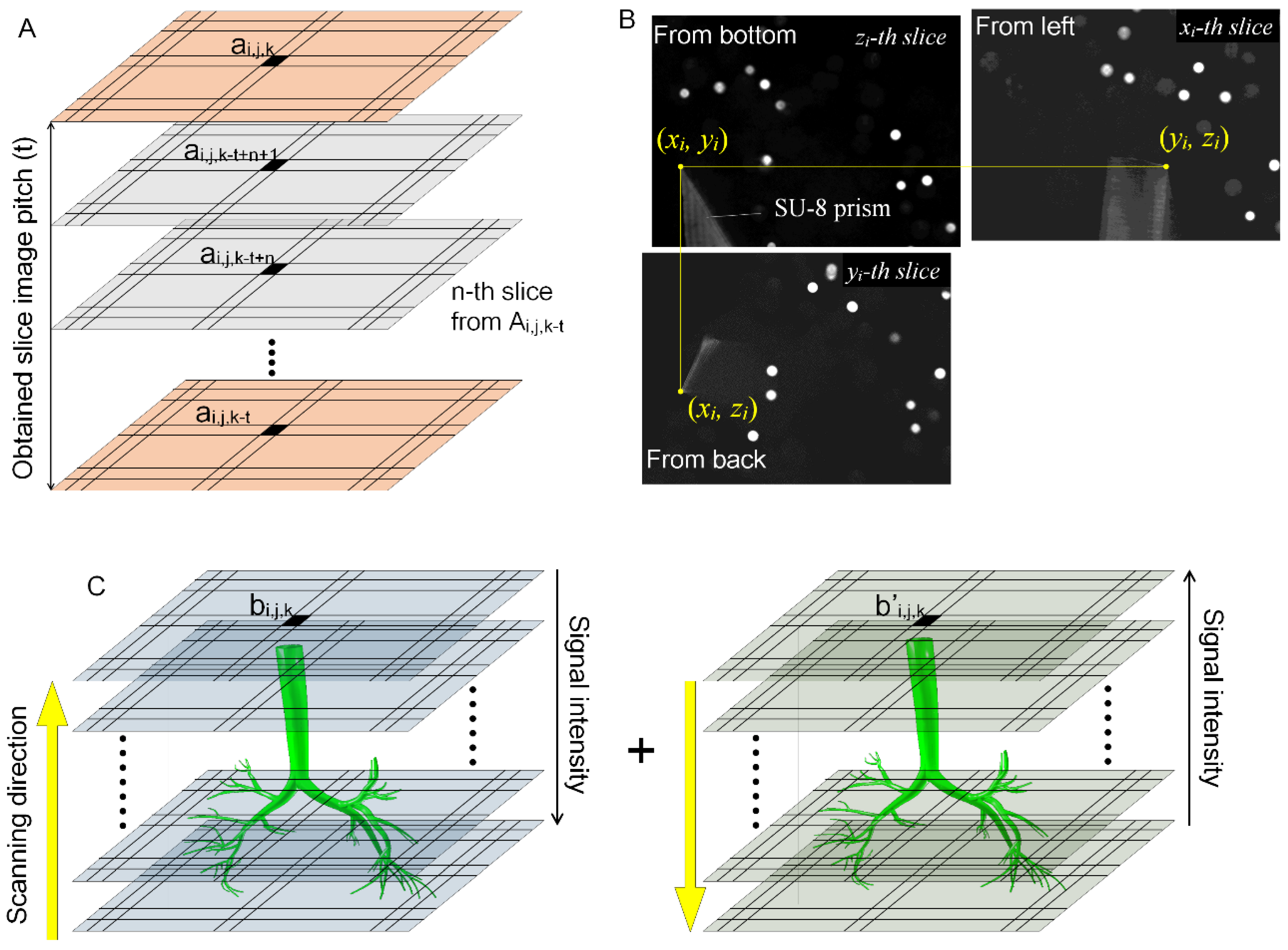
2.4. Spatial Alignments of Scanning Dataset by Self-Fluorescent Microstructure
2.5. Synchronizing Projected Section Images of the x-y, y-z and z-x Planes
2.6. Reconstruction of a 3D Image from Multi-Directional Imaging
3. Results
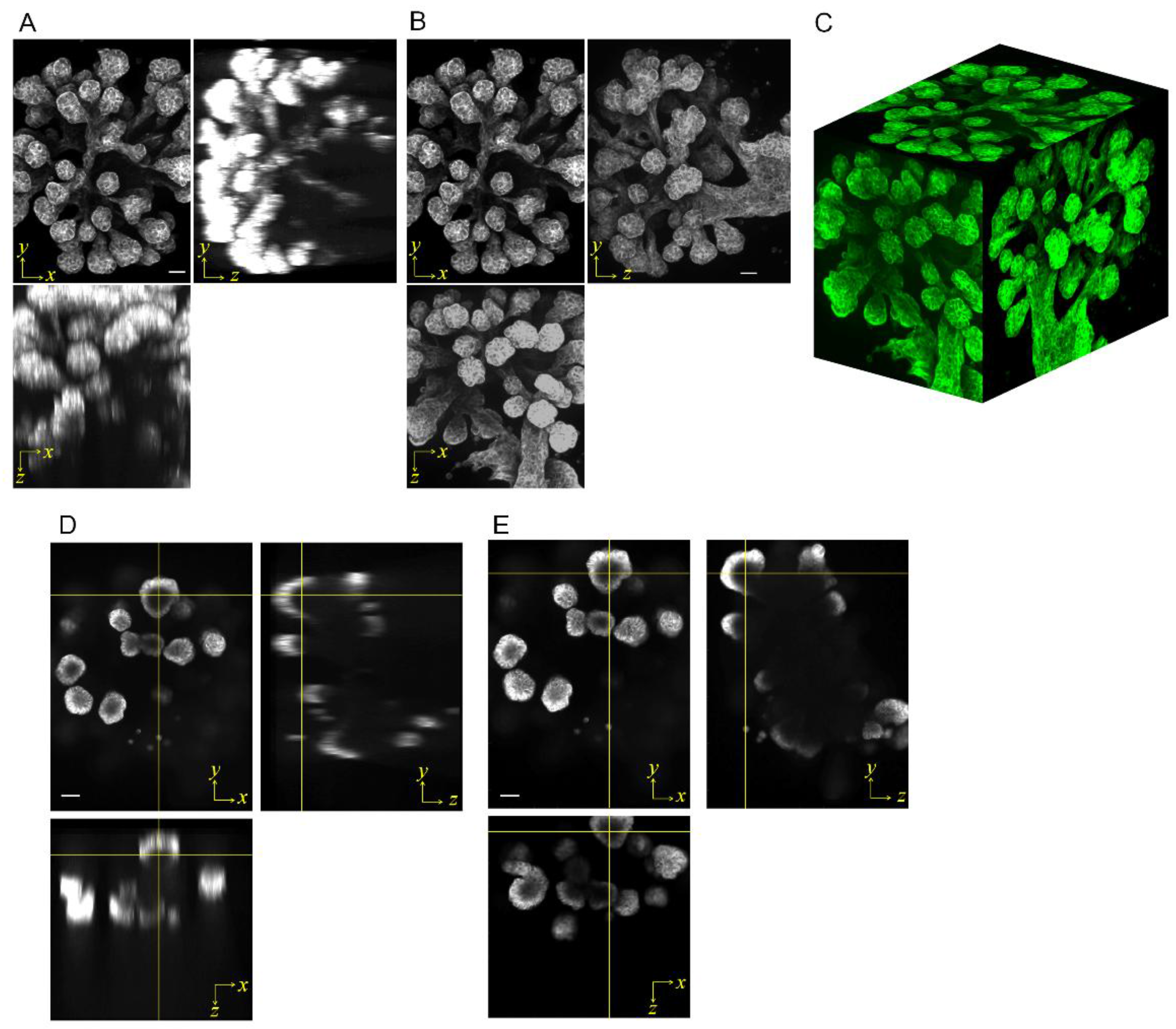
3.1. Large-Scale 3D Imaging of Tissue Sample with a Low Magnification Lens
3.2. Self-Fluorescent Microstructure for Spatial Alignment of Multi-Directional Images
3.3. Multi-Directional Imaging of a Large Branching Structure with a Low Magnification Lens
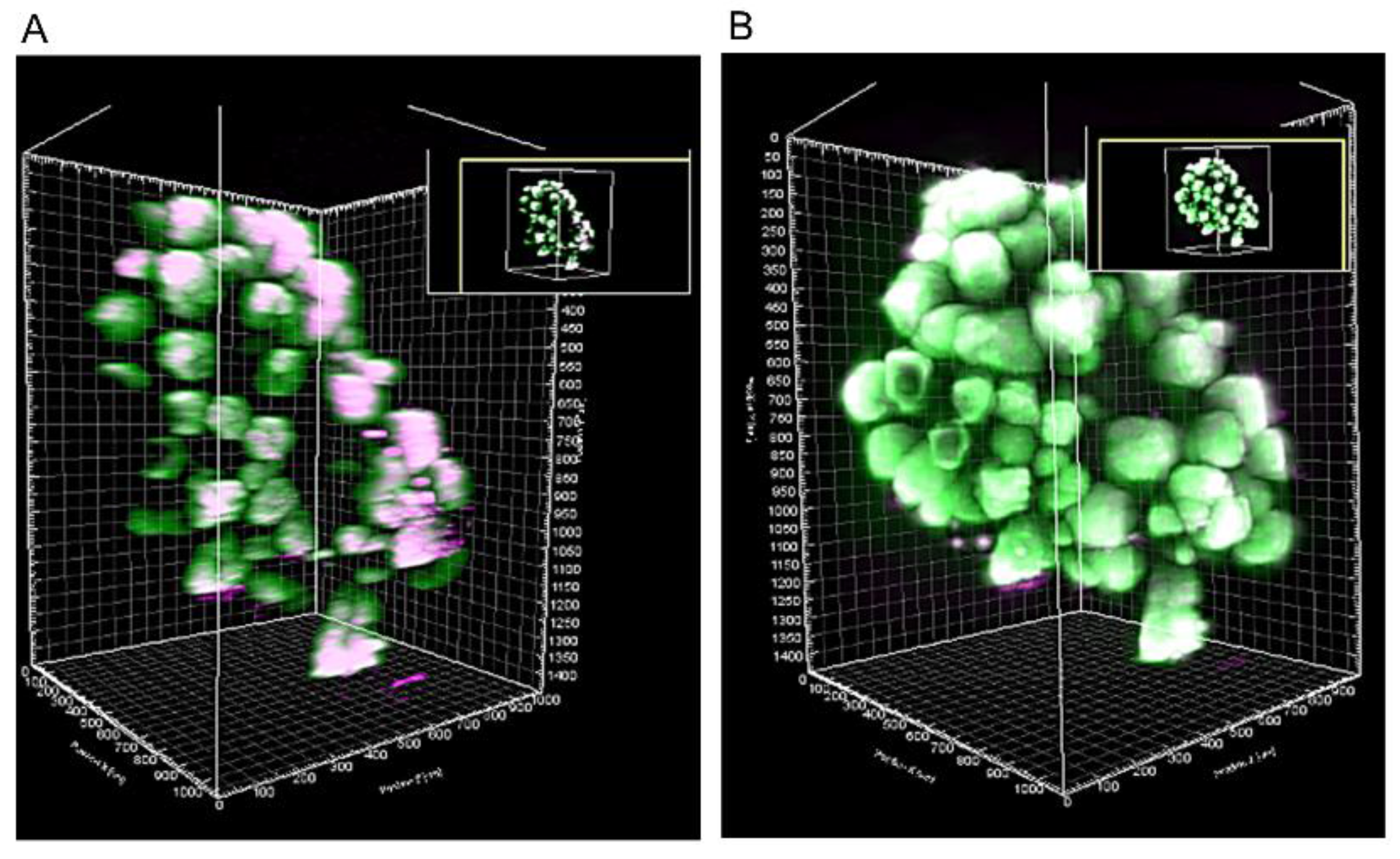
3.4. Reconstructed 3D Images of Large-Sized Branch Structures
4. Discussion and Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
References
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension—How 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Watt, F.M.; Huck, W.T.S. Role of the extracellular matrix in regulating stem cell fate. Nat. Rev. Mol. Cell Biol. 2013, 14, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Gattazzo, F.; Urciuolo, A.; Bonaldo, P. Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta 2014, 1840, 2506–2519. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, T.; Lenne, P.F. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 2007, 8, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Tawk, M.; Araya, C.; Lyons, D.A.; Reugels, A.M.; Girdler, G.C.; Bayley, P.R.; Hyde, D.R.; Tada, M.; Clarke, J.D. A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature 2007, 466, 797–800. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cupmorphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Wyngaarden, L.A.; Vogeli, K.M.; Ciruna, B.G.; Wells, M.; Hadjantonakis, A.K.; Hopyan, S. Oriented cell motility and division underlie early limb bud morphogenesis. Development 2011, 137, 2551–2558. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lacoche, S.; Huang, L.; Xue, B.; Muthuswamy, S.K. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. Proc. Natl. Acad. Sci. USA 2013, 110, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Minsky, M. Memoir on inventing the confocal scanning microscope. Scanning 1988, 10, 128–138. [Google Scholar] [CrossRef]
- Denk, W.; Strickler, D.H.; Webb, W.W. Two-photon laser scanning fluorescence microscopy. Science 1990, 248, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Huisken, J.; Swoger, J.; Del Bene, F.; Wittbrodt, J.; Stelzer, E.H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 2004, 305, 1007–1009. [Google Scholar] [CrossRef] [PubMed]
- Verveer, P.J.; Swoger, J.; Pampaloni, F.; Greger, K.; Marcello, M.; Stelzer, E.H. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat. Methods 2007, 4, 311–313. [Google Scholar] [CrossRef] [PubMed]
- Andilla, J.; Jorand, R.; Olarte, O.E.; Dufour, A.C.; Cazales, M.; Montagner, Y.L.; Ceolato, R.; Riviere, N.; Olivo-Marin, J.C.; Loza-Alvarez, P.; et al. Imaging tissue-mimic with light sheet microscopy: A comparative guideline. Sci. Rep. 2017, 7, 44939. [Google Scholar] [CrossRef] [PubMed]
- Teshima, T.; Onoe, H.; Tottori, S.; Aonuma, H.; Mizutani, T.; Kamiya, K.; Ishihara, H.; Kanuka, H.; Takeuchi, S. High-Resolution Vertical Observation of Intracellular Structure Using Magnetically Responsive Microplates. Small 2013, 12, 3366–3373. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, M.; Kawahara, T.; Nobata, R. Tissue in Cube: In Vitro 3D Culturing Platform with Hybrid Gel Cubes for Multidirectional Observations. Adv. Healthc. Mater. 2016, 5, 1566–1571. [Google Scholar] [CrossRef] [PubMed]
- Preibisch, S.; Saalfeld, S.; Schindelin, J.; Tomancak, P. Software for bead-based registration of selective plane illumination microscopy data. Nat. Methods 2010, 7, 418–419. [Google Scholar] [CrossRef] [PubMed]
- Chhetri, R.K.; Amat, F.; Wan, Y.; Hockendorf, B.; Lemon, W.C.; Keller, P.J. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat. Methods 2015, 12, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Short, K.; Hodson, M.; Smyth, I. Spatial mapping and quantification of developmental branching morphogenesis. Development 2013, 140, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Pallotto, M.; Watkins, P.V.; Fubara, B.; Singer, J.H.; Briggman, K.L. Extracellular space preservation aids the connectomic analysis of neural circuits. eLife 2015, 4, E08206. [Google Scholar] [CrossRef] [PubMed]
- Tainaka, K.; Kubota, S.I.; Suyama, T.Q.; Susaki, E.A.; Perrin, D.; Tadenuma, M.U.; Ukai, H.; Ueda, H.R. Whole-Body Imaging with Single-Cell Resolution by Tissue Decolorization. Cell 2014, 159, 911–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, E.; Cho, J.H.; Goodwin, D.; Ku, T.; Swaney, J.; Kim, S.Y.; Choi, H.; Park, Y.G.; Park, J.Y.; Hubbert, A.; et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 2015, 163, 1500–1514. [Google Scholar] [CrossRef] [PubMed]
- Hell, S.; Reiner, G.; Cremer, C.; Stelzer, E.H.K. Aberrations in confocal fluorescence microscopy induced by mismatches in refractive index. J. Microsc. 1993, 169, 391–405. [Google Scholar] [CrossRef]
- Hagiwara, M.; Peng, F.; Ho, C.M. In vitro reconstruction of branched tubular structures from lung epithelial cells in high cell concentration gradient environment. Sci. Rep. 2014, 5, 8054. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, K.; Shinohara, H.; Hashimoto, T.; Yokoi, T.; Uno, K. An iterative reconstruction using median root prior and anatomical prior from the segmented m-map for count-limited transmission data in PET imaging. Ann. Nucl. Med. 2008, 22, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Kudo, H.; Suzuki, T.; Rashed, E.A. Image reconstruction for sparse-view CT and interior CT—Introduction to compressed sensing and differentiated backprojection. Quant. Imaging Med. Surg. 2013, 3, 147–161. [Google Scholar] [PubMed]
- Hagiwara, M.; Maruta, N.; Marumoto, M. In Vitro Experimental Model for the Long-Term Analysis of Cellular Dynamics during Bronchial Tree Development from Lung Epithelial Cells. Tissue Eng. Part C Methods 2017, 23, 323–332. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hagiwara, M.; Nobata, R.; Kawahara, T. Large Scale Imaging by Fine Spatial Alignment of Multi-Scanning Data with Gel Cube Device. Appl. Sci. 2018, 8, 235. https://doi.org/10.3390/app8020235
Hagiwara M, Nobata R, Kawahara T. Large Scale Imaging by Fine Spatial Alignment of Multi-Scanning Data with Gel Cube Device. Applied Sciences. 2018; 8(2):235. https://doi.org/10.3390/app8020235
Chicago/Turabian StyleHagiwara, Masaya, Rina Nobata, and Tomohiro Kawahara. 2018. "Large Scale Imaging by Fine Spatial Alignment of Multi-Scanning Data with Gel Cube Device" Applied Sciences 8, no. 2: 235. https://doi.org/10.3390/app8020235