Differentiating Authentic Adenophorae Radix from Its Adulterants in Commercially-Processed Samples Using Multiplexed ITS Sequence-Based SCAR Markers
Abstract
:1. Introduction
2. Materials and Methods
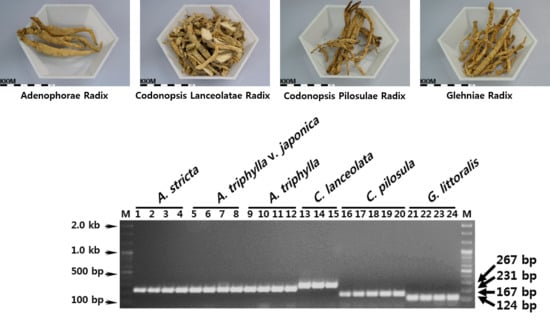
2.1. Plant and Herbal Materials
2.2. Preparation of Genomic DNA and PCR Amplification
2.3. Analysis of Nucleotide Sequences and Phylogenetic Relationships
2.4. Development of the Multiplexed SCAR Marker Assay and Monitoring of Commercial Herbal Medicines
3. Results
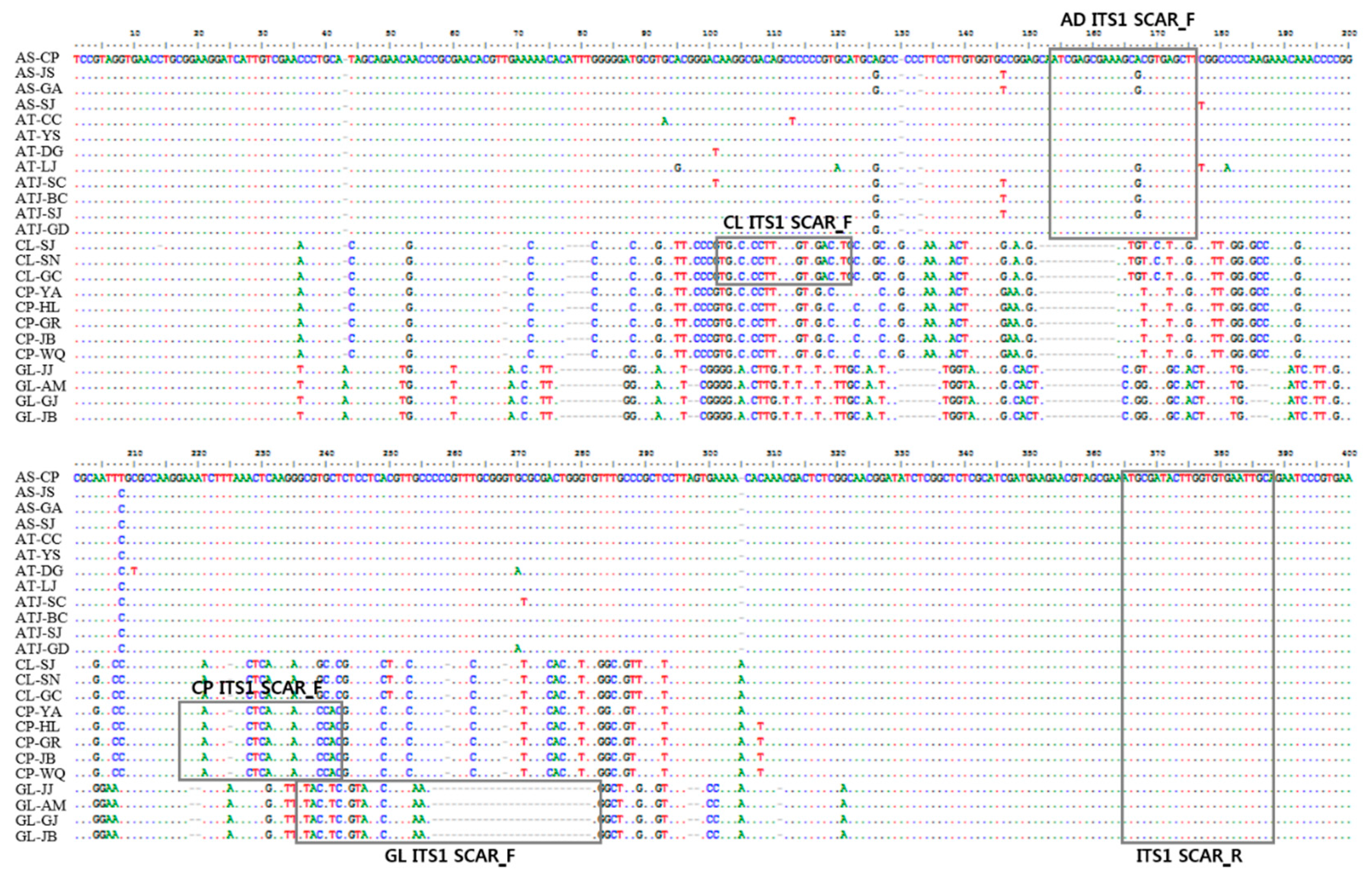
3.1. Analysis of nrDNA-ITS Sequences
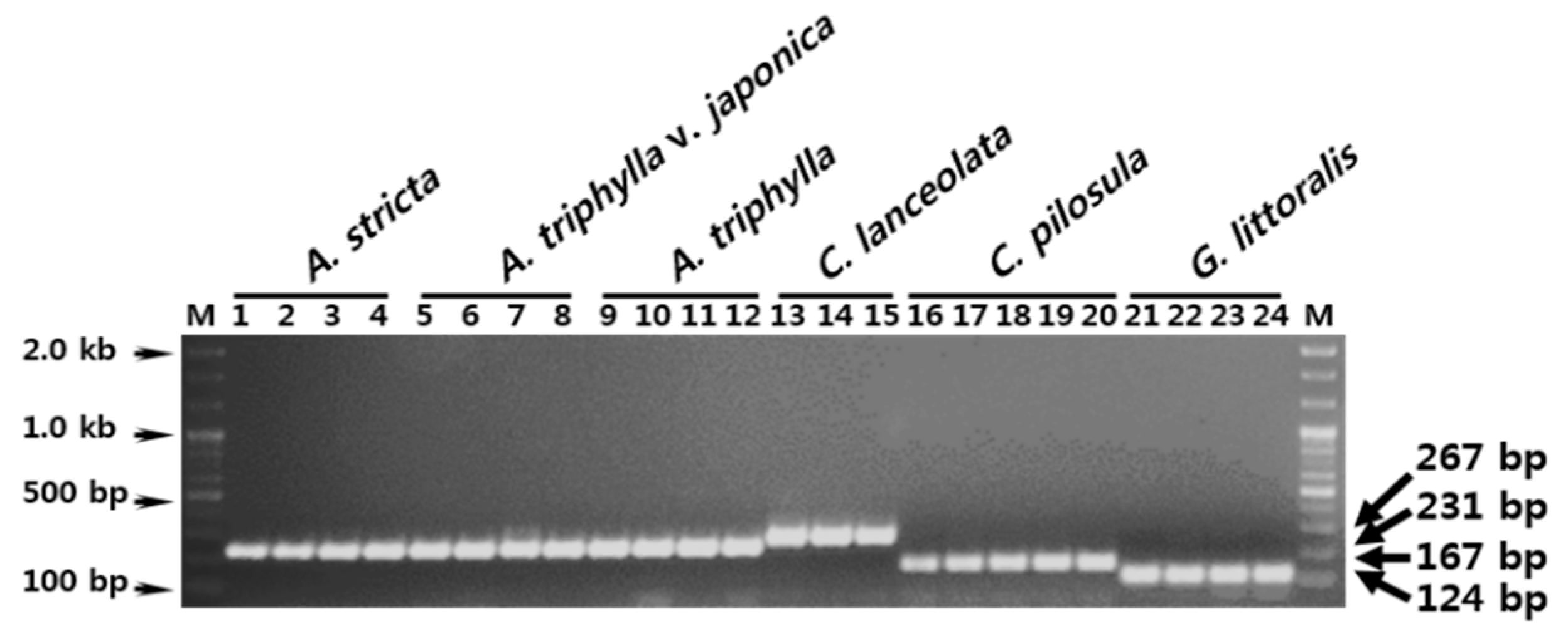
3.2. Development of SCAR Markers for Distinguishing Herbal Medicines
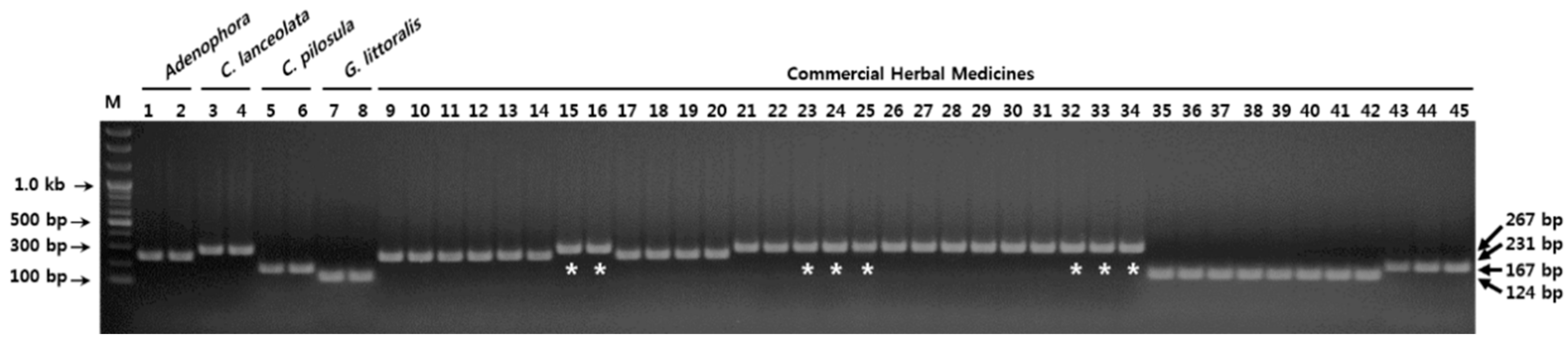
3.3. Establishment of a Multiplex SCAR Amplification Method and Monitoring of Commercially-Processed Herbal Medicines
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ali, M.A.; Gyulai, G.; Al-Hermaid, F. Plant DNA Barcoding and Phyogenetics; LAP LAMBERT Academic Publishing: Saarbrücken, Germany, 2015; pp. 109–130. [Google Scholar]
- Mitra, S.; Kannan, R. A Note on Unintentional Adulterations in Ayurvedic Herbs. Ethnobot. Leafl. 2007, 2007, 3. [Google Scholar]
- Zhu, X.; Zhang, Y.; Liu, X.; Hou, D.; Gao, T. Authentication of Commercial Processed Glehniae Radix (Beishashen) by DNA Barcodes. Chin. Med. 2015, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.J.; Dong, T.T.X.; Cui, X.M.; Tu, P.F.; Tsim, K.W.K. Genetic Distinction of Radix Adenophorae from Its Adulterants by the DNA Sequence of 5S-rRNA Spacer Domains. Am. J. Chin. Med. 2003, 31, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.I.; Zhao, B.T.; Lee, J.H.; Lee, D.U.; Kim, Y.S.; Min, B.S.; Son, J.K.; Woo, M.H. Quantitative and Classification Analyses of Lupenone and β-Sitosterol by GC-FID in Adenophora triphylla var. japonica Hara and Codonopsis lanceolata. Nat. Prod. Sci. 2014, 20, 243–250. [Google Scholar]
- The Plant List. Available online: http://www.theplantlist.org/tpl1.1/search?q=adenophora (accessed on 10 April 2017).
- Defining Dictionary for Medicinal Herbs. Available online: http://boncho.kiom.re.kr/herbarium/codex.php (accessed on 10 April 2017).
- Lee, M.Y.; Mo, S.Y.; Kim, D.W.; Oh, S.E.; Ko, B.S. Discrimination and Genetic Relationship of Adenophora triphylla (Thunb) A. DC. var. japonica Hara and Codonopsis lanceolata Trauty Using RAPD Analysis. Korean J. Med. Crop. Sci. 2001, 9, 205–210. [Google Scholar]
- Kim, J.Y.; Lee, Y.J. A Study on a Morphological Identification of Adenophora triphylla var. japonica, Codonopsis lanceolata, Adenophora remotiflora and Codonopsis pilosula. Korea J. Herbol. 2007, 22, 121–126. [Google Scholar]
- Bensky, D.; Clavey, S.; Stoger, E.; Gamble, A. Chinese Herbal Medicine Materia Medica; Eastland Press: Seattle, WA, USA, 1993; pp. 714–717, 818–822. [Google Scholar]
- Min, S.H.; Han, H.S.; Lee, Y.J. Study on the Anti-oxidative Effects of Adenophorae Radix, Codonopsis lanceolatae Radix and Glehniae Radix cum Rhizoma on Liver Cells Isolated from Oxidatively Stressed Rat. Korea J. Herbol. 2009, 24, 109–119. [Google Scholar]
- Austerlitz, F.; David, O.; Schaeffer, B.; Bleakley, K.; Olteanu, M.; Leblois, R.; Veuille, M.; Laredo, C. DNA Barcode Analysis: A Comparison of Phylogenetic and Statistical Classification Methods. BMC Bioinform. 2009, 10 (Suppl. 14), S10. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA Barcoding: An Efficient Tool to Overcome Authentication Challenges in the Herbal Market. Plant Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Pang, X.; Song, J.; Shi, L.; Yao, H.; Han, J.; Leon, C. A Renaissance in Herbal Medicine Identification: From Morphology to DNA. Biotechnol. Adv. 2014, 32, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Sucher, N.J.; Carles, M.C. Genome-based Approaches to the Authentication of Medicinal Plants. Planta Med. 2008, 74, 603–623. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and Using a Plant DNA Barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- Moon, B.C.; Kim, W.J.; Ji, Y.; Lee, Y.M.; Kang, Y.M.; Choi, G. Molecular Identification of the Traditional Herbal Medicines, Arisaematis Rhizoma and Pinelliae Tuber, and Common Adulterants via Universal DNA Barcode Sequences. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Group, C.P.W.; Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L. A DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M. Refining the DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19451–19452. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Song, J.; Liu, C.; Luo, K.; Han, J.; Li, Y.; Pang, X.; Xu, H.; Zhu, Y.; Xiao, P.; et al. Use of ITS2 Region as the Universal DNA Barcode for Plants and Animals. PLoS ONE 2010, 5, e13102. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.Y.; Chan, G.K.; Xin, G.Z.; Xu, H.; Ku, F.; Chen, J.P.; Yao, P.; Lin, H.Q.; Dong, T.T.; Tsim, K.W. Comparison of ITS Sequence Analysis and RAPD-Derived Molecular Markers. Molecules 2015, 20, 22454–22462. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Moon, B.C.; Yang, S.; Han, K.S.; Choi, G.; Lee, A.Y. Rapid Authentication of the Herbal Medicine Plant Species Aralia continentalis Kitag. and Angelica biserrata CQ Yuan and RH Shan Using ITS2 Sequences and Multiplex-SCAR Markers. Molecules 2016, 21, 270. [Google Scholar] [CrossRef] [PubMed]
- Xin, G.Z.; Lam, Y.C.; Maiwulanjiang, M.; Chan, G.K.; Zhu, K.Y.; Tang, W.L.; Dong, T.T.X.; Shi, Z.Q.; Li, P.; Tsim, K.W. Authentication of Bulbus Fritillariae Cirrhosae by RAPD-derived DNA Markers. Molecules 2014, 19, 3450–3459. [Google Scholar] [CrossRef] [PubMed]
- Bhagyawant, S.S. RAPD-SCAR Markers: An Interface Tool for Authentication of Traits. J. Biosci. Med. 2016, 4, 1–9. [Google Scholar] [CrossRef]
- Lee, Y.M.; Ji, Y.; Kang, Y.M.; Kim, W.J.; Choi, G.; Moon, B.C. Molecular Authentication of Pinelliae Tuber and Its Common Adulterants Using RAPD-derived Multiplex Sequence Characterized Amplified Region (multiplex-SCAR) Markers. Int. J. Clin. Exp. Med. 2016, 9, 40–50. [Google Scholar]
- Moon, B.C.; Lee, Y.M.; Kim, W.J.; Ji, Y.; Kang, Y.M.; Choi, G. Development of Molecular Markers for Authentication of the Medicinal Plant Species Patrinia by Random Smplified Polymorphic DNA (RAPD) Analysis and Multiplex-PCR. Hortic. Environ. Biotechnol. 2016, 57, 182–190. [Google Scholar] [CrossRef]
- Kim, W.J.; Ji, Y.; Choi, G.; Kang, Y.M.; Yang, S.; Moon, B.C. Molecular Identification and Phylogenetic Analysis of Important Medicinal Plant Species in Genus Paeonia Based on rDNA-ITS, matK, and rbcL DNA Barcode Sequences. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Hall, T.A. BioEdit: A User-friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D. BOLD: The Barcode of Life Data. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Särkinen, T.; Staats, M.; Richardson, J.E.; Cowan, R.S.; Bakker, F.T. How to Open the Treasure Chest? Optimising DNA Extraction from Herbarium Dpecimens. PLoS ONE 2012, 7, e43808. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Moon, B.C.; Lee, A.Y.; Chun, J.M.; Choo, B.K.; Kim, H.K. Molecular Phylogenetic Position of Adenophora racemosa, an Endemic Species in Korea. Korean J. Med. Crop. Sci. 2010, 18, 379–388. [Google Scholar]

| Name | Habitat Information | Voucher Number | Abbreviation | Lane in Gel | |
|---|---|---|---|---|---|
| Scientific Name (Family) | Herbal Name | ||||
| Adenophora stricta Miq. (Campanulaceae) | Adenophorae Radix | Caoping, Li, Longnan, Gansu, China | KIOM200701000519 | AS-CP | 1 |
| Jisan, Jindo, Jeonnam, Korea | KIOM200701000873 | AS-JS | 2 | ||
| Guyuan, Ningxia, China | KIOM201201005303 | AS-GA | 3 | ||
| Sangju, Gyeongbuk, Korea | KIOM201301006488 | AS-SJ | 4 | ||
| Adenophora triphylla (Thunb.) A. DC. (Campanulaceae) | Adenophorae Radix | Cheoncheon, Jangsu, Jeonbuk, Korea | KIOM200701000682 | AT-CC | 5 |
| Yaksan, Wando, Jeonnam, Korea | KIOM200701000741 | AT-YS | 6 | ||
| Daegang, Danyang, Chungbuk, Korea | KIOM201101004148 | AT-DG | 7 | ||
| Longjing, Yanbian, Jilin, China | KIOM201201005563 | AT-LJ | 8 | ||
| Adenophora triphylla var. japonica (Regel) Hara (Campanulaceae) | Adenophorae Radix | Sancheong, Gyeongnam, Korea | KIOM201201004951 | ATJ-SC | 9 |
| Beichuan, Mianyang, Sichuan, China | KIOM201201005359 | ATJ-BC | 10 | ||
| Sangju, Gyeongbuk, Korea | KIOM201301006312 | ATJ-SJ | 11 | ||
| Gandong, Hwacheon, Gangwon, Korea | KIOM201401009347 | ATJ-GD | 12 | ||
| Codonopsis lanceolata (Siebold & Zucc.) Benth. & Hook. f. ex Trautv. (Campanulaceae) | Codonopsis Lanceolatae Radix | Sangju, Namhae, Gyeongnam, Korea | KIOM201001002725 | CL-SJ | 13 |
| Sinan, Jeonnam, Korea | KIOM201101003615 | CL-SN | 14 | ||
| Geochang, Gyeongnam, Korea | KIOM201201004752 | CL-GC | 15 | ||
| Codonopsis pilosula (Franch.) Nannf. (Campanulaceae) | Codonopsis Pilosulae Radix | Ya’an, Sichuan, China | KIOM201101004238 | CP-YA | 16 |
| Helong, Yanbian, Jilin, China | KIOM201101004335 | CP-HL | 17 | ||
| Girin, Inje, Gangwon, Korea | KIOM200701000339 | CP-GR | 18 | ||
| Jinbu, Pyeongchang, Gangwon, Korea | KIOM200801001104 | CP-JB | 19 | ||
| Wangqing, Yanbian, Jilin, China | KIOM201101004270 | CP-WQ | 20 | ||
| Glehnia littoralis F. Schmidt ex Miq. (Apiaceae) | Glehniae Radix | Gujwa, Jeju, Jeju, Korea | KIOM200601000041 | GL-JJ | 21 |
| Anmyeon, Taean, Chungnam, Korea | KIOM200701000230 | GL-AM | 22 | ||
| Geojin, Goseong, Gangwon, Korea | KIOM200701000231 | GL-GJ | 23 | ||
| Jukbyeon, Uljin, Gyeongbuk, Korea | KIOM200601000062 | GL-JB | 24 | ||
| Species | Sequence Length (bp) | Intra-Species Variability (%) | Inter-Species Variability (%) | |||
|---|---|---|---|---|---|---|
| Constant | Aligned | ITS1 | ITS2 | ITS1 | ITS2 | |
| A. stricta | 793–795 | 799 | 0.0205 ± 0.0104 | 0.0198 ± 0.0114 | 0.2840 ± 0.2322 | 0.2657 ± 0.2132 |
| A. triphylla | 793–795 | 799 | 0.0111 ± 0.0062 | 0.0198 ± 0.0101 | 0.2760 ± 0.2279 | 0.2661 ± 0.2169 |
| A. triphylla var. japonica | 793–795 | 799 | 0.0092 ± 0.0061 | 0.0192 ± 0.0094 | 0.2778 ± 0.2292 | 0.2686 ± 0.2184 |
| C. lanceolata | 743 | 799 | 0.0000 ± 0.0000 | 0.0029 ± 0.0025 | 0.3566 ± 0.1824 | 0.3233 ± 0.1893 |
| C. pilosula | 743 | 799 | 0.0047 ± 0.0061 | 0.0035 ± 0.0045 | 0.3480 ± 0.1560 | 0.3375 ± 0.1678 |
| G. littoralis | 689 | 799 | 0.0023 ± 0.0025 | 0.0000 ± 0.0000 | 0.5810 ± 0.0122 | 0.5717 ± 0.0133 |
| Primer Direction | Primer Name | Primer Sequence (5′ → 3′) | Specificity | Amplicon Size (bp) |
|---|---|---|---|---|
| Forward | AD ITS1 SCAR F | ATC GAG CGA AAG CGC GTG AGC T | Adenophora sp. | 231 |
| CL ITS1 SCAR F | TGG CCC CTT GCC GTC GAC CT | C. lanceolata | 267 | |
| CP ITS1 SCAR F | AAA ACT TAA CTC AAA GAG CGC GA | C. pilosula | 167 | |
| GL ITS1 SCAR F | GTA CGT CCG TAT CCC GTT AAG G | G. littoralis | 124 | |
| Reverse | ITS1 SCAR R | GCA ATT CAC ACC AAG TAT CGC AT | All samples | - |
| No. | Voucher No. | Product Name | Multiplex-SCAR Result | Country |
|---|---|---|---|---|
| 1 | 2-15-0678 | Adenophorae Radix | Adenophorae Radix | Korea |
| 2 | 2-15-0679 | Adenophorae Radix | Adenophorae Radix | Korea |
| 3 | 2-15-0680 | Adenophorae Radix | Adenophorae Radix | Korea |
| 4 | 2-15-0681 | Adenophorae Radix | Adenophorae Radix | Korea |
| 5 | 2-15-0682 | Adenophorae Radix | Adenophorae Radix | Korea |
| 6 | 2-15-0683 | Adenophorae Radix | Adenophorae Radix | Korea |
| 7 | 2-15-0684 | Adenophorae Radix | Codonopsis Lanceolatae Radix | Korea |
| 8 | 2-15-0685 | Adenophorae Radix | Codonopsis Lanceolatae Radix | Korea |
| 9 | 2-15-0705 | Adenophorae Radix | Adenophorae Radix | China |
| 10 | 2-15-0706 | Adenophorae Radix | Adenophorae Radix | China |
| 11 | 2-15-0707 | Adenophorae Radix | Adenophorae Radix | China |
| 12 | 2-15-0708 | Adenophorae Radix | Adenophorae Radix | China |
| 13 | 2-15-0711 | Codonopsis Lanceolatae Radix | Codonopsis Lanceolatae Radix | Korea |
| 14 | 2-15-0712 | Codonopsis Lanceolatae Radix | Codonopsis Lanceolatae Radix | Korea |
| 15 | 2-15-0713 | Adenophorae Radix | Codonopsis Lanceolatae Radix | Korea |
| 16 | 2-15-0714 | Adenophorae Radix | Codonopsis Lanceolatae Radix | Korea |
| 17 | 2-15-0715 | Adenophorae Radix | Codonopsis Lanceolatae Radix | Korea |
| 18 | 2-15-0716 | Codonopsis Lanceolatae Radix | Codonopsis Lanceolatae Radix | Korea |
| 19 | 2-15-0717 | Codonopsis Lanceolatae Radix | Codonopsis Lanceolatae Radix | Korea |
| 20 | 2-15-0718 | Codonopsis Lanceolatae Radix | Codonopsis Lanceolatae Radix | Korea |
| 21 | 2-15-0719 | Codonopsis Lanceolatae Radix | Codonopsis Lanceolatae Radix | Korea |
| 22 | 2-15-0720 | Codonopsis Lanceolatae Radix | Codonopsis Lanceolatae Radix | Korea |
| 23 | 2-15-0721 | Codonopsis Lanceolatae Radix | Codonopsis Lanceolatae Radix | Korea |
| 24 | 2-15-0722 | Adenophorae Radix | Codonopsis Lanceolatae Radix | Korea |
| 25 | 2-15-0723 | Adenophorae Radix | Codonopsis Lanceolatae Radix | Korea |
| 26 | 2-16-0302 | Adenophorae Radix | Codonopsis Lanceolatae Radix | Korea |
| 27 | 2-15-0730 | Glehniae Radix | Glehniae Radix | Korea |
| 28 | 2-15-0731 | Glehniae Radix | Glehniae Radix | Korea |
| 29 | 2-15-0732 | Glehniae Radix | Glehniae Radix | Korea |
| 30 | 2-15-0733 | Glehniae Radix | Glehniae Radix | Korea |
| 31 | 2-15-0734 | Glehniae Radix | Glehniae Radix | Korea |
| 32 | 2-15-0735 | Glehniae Radix | Glehniae Radix | Korea |
| 33 | 2-15-0736 | Glehniae Radix | Glehniae Radix | Korea |
| 34 | 2-15-0737 | Glehniae Radix | Glehniae Radix | Korea |
| 35 | 2-17-0017 | Codonopsis Pilosulae Radix | Codonopsis Pilosulae Radix | China |
| 36 | 2-16-0025 | Codonopsis Pilosulae Radix | Codonopsis Pilosulae Radix | China |
| 37 | 2-15-0025 | Codonopsis Pilosulae Radix | Codonopsis Pilosulae Radix | China |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, B.C.; Kim, W.J.; Han, K.S.; Yang, S.; Kang, Y.; Park, I.; Piao, R. Differentiating Authentic Adenophorae Radix from Its Adulterants in Commercially-Processed Samples Using Multiplexed ITS Sequence-Based SCAR Markers. Appl. Sci. 2017, 7, 660. https://doi.org/10.3390/app7070660
Moon BC, Kim WJ, Han KS, Yang S, Kang Y, Park I, Piao R. Differentiating Authentic Adenophorae Radix from Its Adulterants in Commercially-Processed Samples Using Multiplexed ITS Sequence-Based SCAR Markers. Applied Sciences. 2017; 7(7):660. https://doi.org/10.3390/app7070660
Chicago/Turabian StyleMoon, Byeong Cheol, Wook Jin Kim, Kyeong Suk Han, Sungyu Yang, Youngmin Kang, Inkyu Park, and Renzhe Piao. 2017. "Differentiating Authentic Adenophorae Radix from Its Adulterants in Commercially-Processed Samples Using Multiplexed ITS Sequence-Based SCAR Markers" Applied Sciences 7, no. 7: 660. https://doi.org/10.3390/app7070660