Novel Fluorinated Indanone, Tetralone and Naphthone Derivatives: Synthesis and Unique Structural Features
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Instrumentation
2.3. General Procedure for the Preparation of Trifluoromethyl-β–Diketones and Triketones [12]
2.4. General Procedure for the Preparation of Selectively Fluorinated Ketones [10,13]
3. Results and Discussion
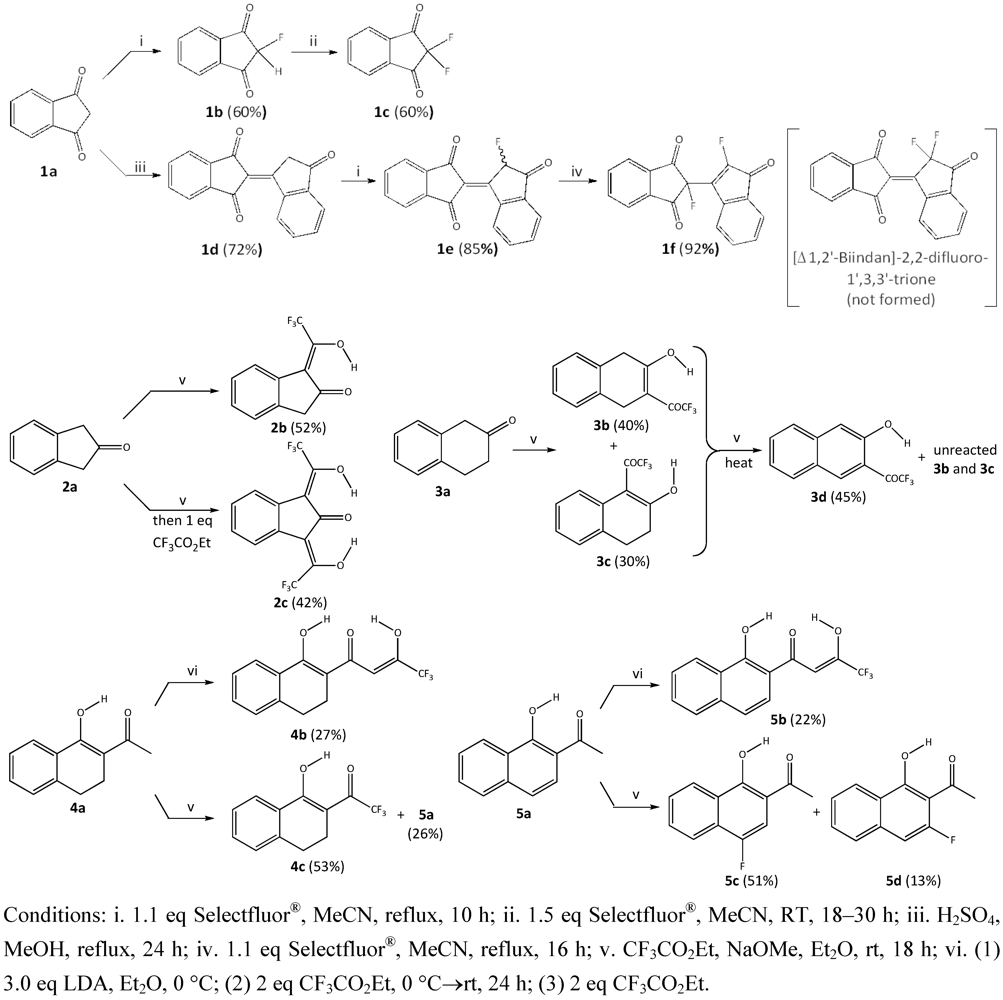
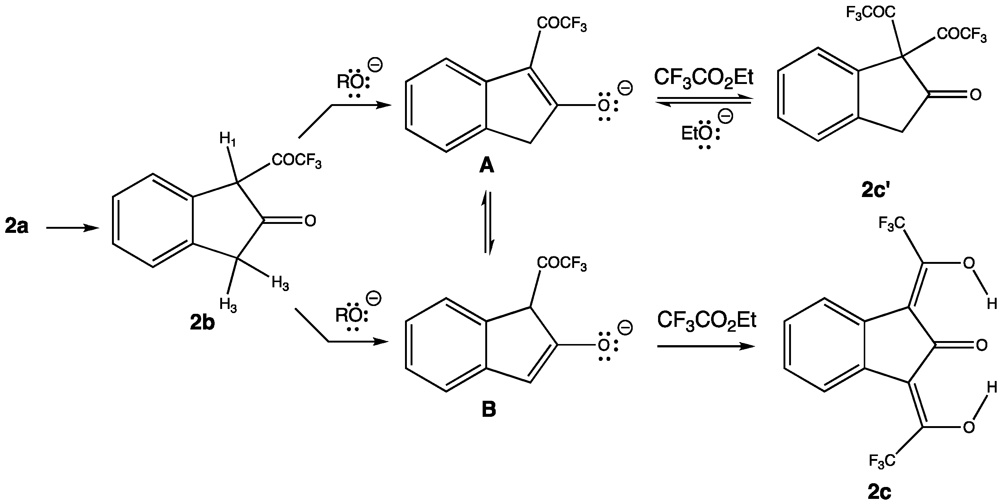
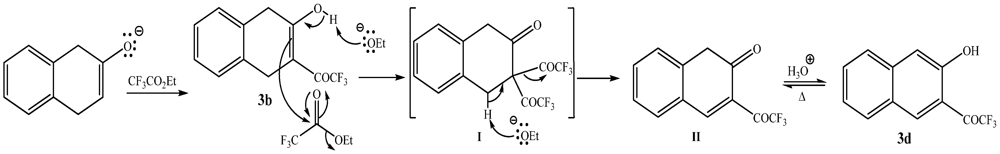
3.1. Synthesis
3.2. Solid State Structural Features: X-ray Crystallography

| Compound | 1d | 2c |
| Formula | C18H10O3 | C13H6F6O3 |
| Formula Weight (g/mol) | 274.26 | 324.18 |
| Crystal Dimensions (mm) | 0.30 × 0.24 × 0.20 | 1.20 × 0.10 × 0.06 |
| Crystal Color and Habit | clear prism | yellow needle |
| Crystal System | orthorhombic | monoclinic |
| Space Group | F d d 2 | P 21/c |
| Temperature, K | 173 | 173 |
| a, Å | 18.0996(6) | |
| b, Å | 20.9271(7) | 18.6978(12) |
| c, Å | 26.0789(8) | 13.8431(9) |
| α,° | 90.00 | 90.0 |
| β,° | 90.00 | 98.964(3) |
| Compound | 1d | 2c |
| γ,° | 90.00 | 90.0 |
| V, Å3 | 9878.0(6) | 1218.11(14) |
| Reflections to determine final unit cell | 9975 | 9959 |
| 2θ range, ° | 5.0, 56.84 | 5.28–57.7 |
| Z | 32 | 4 |
| F(000) | 4544 | 648.71 |
| ρ (g/cm) | 1.475 | 1.768 |
| λ, Å, (MoKα) | 0.71070 | 0.71073 |
| μ, (cm−1) | 0.101 | 0.18 |
| Reflections collected | 103146 | 26516 |
| Unique reflections | 6360 | 3195 |
| Rmerge | 0.0403 | 0.027 |
| Cut off Threshold Expression | >2sigma(I) | Inet > 1.0sigma(Inet) |
| Refinement method | full matrix least-sqs using F2 | full matrix least-sqs using F |
| Weighting Scheme | 1/[sigma2 (Fo2) + (0.0555P)2+3.0465P] where P = (Fo2 + 2Fc2)/3 | 1/(sigma2 (F) + 0.0005F2) |
| R1a | 0.0342 | 0.038 |
| wR2 | 0.0846 b | 0.053 c |
| R1 (all data) | 0.0400 | 0.046 |
| wR2 (all data) | 0.0880 | 0.054 |
| GOF | 1.038 d | 1.74 e |

| Compound | 3d | 4c |
| Formula | C12H7F3O2 | C12H9F3O2 |
| Formula Weight (g/mol) | 240.18 | 242.19 |
| crystal size (mm) | 0.46 × 0.08 × 0.04 | 0.38 × 0.28 × 0.04 |
| crystal color/shape | orange yellow needle | colourless plate |
| cryst syst | orthorhombic | triclinic |
| space group | P n a 21 | P-1 |
| temp, K | 110 | 110 |
| a, Å | 13.5923(5) | 7.3528(2) |
| b, Å | 14.9695(5) | 7.9165(2) |
| c, Å | 4.8381(2) | 9.7991(2) |
| α,° | 90.00 | 73.0533(11) |
| β,° | 90.00 | 85.3968(12) |
| γ,° | 90.00 | 68.3581(11) |
| V, Å3 | 984.41(6) | 506.92(2) |
| Reflections to final unit cell | 5859 | 6416 |
| 2θ range, ° | 5.44–52.66 | 5.78–71.38 |
| Z | 4 | 2 |
| F(000) | 488 | 248 |
| ρ (g/cm) | 1.621 | 1.587 |
| λ, Å, (MoKα) | 0.71070 | 0.71073 |
| μ, (cm−1) | 0.147 | 0.143 |
| Reflections collected | 21568 | 20479 |
| Unique reflections | 2632 | 4691 |
| Rmerge | 0.0444 | 0.0265 |
| Cut off Threshold Expression | >2sigma(I) | >2sigma(I) |
| refinement method | full matrix least-sqs using F2 | full matrix least-sqs using F2 |
| Weighting Scheme | 1/[sigma2 (Fo2) + (0.0406P)2 + 0.0000P] where P = (Fo2 + 2Fc2)/3 | 1/[sigma2 (Fo2) + (0.0707P)2 + 0.0436P] where P = (Fo2 + 2Fc2)/3 |
| R1a | 0.0370 | 0.0382 |
| wR2 | 0.0712 | 0.1082 |
| R1 (all data)b | 0.0538 | 0.0525 |
| wR2 (all data)a | 0.0762 | 0.1220 |
| GOF | 1.035 | 1.048 |
| Interatomic Distances (Å) | Bond Lengths (Å) | Dihedral  (◦) (◦) | ||||
|---|---|---|---|---|---|---|
| O….O | O….H | O-H | C-O | C-C | ||
| 1d | NA | NA | NA | C1A-O1A | C3A-C10A | O3-C18-C10-C3 |
| 1.2143(17) | 1.3574(19) | -2.4(11) | ||||
| C11A-O2A | ||||||
| 1.2212(17) | ||||||
| C18A-O3A | ||||||
| 1.2143(17) | ||||||
| 2c | O1….O2 | O1….H2 | O2-H2 | C1-O1 | C1-C2 | O1-C1-C2-C10 |
| 2.5781(13) | 1.75(2) | 0.90(2) | 1.2576(15) | 1.4547(15) | -1.91(11) | |
| O1….O3 | O1….H3 | O3-H3 | C10-O2 | C2-C10 | ||
| 2.5926(14) | 1.81(2) | 0.88(2) | 1.3308(15) | 1.3593(17) | ||
| C12-O3 | C9-C12 | |||||
| 1.3242(17) | 1.3602(17) | |||||
| 3d | O1….O2 | O2….H | O1-H | C1-O1 | C1-C2 | O1-C1-C2-C11 |
| 2.6142(17) | 1.75(3) | 0.97(3) | 1.3588(19) | 1.438(2) | 1.9(2) | |
| C11-O2 | C2-C11 | |||||
| 1.2199(18) | 1.459(2) | |||||
| 4c | O1….O2 | O2….H | O1-H | C1-O1 | C1-C2 | O1-C1-C2-C11 |
| 2.5063(9) | 1.72(2) | 0.855(19) | 1.3215(9) | 1.3895(10) | -2.04(11) | |
| C11-O2 | C2-C11 | |||||
| 1.2476(10) | 1.4193(10) | |||||
Acknowledgments
References
- Biological Activities of 1,3-Indandiones. Pharmacochemistry of 1,3-Indanediones; Nauta, W.T.; Rekker, R.F. (Eds.) Elsevier Scientific Publishing Co.: New York, NY, USA, 1981; pp. 187–269.
- Wiesner, S.; Springer, E.; Sasson, Y.; Almog, J. Chemical development of latent fingerprints: 1,2-Indanedione has come of age. J. For. Sci. 2001, 46, 1082–1084. [Google Scholar]
- Daehne, S. Near-Infrared Dyes for high Technology Applications. In Proceedings of the NATO Advanced Research Workshop on Syntheses, Optical Properties and Applications of Near Infrared (NIR) Dyes in High Technology Fields; Daehne, S., Resch-Genger, U., Wolfbeis, O.S., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1997; 52, pp. 363–364. [Google Scholar]
- van Klink, J.W.; Larsen, L.; Perry, N.B.; Weavers, R.T.; Cook, G.M.; Bremer, P.J.; MacKenzie, A.D.; Kirikae, T. Triketones active against antibiotic-resistant bacteria: Synthesis, structure-activity relationships, and mode of action. Bioorg. Med. Chem. 2005, 13, 6651–6662. [Google Scholar]
- An, T.Y.; Hu, L.H.; Chen, R.M.; Chen, Z.L.; Li, J.; Shen, Q. Anti-diabetes agents—1. Tetralone derivative from Juglans regia. Chin. Chem. Lett. 2003, 14, 489–490. [Google Scholar]
- Jain, R.; Jain, S.C.; Arora, R. A new cholestane derivative of Abutilon bidentatum Hochst. And ist bioactivity. Pharmazie 1996, 51, 253–254. [Google Scholar]
- Mentré, F.; Pousset, F.; Comets, E.; Plaud, B.; Diquet, B.; Montalescot, G.; Ankri, A.; Mallet, A.; Lechat, P. Population pharmacokinetic-pharmacodynamic analysis of fluindione in patients. Clin. Pharmacol. Ther. 1998, 63, 64–78. [Google Scholar] [CrossRef]
- Sloop, J.C.; Bumgardner, C.; Washington, G.; Loehle, W.D.; Sankar, S.; Lewis, A. Keto-Enol and Enol-Enol Tautomerism in Trifluoromethyl
Diketones. J. Fluorine Chem. 2006, 127, 780–786. [Google Scholar] [CrossRef]
- Sloop, J.C.; Bumgardner, C.; Washington, G.; Loehle, W.D. Synthesis of fluorinated heterocycles. J. Fluorine Chem. 2002, 118, 135–147. [Google Scholar] [CrossRef]
- Sloop, J.C.; Boyle, P.; Fountain, A.W.; Pearman, W.; Swann, J. Electron deficient aryl β-diketones: synthesis and novel tautomeric preferences. Eur. J. Org. Chem. 2011, 5, 936–941. [Google Scholar]
- Bolvig, S.; Hansen, P.E. Deuterium-Induced Isotope Effects on 13C Chemical Shifts as a Probe for Tautomerism in enolic β-Diketones. Mag. Res. Chem. 1996, 34, 467–478. [Google Scholar] [CrossRef]
- Reid, J.C.; Calvin, M. Some New β-Diketones Containing the Trifluoromethyl Group. J. Am. Chem. Soc. 1950, 72, 2948–2952. [Google Scholar] [CrossRef]
- Stavber, G.; Zupan, M.; Stavber, S. Micellar-System-Mediated direct fluorination of ketones in water. Synlett 2009, 4, 589–594. [Google Scholar]
- Sloop, J.C. Synthesis of Fluorinated Pyrazoles and Isoxazoles. The Effect of 2-Fluoro and 2-Chloro Substituents on the Keto-Enol Equilibria of 1,3-Diketones; DOD Technical Report; Defense Technical Information Center: Fort Belvoir, VA, USA, 18 May 1990; pp. 1–32.
- Zajc, B.; Zupan, M. Fluorination with xenon difluoride. 27. The effect of catalyst on fluorination of 1,3-diketones and enol acetates. J. Org. Chem. 1982, 47, 573–575. [Google Scholar] [CrossRef]
- MSDS. Bindone. Available online: http://www.chemcas.org/chemical/msds/cas/AA_M/AAB24557-03.asp (accessed on 20 November 2011). Mp: 205–208 °C. See Carey, F.J.; Sundberg, R.J. (Eds.) Advanced Organic Chemistry, Part B: Reactions and Synthesis, 2nd; Plenum Press: New York, NY, USA, 1983; pp. 43–45.
- Light, R.J.; Hauser, C.R. Aroylations of β-diketones at the terminal methyl group to form 1,3,5-Triketones. Cyclizations to 4-Pyrones and 4-Pyridones. J. Org. Chem. 1960, 25, 538–546. [Google Scholar] [CrossRef]
- Park, J.D.; Brown, H.A.; Lacher, J.R. A Study of Some Fluorine-containing β-Diketones. J. Am. Chem. Soc. 1953, 75, 4753–4756. [Google Scholar]
- Sloop, J.C.; Jackson, J.; Schmidt, R. Microwave-Mediated pyrazole fluorinations using Selectfluor®. Heteroatom Chem. 2009, 20, 341–345. [Google Scholar] [CrossRef]
- Riofski, M.V.; John, J.P.; Zheng, M.M.; Kirshner, J.; Colby, D.A. Exploiting the facile release of trifluoroacetate for the α-Methylation of the sterically hindered carbonyl groups on (+)-Sclareolide and (−)-Eburnamonine. J. Org. Chem. 2011, 76, 3676–3683. [Google Scholar]
- Kirsch, P. Modern Fluoroorganic Chemistry: Synthesis, Reactivity, Applications; Wiley-VCH VerlagGmbH & Co.: Darmstadt, Germany, 2004; pp. 78–79. [Google Scholar]
- Crouse, D.J.; Hurlbut, S.L.; Wheeler, D.M. Photo Fries rearrangements of 1-naphthyl esters in the synthesis of 2-acylnaphthoquinones. J. Org. Chem. 1981, 46, 374–378. [Google Scholar]
- Murdock, K.C. Triacylhalomethanes: 2-Halo-2-acyl-1,3-indanediones. J. Org. Chem. 1959, 24, 845–849. [Google Scholar] [CrossRef]
- Forsen, S.; Nilsson, M. Proton magnetic resonance studies of enolised β-Triketones. Acta Chem. Scand. 1959, 13, 1383–1394. [Google Scholar] [CrossRef]
- Hunig, S.; Hoch, H. 2-Acetyl-cyclanone und Cyclandione-(1,3), ein Vergleich. Justus Liebigs Ann. Chem. 1968, 716, 68–77. [Google Scholar] [CrossRef]
- Ebraheem, K.A. 1H, 13C and 19F NMR Studies on the Structure of the Intramolecularly Hydrogen Bonded cis-Enols of 2-Trifluoroacetylcycloalkanones. Monatsh. Chem. 1991, 122, 157–163. [Google Scholar] [CrossRef]
- Hansen, P.E.; Ibsen, S.N.; Kristensen, T.; Bolvig, S. Deuterium and 18O Isotope Effects on 13C Chemical Shifts of Sterically Hindered and/or Intramolecularly Hydrogen-Bonded o-Hydroxy Acyl Aromatics. Mag. Res. Chem. 1994, 32, 399–408. [Google Scholar] [CrossRef]
- Dolbier, W.R. Guide to Fluorine NMR for Organic Chemists; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2009; Volume 70–81, pp. 152–158. [Google Scholar]
- Bruker-Nonius. SAINT version 7.34A, Bruker-Nonius: Madison, WI, USA, 2006.
- Bruker-Nonius. SADABS version 2.10, Bruker-Nonius: Madison, WI, USA, 2004.
- Altomare, A.; Cascarano, G.; Giacovazzo, C.; Guagliardi, A.; Burla, M. C.; Polidori, G.; Camalli, M. SIR92-a program for automatic solution of crystal structures by direct methods. J. Appl. Cryst. 1994, 27, 435. [Google Scholar]
- Bruker-AXS. XL version 6.12, Bruker-AXS: Madison, WI, USA.
- Gabe, E.J.; Le Page, Y.; Charland, J.P.; Lee, F.L.; White, P.S. NCRVAX-an interactive program system for structure analysis. J. Appl. Cryst. 1989, 22, 384–387. [Google Scholar] [CrossRef]
Appendix. X-Ray Experimental Procedures and Data
1. Compound 1d
Experimental for C18H10O3 (1d)


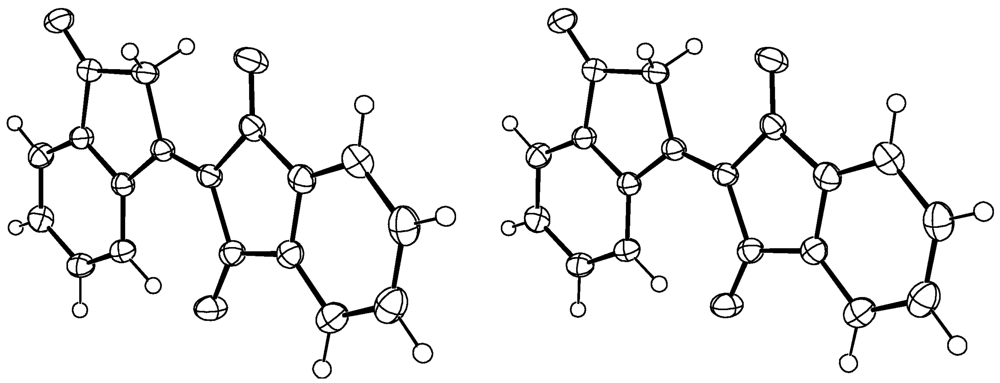
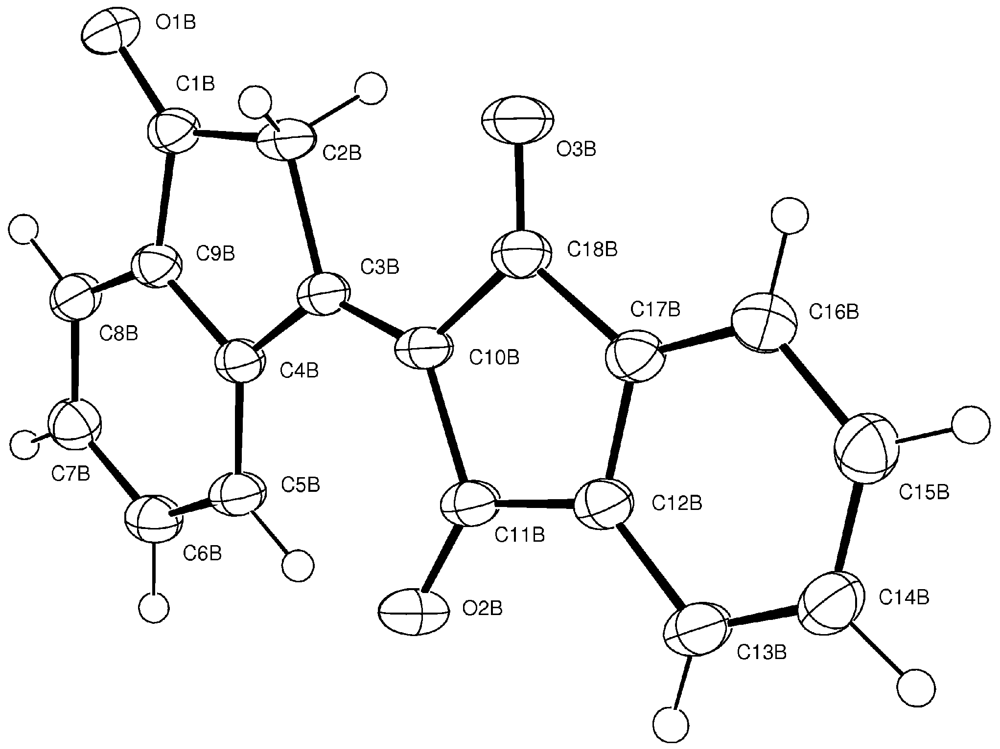
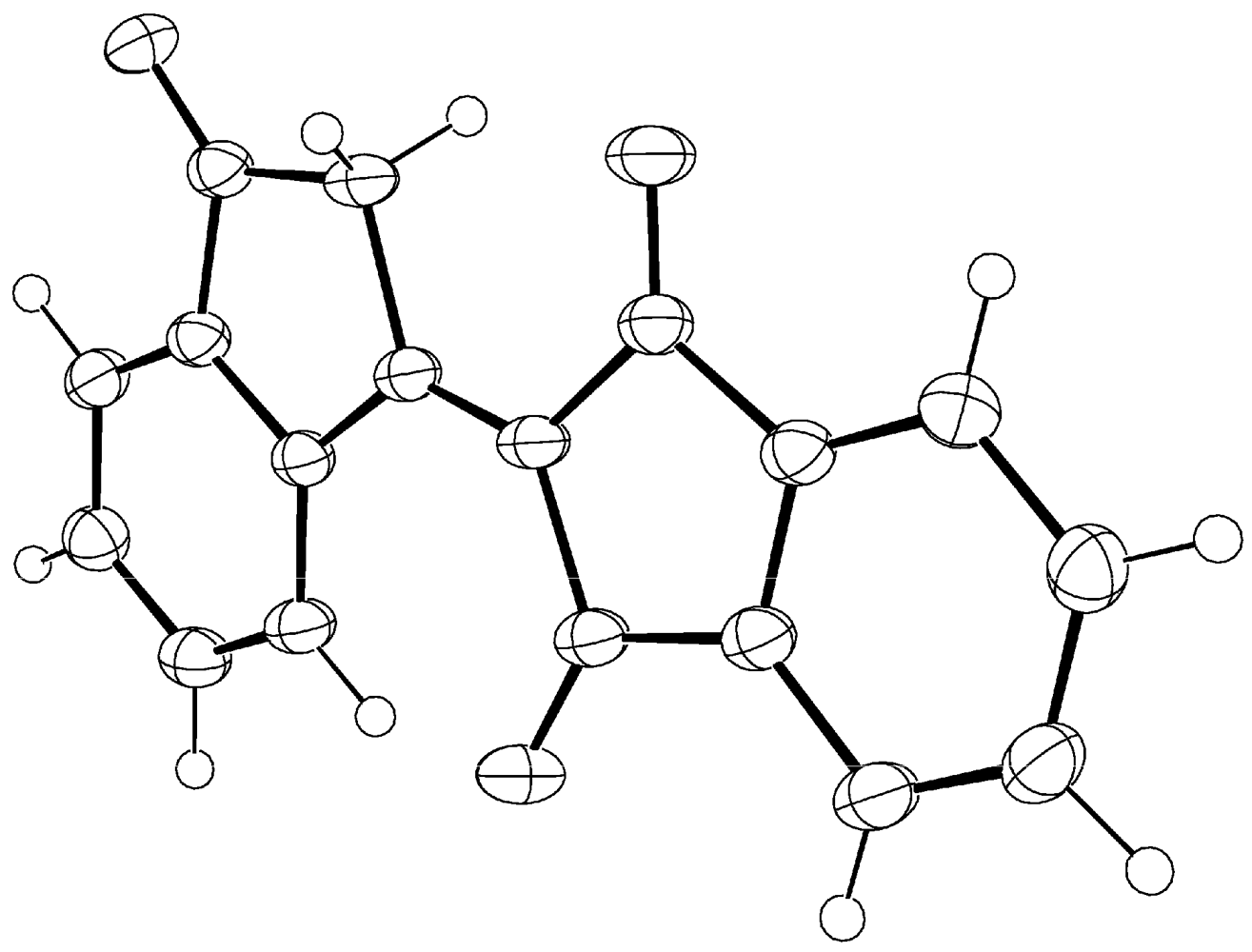
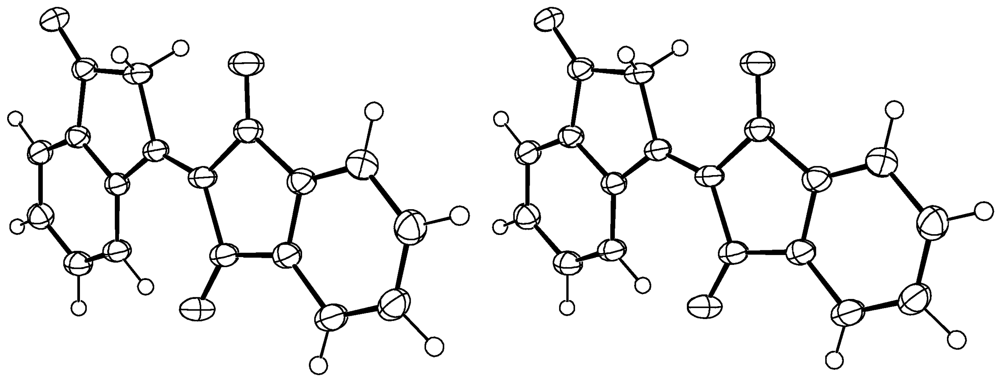
| Formula | C18H10O3 |
| Formula Weight (g/mol) | 274.26 |
| Crystal Dimensions (mm ) | 0.30 × 0.24 × 0.20 |
| Crystal Color and Habit | clear prism |
| Crystal System | orthorhombic |
| Space Group | F d d 2 |
| Temperature, K | 173 |
| a, Å | 18.0996(6) |
| b, Å | 20.9271(7) |
| c, Å | 26.0789(8) |
| α,° | 90.00 |
| β,° | 90.00 |
| γ,° | 90.00 |
| V, Å3 | 9878.0(6) |
| Number of reflections to determine final unit cell | 9975 |
| Min and Max 2θ for cell determination, ° | 5.0, 56.84 |
| Z | 32 |
| F(000) | 4544 |
| ρ (g/cm) | 1.475 |
| λ, Å, (MoKα) | 0.71070 |
| μ, (cm−1) | 0.101 |
| Diffractometer Type | Bruker-Nonius X8 Apex2 |
| Scan Type(s) | omega and phi scans |
| Max 2θ for data collection, ° | 58.24 |
| Measured fraction of data | 1.000 |
| Number of reflections measured | 103146 |
| Unique reflections measured | 6360 |
| Rmerge | 0.0403 |
| Number of reflections included in refinement | 6360 |
| Cut off Threshold Expression | >2sigma(I) |
| Structure refined using | full matrix least-squares using F2 |
| Weighting Scheme | calc w = 1/[sigma2(Fo2) + (0.0555P)2+ 3.0465P] where P=(Fo2+ 2Fc2)/3 |
| Number of parameters in least-squares | 458 |
| R1 | 0.0342 |
| wR2 | 0.0846 |
| R1 (all data) | 0.0400 |
| wR2 (all data) | 0.0880 |
| GOF | 1.038 |
| Maximum shift/error | 0.000 |
| Min & Max peak heights on final ΔF Map (e-/Å) | −0.219, 0.216 |
| Atom | x | y | z | Uiso/equiv |
|---|---|---|---|---|
| O1A | 0.20028(6) | 0.13639(6) | 0.62634(6) | 0.0362(3) |
| O2A | 0.36237(6) | 0.01557(5) | 0.4220 | 0.0310(2) |
| O3A | 0.40033(6) | −0.02683(5) | 0.59892(6) | 0.0329(2) |
| C1A | 0.23461(8) | 0.12010(7) | 0.58853(6) | 0.0257(3) |
| C2A | 0.29550(8) | 0.07078(7) | 0.58765(6) | 0.0265(3) |
| C3A | 0.31404(7) | 0.06171(6) | 0.53149(6) | 0.0215(2) |
| C4A | 0.27141(7) | 0.10937(6) | 0.50228(6) | 0.0214(2) |
| C5A | 0.27082(8) | 0.12548(7) | 0.45022(6) | 0.0246(3) |
| C6A | 0.22256(8) | 0.17252(7) | 0.43340(7) | 0.0271(3) |
| C7A | 0.17513(8) | 0.20411(7) | 0.46700(7) | 0.0283(3) |
| C8A | 0.17630(8) | 0.19033(7) | 0.51873(7) | 0.0279(3) |
| C9A | 0.22477(7) | 0.14322(7) | 0.53572(6) | 0.0234(3) |
| C10A | 0.36116(7) | 0.01514(6) | 0.51589(6) | 0.0217(2) |
| C11A | 0.38319(7) | −0.00519(6) | 0.46328(6) | 0.0228(3) |
| C12A | 0.43567(7) | −0.05931(6) | 0.46890(7) | 0.0231(3) |
| C13A | 0.47199(8) | −0.09342(7) | 0.43109(7) | 0.0289(3) |
| C14A | 0.51888(9) | −0.14212(8) | 0.44613(8) | 0.0328(3) |
| C15A | 0.52844(9) | −0.15676(8) | 0.49786(8) | 0.0331(3) |
| C16A | 0.49169(8) | −0.12272(7) | 0.53601(7) | 0.0293(3) |
| C17A | 0.44557(7) | −0.07332(6) | 0.52064(7) | 0.0242(3) |
| C18A | 0.40165(7) | −0.02813(7) | 0.55238(7) | 0.0237(3) |
| O1B | 0.14120(7) | 0.04113(5) | 0.35875(6) | 0.0349(2) |
| O2B | 0.25973(7) | −0.11687(6) | 0.56110(5) | 0.0351(2) |
| O3B | 0.29954(7) | −0.15569(6) | 0.38414(6) | 0.0367(3) |
| C1B | 0.15538(8) | 0.00789(7) | 0.39573(6) | 0.0270(3) |
| C2B | 0.20581(8) | −0.04922(7) | 0.39627(6) | 0.0267(3) |
| C3B | 0.21160(8) | −0.06899(7) | 0.45205(6) | 0.0233(3) |
| C4B | 0.16128(8) | −0.02763(7) | 0.48137(7) | 0.0241(3) |
| C5B | 0.14109(8) | −0.02690(8) | 0.53345(7) | 0.0286(3) |
| C6B | 0.09013(9) | 0.01792(8) | 0.54976(7) | 0.0314(3) |
| C7B | 0.05876(9) | 0.06238(8) | 0.51670(7) | 0.0318(3) |
| C8B | 0.07692(9) | 0.06178(7) | 0.46499(7) | 0.0298(3) |
| C9B | 0.12772(8) | 0.01681(7) | 0.44855(6) | 0.0255(3) |
| C10B | 0.25683(8) | −0.11716(7) | 0.46724(6) | 0.0245(3) |
| C11B | 0.27541(8) | −0.14057(7) | 0.52003(7) | 0.0259(3) |
| C12B | 0.32241(8) | −0.19790(7) | 0.51345(7) | 0.0262(3) |
| C13B | 0.35157(9) | −0.23821(8) | 0.55099(7) | 0.0322(3) |
| C14B | 0.39368(10) | −0.28954(8) | 0.53526(8) | 0.0352(3) |
| C15B | 0.40794(9) | −0.30056(8) | 0.48342(8) | 0.0352(3) |
| C16B | 0.37983(9) | −0.25975(7) | 0.44609(7) | 0.0322(3) |
| C17B | 0.33643(8) | −0.20886(7) | 0.46172(7) | 0.0264(3) |
| C18B | 0.29790(8) | −0.15934(7) | 0.43065(7) | 0.0269(3) |
| H2A1 | 0.2800(10) | 0.0306(10) | 0.6041(7) | 0.038(5) |
| H2A2 | 0.3395(11) | 0.0877(9) | 0.6072(7) | 0.037(5) |
| H5A | 0.3041(10) | 0.1074(9) | 0.4261(8) | 0.033(4) |
| H6A | 0.2211(10) | 0.1858(9) | 0.3965(7) | 0.034(5) |
| H7A | 0.1431(11) | 0.2367(9) | 0.4567(7) | 0.036(5) |
| H8A | 0.1442(12) | 0.2130(10) | 0.5435(9) | 0.045(6) |
| H13A | 0.4637(10) | −0.0835(9) | 0.3957(8) | 0.036(5) |
| H14A | 0.5436(10) | −0.1635(9) | 0.4193(8) | 0.035(5) |
| H15A | 0.5585(12) | −0.1908(10) | 0.5071(8) | 0.044(6) |
| H16A | 0.4962(11) | −0.1324(10) | 0.5727(8) | 0.041(5) |
| H2B1 | 0.2541(12) | −0.0384(10) | 0.3821(9) | 0.052(6) |
| H2B2 | 0.1831(10) | −0.0855(9) | 0.3765(8) | 0.035(5) |
| H5B | 0.1623(10) | −0.0581(9) | 0.5574(7) | 0.033(5) |
| H6B | 0.0744(12) | 0.0181(10) | 0.5865(9) | 0.049(6) |
| H7B | 0.0234(10) | 0.0921(9) | 0.5291(8) | 0.035(5) |
| H8B | 0.0572(10) | 0.0917(9) | 0.4398(8) | 0.038(5) |
| H13B | 0.3432(11) | −0.2287(9) | 0.5847(8) | 0.036(5) |
| H14B | 0.4143(11) | −0.3227(10) | 0.5586(8) | 0.039(5) |
| H15B | 0.4354(11) | −0.3383(9) | 0.4722(8) | 0.043(5) |
| H16B | 0.3894(10) | −0.2680(9) | 0.4103(7) | 0.029(4) |
| Atom | u11 | u22 | u33 | u12 | u13 | u23 |
|---|---|---|---|---|---|---|
| O1A | 0.0362(6) | 0.0485(7) | 0.0238(5) | 0.0094(5) | 0.0051(4) | −0.0015(5) |
| O2A | 0.0394(6) | 0.0338(6) | 0.0198(5) | 0.0040(4) | 0.0011(4) | 0.0012(4) |
| O3A | 0.0398(6) | 0.0369(6) | 0.0221(5) | 0.0069(5) | −0.0075(4) | −0.0029(4) |
| C1A | 0.0256(6) | 0.0301(7) | 0.0215(6) | 0.0000(5) | −0.0008(5) | −0.0014(5) |
| C2A | 0.0291(7) | 0.0326(7) | 0.0179(6) | 0.0040(6) | −0.0015(5) | −0.0015(5) |
| C3A | 0.0217(6) | 0.0238(6) | 0.0191(6) | −0.0048(5) | −0.0004(4) | −0.0006(5) |
| C4A | 0.0218(6) | 0.0212(6) | 0.0213(6) | −0.0030(5) | −0.0006(4) | −0.0007(5) |
| C5A | 0.0276(7) | 0.0253(7) | 0.0210(6) | −0.0012(5) | 0.0020(5) | 0.0006(5) |
| C6A | 0.0332(7) | 0.0251(7) | 0.0230(6) | −0.0029(5) | 0.0008(5) | 0.0032(5) |
| C7A | 0.0299(7) | 0.0245(7) | 0.0306(7) | 0.0028(5) | −0.0008(6) | 0.0047(6) |
| C8A | 0.0283(7) | 0.0272(7) | 0.0282(7) | 0.0019(5) | 0.0025(5) | −0.0001(6) |
| C9A | 0.0244(6) | 0.0250(6) | 0.0209(6) | −0.0010(5) | 0.0003(5) | −0.0001(5) |
| C10A | 0.0227(6) | 0.0235(6) | 0.0190(6) | −0.0021(5) | −0.0016(5) | −0.0008(5) |
| C11A | 0.0232(6) | 0.0235(6) | 0.0217(6) | −0.0034(5) | 0.0019(5) | −0.0011(5) |
| C12A | 0.0223(6) | 0.0225(6) | 0.0247(6) | −0.0031(5) | 0.0014(5) | −0.0011(5) |
| C13A | 0.0300(7) | 0.0286(7) | 0.0282(7) | −0.0032(6) | 0.0055(5) | −0.0030(5) |
| C14A | 0.0305(7) | 0.0284(7) | 0.0396(8) | −0.0010(6) | 0.0067(6) | −0.0076(6) |
| C15A | 0.0266(7) | 0.0284(7) | 0.0443(9) | 0.0033(6) | −0.0025(6) | −0.0028(6) |
| C16A | 0.0279(7) | 0.0279(7) | 0.0321(8) | 0.0000(5) | −0.0052(6) | −0.0001(6) |
| C17A | 0.0225(6) | 0.0235(6) | 0.0264(6) | −0.0046(5) | −0.0021(5) | −0.0016(5) |
| C18A | 0.0225(6) | 0.0249(7) | 0.0237(6) | −0.0015(5) | −0.0034(5) | −0.0017(5) |
| O1B | 0.0463(6) | 0.0351(6) | 0.0235(5) | −0.0023(5) | −0.0037(4) | 0.0079(4) |
| O2B | 0.0415(6) | 0.0444(6) | 0.0192(5) | 0.0011(5) | −0.0023(4) | 0.0013(4) |
| O3B | 0.0480(6) | 0.0425(6) | 0.0196(5) | 0.0073(5) | −0.0015(4) | 0.0010(4) |
| C1B | 0.0292(7) | 0.0303(7) | 0.0214(6) | −0.0089(5) | −0.0038(5) | 0.0019(5) |
| C2B | 0.0287(7) | 0.0344(7) | 0.0168(6) | −0.0024(6) | −0.0015(5) | 0.0028(5) |
| C3B | 0.0240(6) | 0.0277(7) | 0.0181(6) | −0.0080(5) | −0.0024(5) | 0.0031(5) |
| C4B | 0.0252(6) | 0.0271(6) | 0.0200(6) | −0.0078(5) | −0.0020(5) | 0.0014(5) |
| C5B | 0.0309(7) | 0.0349(8) | 0.0200(6) | −0.0068(6) | −0.0009(5) | 0.0039(5) |
| C6B | 0.0326(8) | 0.0379(8) | 0.0238(7) | −0.0048(6) | 0.0018(5) | −0.0002(6) |
| C7B | 0.0329(7) | 0.0330(7) | 0.0296(7) | −0.0032(6) | 0.0011(6) | −0.0008(6) |
| C8B | 0.0328(7) | 0.0306(7) | 0.0261(7) | −0.0037(6) | −0.0026(6) | 0.0045(6) |
| C9B | 0.0270(7) | 0.0288(7) | 0.0207(6) | −0.0068(5) | −0.0034(5) | 0.0014(5) |
| C10B | 0.0261(7) | 0.0310(7) | 0.0163(6) | −0.0071(5) | −0.0015(5) | 0.0022(5) |
| C11B | 0.0273(7) | 0.0313(7) | 0.0190(6) | −0.0078(5) | −0.0020(5) | 0.0040(5) |
| C12B | 0.0268(6) | 0.0289(7) | 0.0227(7) | −0.0089(5) | −0.0046(5) | 0.0034(5) |
| C13B | 0.0367(8) | 0.0351(8) | 0.0247(7) | −0.0091(6) | −0.0067(6) | 0.0076(6) |
| C14B | 0.0410(9) | 0.0304(8) | 0.0344(8) | −0.0063(6) | −0.0125(7) | 0.0085(6) |
| C15B | 0.0378(8) | 0.0293(7) | 0.0386(9) | −0.0001(6) | −0.0093(6) | 0.0010(6) |
| C16B | 0.0373(8) | 0.0315(8) | 0.0279(8) | −0.0012(6) | −0.0054(6) | −0.0008(6) |
| C17B | 0.0284(7) | 0.0281(7) | 0.0228(6) | −0.0064(5) | −0.0043(5) | 0.0020(5) |
| C18B | 0.0301(7) | 0.0304(7) | 0.0203(6) | −0.0048(6) | −0.0030(5) | 0.0016(5) |
| O1A-C1A | 1.2143(17) | O1B-C1B | 1.2166(17) |
| O2A-C11A | 1.2212(17) | O2B-C11B | 1.2140(18) |
| O3A-C18A | 1.2143(17) | O3B-C18B | 1.2159(17) |
| C1A-C9A | 1.4705(19) | C1B-C9B | 1.4775(19) |
| C1A-C2A | 1.510(2) | C1B-C2B | 1.504(2) |
| C2A-C3A | 1.5145(18) | C2B-C3B | 1.5159(17) |
| C2A-H2A1 | 0.99(2) | C2B-H2B1 | 0.98(2) |
| C2A-H2A2 | 1.01(2) | C2B-H2B2 | 1.006(19) |
| C3A-C10A | 1.3574(19) | C3B-C10B | 1.358(2) |
| C3A-C4A | 1.4733(19) | C3B-C4B | 1.471(2) |
| C4A-C5A | 1.3989(18) | C4B-C9B | 1.4025(19) |
| C4A-C9A | 1.4052(19) | C4B-C5B | 1.4065(19) |
| C5A-C6A | 1.387(2) | C5B-C6B | 1.383(2) |
| C5A-H5A | 0.95(2) | C5B-H5B | 0.982(19) |
| C6A-C7A | 1.394(2) | C6B-C7B | 1.390(2) |
| C6A-H6A | 1.001(19) | C6B-H6B | 1.00(2) |
| C7A-C8A | 1.380(2) | C7B-C8B | 1.388(2) |
| C7A-H7A | 0.94(2) | C7B-H7B | 0.95(2) |
| C8A-C9A | 1.392(2) | C8B-C9B | 1.384(2) |
| C8A-H8A | 0.99(2) | C8B-H8B | 0.98(2) |
| C10A-C11A | 1.4908(18) | C10B-C18B | 1.497(2) |
| C10A-C18A | 1.5043(19) | C10B-C11B | 1.4995(18) |
| C11A-C12A | 1.4855(19) | C11B-C12B | 1.481(2) |
| C12A-C13A | 1.383(2) | C12B-C17B | 1.3916(19) |
| C12A-C17A | 1.3924(19) | C12B-C13B | 1.396(2) |
| C13A-C14A | 1.383(2) | C13B-C14B | 1.379(3) |
| C13A-H13A | 0.96(2) | C13B-H13B | 0.91(2) |
| C14A-C15A | 1.394(2) | C14B-C15B | 1.395(2) |
| C14A-H14A | 0.94(2) | C14B-H14B | 1.00(2) |
| C15A-C16A | 1.393(2) | C15B-C16B | 1.392(2) |
| C15A-H15A | 0.93(2) | C15B-H15B | 0.98(2) |
| C16A-C17A | 1.388(2) | C16B-C17B | 1.385(2) |
| C16A-H16A | 0.98(2) | C16B-H16B | 0.965(19) |
| C17A-C18A | 1.4870(19) | C17B-C18B | 1.489(2) |
| O1A-C1A-C9A | 127.32(14) | O1B-C1B-C9B | 126.54(14) |
| O1A-C1A-C2A | 125.28(13) | O1B-C1B-C2B | 126.15(13) |
| C9A-C1A-C2A | 107.40(11) | C9B-C1B-C2B | 107.30(11) |
| C1A-C2A-C3A | 105.20(11) | C1B-C2B-C3B | 105.54(11) |
| C1A-C2A-H2A1 | 111.6(11) | C1B-C2B-H2B1 | 110.9(13) |
| C3A-C2A-H2A1 | 112.2(11) | C3B-C2B-H2B1 | 111.4(13) |
| C1A-C2A-H2A2 | 109.2(11) | C1B-C2B-H2B2 | 110.3(11) |
| C3A-C2A-H2A2 | 110.9(11) | C3B-C2B-H2B2 | 108.3(11) |
| H2A1-C2A-H2A2 | 107.7(16) | H2B1-C2B-H2B2 | 110.2(18) |
| C10A-C3A-C4A | 131.31(12) | C10B-C3B-C4B | 131.19(12) |
| C10A-C3A-C2A | 121.27(12) | C10B-C3B-C2B | 121.63(13) |
| C4A-C3A-C2A | 107.40(11) | C4B-C3B-C2B | 107.18(12) |
| C5A-C4A-C9A | 118.44(12) | C9B-C4B-C5B | 118.01(13) |
| C5A-C4A-C3A | 131.98(13) | C9B-C4B-C3B | 109.95(12) |
| C9A-C4A-C3A | 109.58(12) | C5B-C4B-C3B | 132.01(13) |
| C6A-C5A-C4A | 118.86(13) | C6B-C5B-C4B | 118.54(14) |
| C6A-C5A-H5A | 118.3(12) | C6B-C5B-H5B | 121.1(11) |
| C4A-C5A-H5A | 122.8(12) | C4B-C5B-H5B | 120.3(11) |
| C5A-C6A-C7A | 121.72(13) | C5B-C6B-C7B | 122.40(14) |
| C5A-C6A-H6A | 121.2(11) | C5B-C6B-H6B | 119.2(12) |
| C7A-C6A-H6A | 117.1(11) | C7B-C6B-H6B | 118.4(12) |
| C8A-C7A-C6A | 120.41(14) | C8B-C7B-C6B | 120.00(15) |
| C8A-C7A-H7A | 116.4(12) | C8B-C7B-H7B | 119.7(12) |
| C6A-C7A-H7A | 123.2(12) | C6B-C7B-H7B | 120.2(12) |
| C7A-C8A-C9A | 117.96(14) | C9B-C8B-C7B | 117.68(14) |
| C7A-C8A-H8A | 121.9(13) | C9B-C8B-H8B | 118.1(12) |
| C9A-C8A-H8A | 120.2(13) | C7B-C8B-H8B | 124.2(12) |
| C8A-C9A-C4A | 122.55(13) | C8B-C9B-C4B | 123.35(13) |
| C8A-C9A-C1A | 127.40(13) | C8B-C9B-C1B | 126.87(13) |
| C4A-C9A-C1A | 110.03(12) | C4B-C9B-C1B | 109.78(13) |
| C3A-C10A-C11A | 130.45(12) | C3B-C10B-C18B | 123.44(12) |
| C3A-C10A-C18A | 123.28(12) | C3B-C10B-C11B | 130.22(13) |
| C11A-C10A-C18A | 106.27(11) | C18B-C10B-C11B | 106.33(12) |
| O2A-C11A-C12A | 123.76(13) | O2B-C11B-C12B | 124.59(13) |
| O2A-C11A-C10A | 128.89(12) | O2B-C11B-C10B | 128.62(14) |
| C12A-C11A-C10A | 107.32(11) | C12B-C11B-C10B | 106.68(12) |
| C13A-C12A-C17A | 121.39(13) | C17B-C12B-C13B | 120.75(14) |
| C13A-C12A-C11A | 128.82(13) | C17B-C12B-C11B | 110.53(12) |
| C17A-C12A-C11A | 109.79(11) | C13B-C12B-C11B | 128.72(14) |
| C14A-C13A-C12A | 118.02(15) | C14B-C13B-C12B | 118.09(15) |
| C14A-C13A-H13A | 121.9(11) | C14B-C13B-H13B | 123.2(12) |
| C12A-C13A-H13A | 120.1(11) | C12B-C13B-H13B | 118.7(12) |
| C13A-C14A-C15A | 120.83(14) | C13B-C14B-C15B | 121.28(15) |
| C13A-C14A-H14A | 115.4(12) | C13B-C14B-H14B | 124.6(12) |
| C15A-C14A-H14A | 123.7(12) | C15B-C14B-H14B | 114.1(12) |
| C16A-C15A-C14A | 121.30(15) | C16B-C15B-C14B | 120.57(16) |
| C16A-C15A-H15A | 119.2(13) | C16B-C15B-H15B | 118.2(12) |
| C14A-C15A-H15A | 119.5(13) | C14B-C15B-H15B | 121.2(12) |
| C17A-C16A-C15A | 117.51(15) | C17B-C16B-C15B | 118.27(15) |
| C17A-C16A-H16A | 119.1(12) | C17B-C16B-H16B | 121.7(11) |
| C15A-C16A-H16A | 123.4(12) | C15B-C16B-H16B | 120.0(11) |
| C16A-C17A-C12A | 120.94(13) | C16B-C17B-C12B | 121.03(14) |
| C16A-C17A-C18A | 129.38(13) | C16B-C17B-C18B | 129.85(14) |
| C12A-C17A-C18A | 109.68(11) | C12B-C17B-C18B | 109.11(13) |
| O3A-C18A-C17A | 125.54(13) | O3B-C18B-C17B | 125.12(14) |
| O3A-C18A-C10A | 127.53(13) | O3B-C18B-C10B | 127.65(14) |
| C17A-C18A-C10A | 106.93(11) | C17B-C18B-C10B | 107.22(11) |
| O1A-C1A-C2A-C3A | 173.35(14) | O1B-C1B-C2B-C3B | 173.77(14) |
| C9A-C1A-C2A-C3A | −5.93(15) | C9B-C1B-C2B-C3B | −5.04(14) |
| C1A-C2A-C3A-C10A | −172.45(12) | C1B-C2B-C3B-C10B | −175.90(12) |
| C1A-C2A-C3A-C4A | 6.20(14) | C1B-C2B-C3B-C4B | 4.41(14) |
| C10A-C3A-C4A-C5A | −6.6(2) | C10B-C3B-C4B-C9B | 178.21(14) |
| C2A-C3A-C4A-C5A | 174.97(14) | C2B-C3B-C4B-C9B | −2.13(15) |
| C10A-C3A-C4A-C9A | 174.20(13) | C10B-C3B-C4B-C5B | −3.9(2) |
| C2A-C3A-C4A-C9A | −4.26(15) | C2B-C3B-C4B-C5B | 175.74(14) |
| C9A-C4A-C5A-C6A | −2.2(2) | C9B-C4B-C5B-C6B | −0.83(19) |
| C3A-C4A-C5A-C6A | 178.62(13) | C3B-C4B-C5B-C6B | −178.57(14) |
| C4A-C5A-C6A-C7A | 0.3(2) | C4B-C5B-C6B-C7B | −0.5(2) |
| C5A-C6A-C7A-C8A | 1.6(2) | C5B-C6B-C7B-C8B | 1.6(2) |
| C6A-C7A-C8A-C9A | −1.5(2) | C6B-C7B-C8B-C9B | −1.3(2) |
| C7A-C8A-C9A-C4A | −0.5(2) | C7B-C8B-C9B-C4B | 0.0(2) |
| C7A-C8A-C9A-C1A | −179.00(14) | C7B-C8B-C9B-C1B | −179.50(14) |
| C5A-C4A-C9A-C8A | 2.4(2) | C5B-C4B-C9B-C8B | 1.1(2) |
| C3A-C4A-C9A-C8A | −178.25(13) | C3B-C4B-C9B-C8B | 179.32(13) |
| C5A-C4A-C9A-C1A | −178.91(12) | C5B-C4B-C9B-C1B | −179.32(12) |
| C3A-C4A-C9A-C1A | 0.45(15) | C3B-C4B-C9B-C1B | −1.12(15) |
| O1A-C1A-C9A-C8A | 2.9(3) | O1B-C1B-C9B-C8B | 4.7(2) |
| C2A-C1A-C9A-C8A | −177.83(14) | C2B-C1B-C9B-C8B | −176.51(14) |
| O1A-C1A-C9A-C4A | −175.70(15) | O1B-C1B-C9B-C4B | −174.87(14) |
| C2A-C1A-C9A-C4A | 3.56(15) | C2B-C1B-C9B-C4B | 3.94(15) |
| C4A-C3A-C10A-C11A | −2.9(2) | C4B-C3B-C10B-C18B | 172.57(13) |
| C2A-C3A-C10A-C11A | 175.35(13) | C2B-C3B-C10B-C18B | −7.0(2) |
| C4A-C3A-C10A-C18A | 178.09(13) | C4B-C3B-C10B-C11B | −6.0(2) |
| C2A-C3A-C10A-C18A | −3.6(2) | C2B-C3B-C10B-C11B | 174.40(13) |
| C3A-C10A-C11A-O2A | −1.1(2) | C3B-C10B-C11B-O2B | −8.6(2) |
| C18A-C10A-C11A-O2A | 177.98(14) | C18B-C10B-C11B-O2B | 172.65(15) |
| C3A-C10A-C11A-C12A | −179.17(13) | C3B-C10B-C11B-C12B | 175.12(14) |
| C18A-C10A-C11A-C12A | −0.06(14) | C18B-C10B-C11B-C12B | −3.62(14) |
| O2A-C11A-C12A-C13A | 2.9(2) | O2B-C11B-C12B-C17B | −173.55(14) |
| C10A-C11A-C12A-C13A | −178.90(13) | C10B-C11B-C12B-C17B | 2.91(15) |
| O2A-C11A-C12A-C17A | −177.23(13) | O2B-C11B-C12B-C13B | 6.2(2) |
| C10A-C11A-C12A-C17A | 0.94(15) | C10B-C11B-C12B-C13B | −177.33(14) |
| C17A-C12A-C13A-C14A | −0.1(2) | C17B-C12B-C13B-C14B | −0.9(2) |
| C11A-C12A-C13A-C14A | 179.72(14) | C11B-C12B-C13B-C14B | 179.39(14) |
| C12A-C13A-C14A-C15A | 0.7(2) | C12B-C13B-C14B-C15B | 1.0(2) |
| C13A-C14A-C15A-C16A | −0.5(2) | C13B-C14B-C15B-C16B | 0.0(3) |
| C14A-C15A-C16A-C17A | −0.5(2) | C14B-C15B-C16B-C17B | −1.2(2) |
| C15A-C16A-C17A-C12A | 1.1(2) | C15B-C16B-C17B-C12B | 1.3(2) |
| C15A-C16A-C17A-C18A | −177.97(13) | C15B-C16B-C17B-C18B | −178.14(15) |
| C13A-C12A-C17A-C16A | −0.8(2) | C13B-C12B-C17B-C16B | −0.3(2) |
| C11A-C12A-C17A-C16A | 179.30(13) | C11B-C12B-C17B-C16B | 179.50(13) |
| C13A-C12A-C17A-C18A | 178.41(12) | C13B-C12B-C17B-C18B | 179.26(13) |
| C11A-C12A-C17A-C18A | −1.45(15) | C11B-C12B-C17B-C18B | −0.96(16) |
| C16A-C17A-C18A-O3A | 1.4(2) | C16B-C17B-C18B-O3B | −0.8(2) |
| C12A-C17A-C18A-O3A | −177.79(14) | C12B-C17B-C18B-O3B | 179.73(14) |
| C16A-C17A-C18A-C10A | −179.45(13) | C16B-C17B-C18B-C10B | 178.11(15) |
| C12A-C17A-C18A-C10A | 1.39(15) | C12B-C17B-C18B-C10B | −1.38(15) |
| C3A-C10A-C18A-O3A | −2.4(2) | C3B-C10B-C18B-O3B | 3.1(2) |
| C11A-C10A-C18A-O3A | 178.39(14) | C11B-C10B-C18B-O3B | −178.05(15) |
| C3A-C10A-C18A-C17A | 178.42(12) | C3B-C10B-C18B-C17B | −175.76(12) |
| C11A-C10A-C18A-C17A | −0.77(14) | C11B-C10B-C18B-C17B | 3.10(14) |
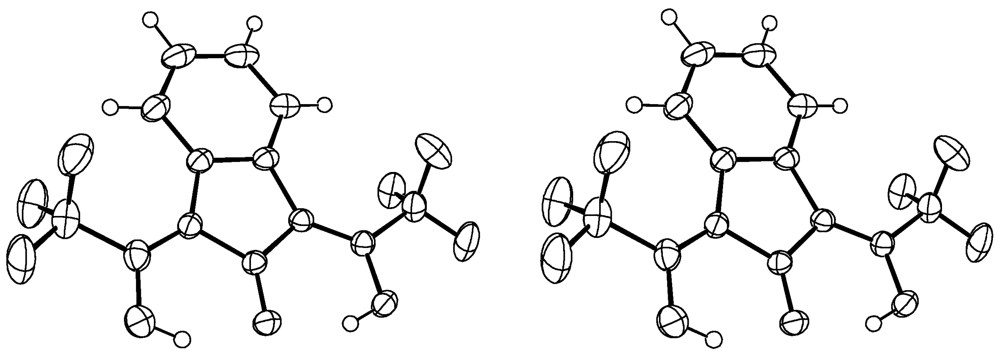
2. Compound 2c
Experimental for C13H6F6O3 (2c)


| Formula | C13H6F6O3 |
| Formula Weight (g/mol) | 324.18 |
| Crystal Dimensions (mm ) | 1.20 × 0.10 × 0.06 |
| Crystal Color and Habit | yellow needle |
| Crystal System | monoclinic |
| Space Group | P 21/c |
| Temperature, K | 173 |
| a, Å | 4.7643(3) |
| b, Å | 18.6978(12) |
| c, Å | 13.8431(9) |
| α,° | 90.0 |
| β,° | 98.964(3) |
| γ,° | 90.0 |
| V, Å3 | 1218.11(14) |
| Number of reflections to determine final unit cell | 9959 |
| Min and Max 2θ for cell determination, ° | 5.28, 57.7 |
| Z | 4 |
| F(000) | 648.71 |
| ρ (g/cm) | 1.768 |
| λ, Å, (MoKα) | 0.71073 |
| μ, (cm−1) | 0.18 |
| Diffractometer Type | Bruker-Nonius X8 Apex2 |
| Scan Type(s) | omega and phi scans |
| Max 2θ for data collection, ° | 57.74 |
| Measured fraction of data | 0.98 |
| Number of reflections measured | 26516 |
| Unique reflections measured | 3195 |
| Rmerge | 0.027 |
| Number of reflections included in refinement | 2755 |
| Cut off Threshold Expression | Inet > 1.0 sigma(Inet) |
| Structure refined using | full matrix least-squares using F |
| Weighting Scheme | 1/(sigma2(F) + 0.0005F2) |
| Number of parameters in least-squares | 223 |
| Rf | 0.038 |
| Rw | 0.053 |
| Rf (all data) | 0.046 |
| Rw (all data) | 0.054 |
| GOF | 1.74 |
| Maximum shift/error | 0.003 |
| Min & Max peak heights on final ΔF Map (e−/Å) | −0.30, 0.35 |
| Atom | x | y | z | Uiso/equiv |
|---|---|---|---|---|
| O1 | 0.81730(18) | 0.99675(5) | 0.07954(6) | 0.0316(4) |
| O2 | 0.77456(19) | 1.10306(5) | −0.04046(7) | 0.0328(5) |
| O3 | 0.8507(2) | 0.90513(5) | 0.22145(8) | 0.0392(5) |
| C1 | 0.6645(2) | 1.03312(6) | 0.12739(8) | 0.0244(5) |
| C2 | 0.5495(2) | 1.10365(6) | 0.10029(9) | 0.0243(5) |
| C3 | 0.3836(2) | 1.12503(7) | 0.17672(8) | 0.0257(5) |
| C4 | 0.2245(3) | 1.18602(8) | 0.18650(10) | 0.0319(6) |
| C5 | 0.0812(3) | 1.19205(8) | 0.26621(11) | 0.0377(7) |
| C6 | 0.0964(3) | 1.13855(8) | 0.33508(11) | 0.0396(7) |
| C7 | 0.2560(3) | 1.07730(8) | 0.32738(10) | 0.0352(6) |
| C8 | 0.4020(2) | 1.07009(7) | 0.24809(9) | 0.0270(5) |
| C9 | 0.5808(2) | 1.01187(6) | 0.22025(8) | 0.0263(5) |
| C10 | 0.6161(2) | 1.13484(7) | 0.01809(9) | 0.0259(5) |
| C11 | 0.5253(3) | 1.20877(7) | −0.01820(11) | 0.0337(6) |
| C12 | 0.6894(3) | 0.95005(7) | 0.26242(9) | 0.0311(6) |
| C13 | 0.6515(3) | 0.92528(8) | 0.36396(11) | 0.0401(7) |
| F1 | 0.61454(20) | 1.22376(5) | −0.10179(7) | 0.0526(5) |
| F2 | 0.24328(16) | 1.21541(5) | −0.03293(6) | 0.0430(4) |
| F3 | 0.62851(20) | 1.25846(5) | 0.04657(7) | 0.0526(5) |
| F4 | 0.8000(2) | 0.86675(5) | 0.39034(7) | 0.0592(6) |
| F5 | 0.7410(2) | 0.97545(6) | 0.43055(6) | 0.0548(5) |
| F6 | 0.38149(19) | 0.91237(6) | 0.37079(7) | 0.0579(5) |
| H2 | 0.822(4) | 1.0603(13) | −0.0133(14) | 0.064(6) |
| H3 | 0.877(5) | 0.9223(11) | 0.1643(16) | 0.072(6) |
| H4 | 0.213(3) | 1.2212(8) | 0.1414(11) | 0.030(4) |
| H5 | −0.019(3) | 1.2318(8) | 0.2728(11) | 0.035(4) |
| H6 | −0.004(4) | 1.1455(9) | 0.3899(12) | 0.045(4) |
| H7 | 0.272(3) | 1.0418(9) | 0.3767(12) | 0.039(4) |
| Atom | u11 | u22 | u33 | u12 | u13 | u23 |
|---|---|---|---|---|---|---|
| O1 | 0.0384(5) | 0.0291(5) | 0.0291(5) | 0.0065(4) | 0.0112(4) | 0.0003(4) |
| O2 | 0.0380(5) | 0.0333(5) | 0.0302(5) | 0.0053(4) | 0.0146(4) | 0.0045(4) |
| O3 | 0.0482(6) | 0.0321(5) | 0.0380(6) | 0.0067(4) | 0.0085(5) | 0.0061(4) |
| C1 | 0.0264(5) | 0.0255(6) | 0.0214(5) | −0.0027(5) | 0.0040(4) | −0.0019(4) |
| C2 | 0.0245(5) | 0.0243(6) | 0.0243(5) | −0.0014(5) | 0.0042(4) | −0.0029(4) |
| C3 | 0.0230(5) | 0.0294(6) | 0.0248(6) | −0.0045(5) | 0.0042(4) | −0.0063(5) |
| C4 | 0.0303(6) | 0.0330(7) | 0.0321(6) | 0.0012(5) | 0.0041(5) | −0.0072(5) |
| C5 | 0.0313(6) | 0.0419(8) | 0.0407(7) | 0.0023(6) | 0.0084(6) | −0.0160(6) |
| C6 | 0.0341(6) | 0.0531(9) | 0.0343(7) | −0.0067(6) | 0.0139(6) | −0.0166(7) |
| C7 | 0.0358(7) | 0.0441(8) | 0.0275(6) | −0.0092(6) | 0.0112(5) | −0.0064(6) |
| C8 | 0.0255(5) | 0.0316(6) | 0.0242(6) | −0.0068(5) | 0.0051(4) | −0.0053(5) |
| C9 | 0.0290(5) | 0.0282(6) | 0.0222(6) | −0.0067(5) | 0.0054(5) | −0.0015(4) |
| C10 | 0.0241(5) | 0.0270(6) | 0.0271(6) | −0.0010(5) | 0.0051(4) | 0.0005(5) |
| C11 | 0.0293(6) | 0.0323(7) | 0.0409(7) | 0.0007(5) | 0.0096(5) | 0.0079(5) |
| C12 | 0.0324(6) | 0.0314(7) | 0.0291(6) | −0.0071(5) | 0.0031(5) | 0.0023(5) |
| C13 | 0.0380(7) | 0.0444(8) | 0.0374(7) | −0.0058(6) | 0.0045(6) | 0.0128(6) |
| F1 | 0.0516(5) | 0.0533(6) | 0.0594(6) | 0.0131(4) | 0.0287(5) | 0.0306(5) |
| F2 | 0.0296(4) | 0.0493(5) | 0.0507(5) | 0.0088(4) | 0.0084(4) | 0.0178(4) |
| F3 | 0.0538(5) | 0.0258(4) | 0.0750(7) | 0.0008(4) | 0.0006(5) | −0.0021(4) |
| F4 | 0.0650(6) | 0.0577(6) | 0.0569(6) | 0.0082(5) | 0.0154(5) | 0.0322(5) |
| F5 | 0.0661(6) | 0.0702(7) | 0.0276(4) | −0.0148(5) | 0.0057(4) | 0.0044(4) |
| F6 | 0.0427(5) | 0.0755(7) | 0.0574(6) | −0.0133(5) | 0.0133(4) | 0.0236(5) |
| O1-C1 | 1.2576(15) | C6-C7 | 1.388(2) |
| O2-C10 | 1.3308(15) | C6-H6 | 0.967(18) |
| O2-H2 | 0.90(2) | C7-C8 | 1.3944(18) |
| O3-C12 | 1.3242(17) | C7-H7 | 0.947(17) |
| O3-H3 | 0.88(2) | C8-C9 | 1.4703(18) |
| C1-C2 | 1.4547(17) | C9-C12 | 1.3602(18) |
| C1-C9 | 1.4590(17) | C10-C11 | 1.5108(18) |
| C2-C3 | 1.4714(17) | C11-F1 | 1.3235(16) |
| C2-C10 | 1.3593(17) | C11-F2 | 1.3329(15) |
| C3-C4 | 1.3876(18) | C11-F3 | 1.3311(17) |
| C3-C8 | 1.4186(18) | C12-C13 | 1.5173(19) |
| C4-C5 | 1.3893(20) | C13-F4 | 1.3233(18) |
| C4-H4 | 0.903(15) | C13-F5 | 1.3375(18) |
| C5-C6 | 1.376(2) | C13-F6 | 1.3267(17) |
| C5-H5 | 0.896(17) |
| C10-O2-H2 | 105.8(12) | C3-C8-C9 | 109.21(10) |
| C12-O3-H3 | 109.1(14) | C7-C8-C9 | 131.18(12) |
| O1-C1-C2 | 125.45(11) | C1-C9-C8 | 106.17(10) |
| O1-C1-C9 | 125.23(11) | C1-C9-C12 | 118.14(11) |
| C2-C1-C9 | 109.30(10) | C8-C9-C12 | 135.54(12) |
| C1-C2-C3 | 106.48(10) | O2-C10-C2 | 123.15(11) |
| C1-C2-C10 | 118.51(11) | O2-C10-C11 | 111.48(11) |
| C3-C2-C10 | 134.99(11) | C2-C10-C11 | 125.36(11) |
| C2-C3-C4 | 131.02(12) | C10-C11-F1 | 111.72(11) |
| C2-C3-C8 | 108.80(10) | C10-C11-F2 | 111.45(10) |
| C4-C3-C8 | 120.17(11) | C10-C11-F3 | 110.99(11) |
| C3-C4-C5 | 119.18(14) | F1-C11-F2 | 107.46(11) |
| C3-C4-H4 | 120.4(9) | F1-C11-F3 | 107.80(11) |
| C5-C4-H4 | 120.4(9) | F2-C11-F3 | 107.21(11) |
| C4-C5-C6 | 120.80(14) | O3-C12-C9 | 124.23(12) |
| C4-C5-H5 | 119.0(10) | O3-C12-C13 | 111.34(12) |
| C6-C5-H5 | 120.2(10) | C9-C12-C13 | 124.39(13) |
| C5-C6-C7 | 121.13(13) | C12-C13-F4 | 111.87(13) |
| C5-C6-H6 | 117.8(10) | C12-C13-F5 | 110.68(11) |
| C7-C6-H6 | 121.0(10) | C12-C13-F6 | 112.22(11) |
| C6-C7-C8 | 119.11(14) | F4-C13-F5 | 106.89(12) |
| C6-C7-H7 | 120.5(9) | F4-C13-F6 | 108.20(12) |
| C8-C7-H7 | 120.3(9) | F5-C13-F6 | 106.69(13) |
| C3-C8-C7 | 119.60(12) |
| O1-C1-C2-C3 | 179.6(2) | C8-C3-C4-C5 | −0.78(13) |
| O1-C1-C2-C10 | −1.91(11) | C2-C3-C8-C7 | −178.6(3) |
| C9-C1-C2-C3 | −1.90(11) | C2-C3-C8-C9 | 0.26(11) |
| C9-C1-C2-C10 | 176.6(2) | C4-C3-C8-C7 | 0.88(13) |
| O1-C1-C9-C8 | −179.4(3) | C4-C3-C8-C9 | 179.7(3) |
| O1-C1-C9-C12 | 4.37(11) | C3-C4-C5-C6 | 0.17(13) |
| C2-C1-C9-C8 | 2.04(11) | C4-C5-C6-C7 | 0.35(13) |
| C2-C1-C9-C12 | −174.2(3) | C5-C6-C7-C8 | −0.25(13) |
| C1-C2-C3-C4 | −178.4(2) | C6-C7-C8-C3 | −0.36(13) |
| C1-C2-C3-C8 | 1.00(11) | C6-C7-C8-C9 | −178.9(3) |
| C10-C2-C3-C4 | 3.44(12) | C3-C8-C9-C1 | −1.41(11) |
| C10-C2-C3-C8 | −177.2(3) | C3-C8-C9-C12 | 173.8(3) |
| C1-C2-C10-O2 | 2.08(10) | C7-C8-C9-C1 | 177.3(3) |
| C1-C2-C10-C11 | −177.3(2) | C7-C8-C9-C12 | −7.50(13) |
| C3-C2-C10-O2 | −179.9(2) | C1-C9-C12-O3 | −4.98(11) |
| C3-C2-C10-C11 | 0.67(11) | C1-C9-C12-C13 | 172.4(3) |
| C2-C3-C4-C5 | 178.6(3) |
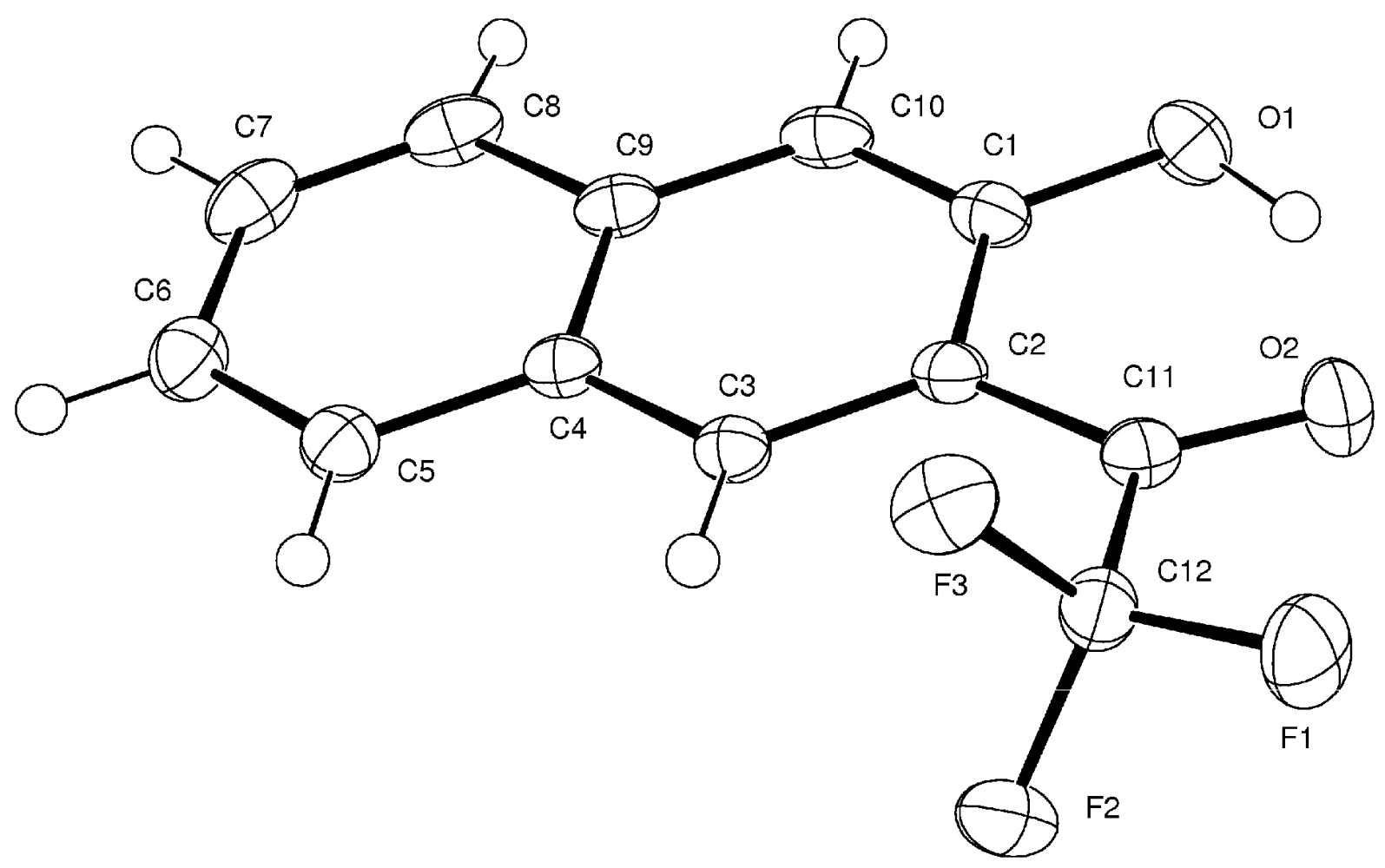
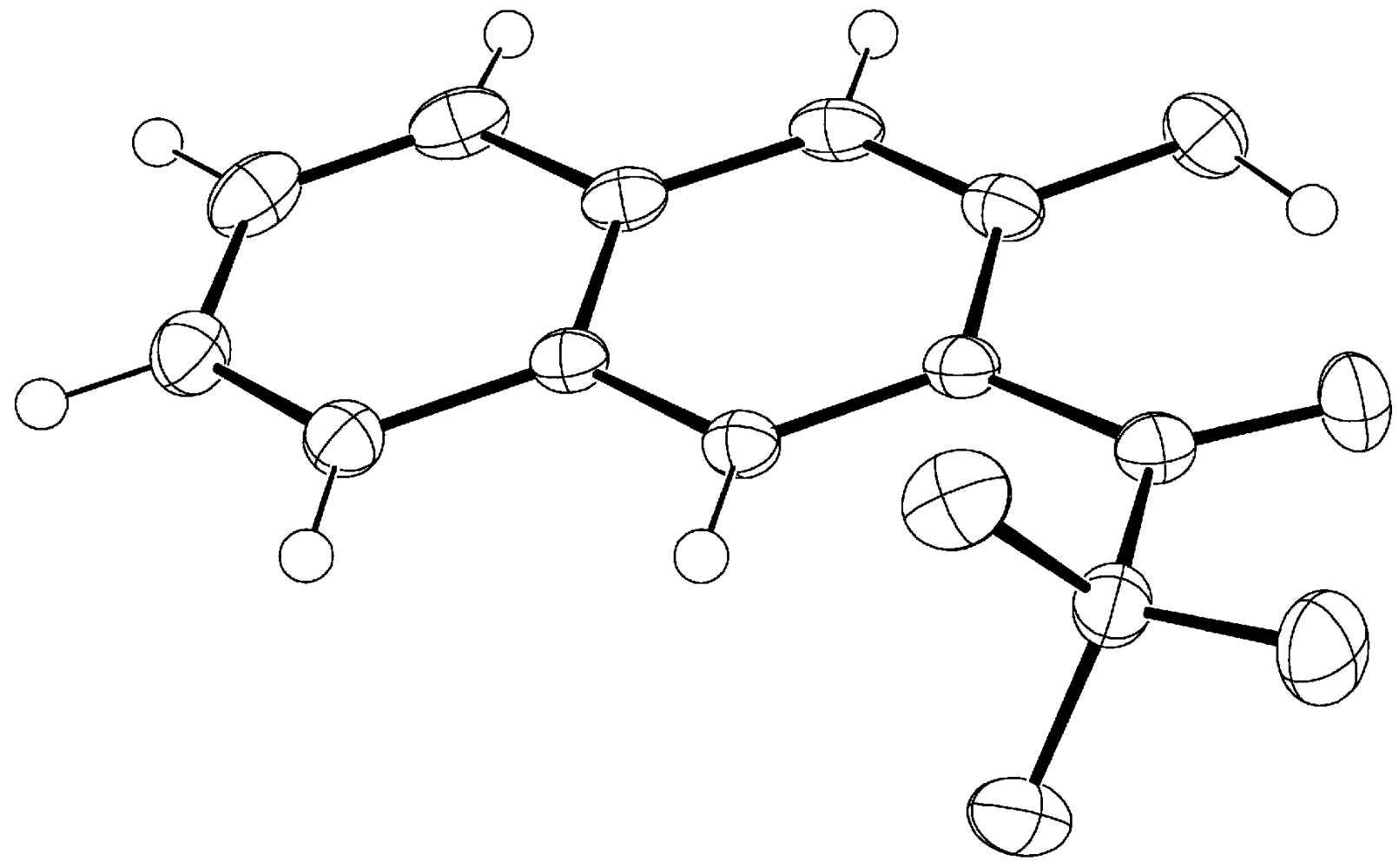
3. Compound 3d
Experimental for C12H7F3O2 (3d)

| Formula | C12H7F3O2 |
| Formula Weight (g/mol) | 240.18 |
| Crystal Dimensions (mm ) | 0.46 × 0.08 × 0.04 |
| Crystal Color and Habit | orange yellow needle |
| Crystal System | orthorhombic |
| Space Group | P n a 21 |
| Temperature, K | 110 |
| a, Å | 13.5923(5) |
| b, Å | 14.9695(5) |
| c, Å | 4.8381(2) |
| α,° | 90.00 |
| β,° | 90.00 |
| γ,° | 90.00 |
| V, Å3 | 984.41(6) |
| Number of reflections to determine final unit cell | 5859 |
| Min and Max 2θ for cell determination, ° | 5.44, 52.66 |
| Z | 4 |
| F(000) | 488 |
| ρ (g/cm) | 1.621 |
| λ, Å, (MoKα) | 0.71070 |
| μ, (cm−1) | 0.147 |
| Diffractometer Type | Bruker-Nonius X8 Apex2 |
| Scan Type(s) | omega and phi scans |
| Max 2θ for data collection, ° | 62.92 |
| Measured fraction of data | 0.874 |
| Number of reflections measured | 21568 |
| Unique reflections measured | 2632 |
| Rmerge | 0.0444 |
| Number of reflections included in refinement | 2632 |
| Cut off Threshold Expression | >2sigma(I) |
| Structure refined using | full matrix least-squares using F2 |
| Weighting Scheme | calc w = 1/[sigma2(Fo2) + (0.0406P)2 + 0.0000P] where P=(Fo2 + 2Fc2)/3 |
| Number of parameters in least-squares | 182 |
| R1 | 0.0370 |
| wR2 | 0.0712 |
| R1 (all data) | 0.0538 |
| wR2 (all data) | 0.0762 |
| GOF | 1.035 |
| Maximum shift/error | 0.000 |
| Min & Max peak heights on final ΔF Map (e-/Å) | −0.229, 0.183 |
| Atom | x | y | z | Uiso/equiv |
|---|---|---|---|---|
| O1 | 0.11391(8) | 0.26937(8) | 0.6424(3) | 0.0315(3) |
| O2 | 0.22469(8) | 0.37533(7) | 0.3464(3) | 0.0294(3) |
| C1 | 0.19885(11) | 0.24126(10) | 0.7623(4) | 0.0227(3) |
| C2 | 0.29250(11) | 0.27865(9) | 0.6863(4) | 0.0190(3) |
| C3 | 0.37673(11) | 0.24539(9) | 0.8101(4) | 0.0193(3) |
| C4 | 0.37205(11) | 0.17792(10) | 1.0120(4) | 0.0192(3) |
| C5 | 0.45775(12) | 0.14260(10) | 1.1386(4) | 0.0230(3) |
| C6 | 0.45137(13) | 0.07873(10) | 1.3373(4) | 0.0269(3) |
| C7 | 0.35837(14) | 0.04578(11) | 1.4184(4) | 0.0300(4) |
| C8 | 0.27466(13) | 0.07695(10) | 1.3003(4) | 0.0275(4) |
| C9 | 0.27804(11) | 0.14416(10) | 1.0917(4) | 0.0213(3) |
| C10 | 0.19325(12) | 0.17720(11) | 0.9629(4) | 0.0247(3) |
| C11 | 0.29517(12) | 0.35023(10) | 0.4818(4) | 0.0217(3) |
| C12 | 0.39217(11) | 0.40275(10) | 0.4367(4) | 0.0224(3) |
| F1 | 0.37724(7) | 0.47125(6) | 0.2675 | 0.0341(3) |
| F2 | 0.46266(6) | 0.35198(6) | 0.3256(3) | 0.0286(2) |
| F3 | 0.42682(6) | 0.43562(6) | 0.6742(3) | 0.0297(2) |
| H1 | 0.1301(19) | 0.3131(16) | 0.500(7) | 0.079(9) |
| H3 | 0.4391(12) | 0.2686(10) | 0.766(4) | 0.022(4) |
| H5 | 0.5197(14) | 0.1674(10) | 1.079(4) | 0.033(5) |
| H6 | 0.5078(13) | 0.0544(10) | 1.422(4) | 0.026(4) |
| H7 | 0.3543(13) | 0.0040(11) | 1.553(4) | 0.030(5) |
| H8 | 0.2120(15) | 0.0566(12) | 1.334(5) | 0.053(6) |
| Atom | u11 | u22 | u33 | u12 | u13 | u23 |
|---|---|---|---|---|---|---|
| O1 | 0.0164(6) | 0.0433(7) | 0.0347(7) | 0.0007(5) | −0.0018(5) | 0.0012(6) |
| O2 | 0.0251(6) | 0.0319(6) | 0.0312(6) | 0.0036(5) | −0.0074(6) | 0.0048(6) |
| C1 | 0.0174(8) | 0.0271(8) | 0.0236(8) | −0.0003(6) | 0.0012(6) | −0.0060(7) |
| C2 | 0.0180(8) | 0.0211(7) | 0.0178(7) | 0.0006(6) | 0.0021(6) | −0.0031(6) |
| C3 | 0.0174(8) | 0.0209(7) | 0.0196(7) | −0.0016(6) | 0.0019(6) | −0.0013(7) |
| C4 | 0.0199(8) | 0.0195(7) | 0.0182(7) | −0.0011(6) | 0.0023(6) | −0.0032(6) |
| C5 | 0.0249(9) | 0.0212(8) | 0.0229(8) | 0.0011(6) | 0.0005(7) | −0.0006(6) |
| C6 | 0.0338(9) | 0.0226(8) | 0.0245(8) | 0.0051(7) | −0.0020(7) | 0.0001(7) |
| C7 | 0.0471(11) | 0.0208(8) | 0.0223(9) | −0.0038(7) | 0.0034(7) | 0.0020(7) |
| C8 | 0.0324(9) | 0.0256(8) | 0.0247(9) | −0.0107(7) | 0.0084(8) | −0.0037(7) |
| C9 | 0.0249(8) | 0.0193(7) | 0.0198(7) | −0.0031(6) | 0.0041(6) | −0.0060(6) |
| C10 | 0.0178(8) | 0.0300(8) | 0.0264(8) | −0.0068(7) | 0.0072(7) | −0.0062(7) |
| C11 | 0.0226(9) | 0.0230(8) | 0.0195(8) | 0.0020(6) | 0.0017(6) | −0.0026(6) |
| C12 | 0.0238(8) | 0.0226(8) | 0.0208(7) | 0.0010(6) | −0.0013(7) | 0.0006(6) |
| F1 | 0.0369(6) | 0.0279(5) | 0.0375(6) | −0.0005(4) | −0.0013(5) | 0.0126(5) |
| F2 | 0.0238(5) | 0.0291(5) | 0.0329(5) | 0.0011(4) | 0.0082(4) | −0.0003(4) |
| F3 | 0.0309(5) | 0.0300(5) | 0.0282(5) | −0.0080(4) | −0.0005(4) | −0.0051(4) |
| O1-C1 | 1.3588(19) | C6-C7 | 1.412(2) |
| O1-H1 | 0.97(3) | C6-H6 | 0.941(18) |
| O2-C11 | 1.2199(18) | C7-C8 | 1.356(2) |
| C1-C10 | 1.367(2) | C7-H7 | 0.905(18) |
| C1-C2 | 1.438(2) | C8-C9 | 1.426(2) |
| C2-C3 | 1.385(2) | C8-H8 | 0.92(2) |
| C2-C11 | 1.459(2) | C9-C10 | 1.400(2) |
| C3-C4 | 1.406(2) | C10-H10 | 0.939(18) |
| C3-H3 | 0.941(16) | C11-C12 | 1.551(2) |
| C4-C5 | 1.418(2) | C12-F1 | 1.3278(18) |
| C4-C9 | 1.427(2) | C12-F3 | 1.3356(19) |
| C5-C6 | 1.359(2) | C12-F2 | 1.3359(18) |
| C5-H5 | 0.963(19) |
| C1-O1-H1 | 108.5(16) | C8-C7-H7 | 119.3(12) |
| O1-C1-C10 | 118.24(14) | C6-C7-H7 | 119.8(12) |
| O1-C1-C2 | 121.49(14) | C7-C8-C9 | 120.94(15) |
| C10-C1-C2 | 120.25(14) | C7-C8-H8 | 126.2(13) |
| C3-C2-C1 | 118.78(13) | C9-C8-H8 | 112.8(13) |
| C3-C2-C11 | 122.46(13) | C10-C9-C8 | 122.53(15) |
| C1-C2-C11 | 118.76(13) | C10-C9-C4 | 119.43(15) |
| C2-C3-C4 | 121.41(13) | C8-C9-C4 | 118.04(15) |
| C2-C3-H3 | 121.0(10) | C1-C10-C9 | 121.21(15) |
| C4-C3-H3 | 117.5(10) | C1-C10-H10 | 116.9(11) |
| C3-C4-C5 | 122.04(13) | C9-C10-H10 | 121.8(11) |
| C3-C4-C9 | 118.85(13) | O2-C11-C2 | 124.81(14) |
| C5-C4-C9 | 119.11(14) | O2-C11-C12 | 115.85(13) |
| C6-C5-C4 | 121.03(15) | C2-C11-C12 | 119.28(13) |
| C6-C5-H5 | 122.5(11) | F1-C12-F3 | 107.45(12) |
| C4-C5-H5 | 116.5(11) | F1-C12-F2 | 107.51(13) |
| C5-C6-C7 | 119.96(16) | F3-C12-F2 | 107.63(12) |
| C5-C6-H6 | 121.7(10) | F1-C12-C11 | 110.39(12) |
| C7-C6-H6 | 118.3(10) | F3-C12-C11 | 111.44(13) |
| C8-C7-C6 | 120.91(16) | F2-C12-C11 | 112.21(12) |
| O1-C1-C2-C3 | −178.15(14) | C3-C4-C9-C8 | −178.87(13) |
| C10-C1-C2-C3 | 3.2(2) | C5-C4-C9-C8 | 1.1(2) |
| O1-C1-C2-C11 | 1.9(2) | O1-C1-C10-C9 | 178.95(14) |
| C10-C1-C2-C11 | −176.80(13) | C2-C1-C10-C9 | −2.3(2) |
| C1-C2-C3-C4 | −1.6(2) | C8-C9-C10-C1 | −179.58(14) |
| C11-C2-C3-C4 | 178.34(13) | C4-C9-C10-C1 | −0.1(2) |
| C2-C3-C4-C5 | 179.37(14) | C3-C2-C11-O2 | 171.95(15) |
| C2-C3-C4-C9 | −0.7(2) | C1-C2-C11-O2 | −8.1(2) |
| C3-C4-C5-C6 | 178.62(14) | C3-C2-C11-C12 | −11.0(2) |
| C9-C4-C5-C6 | −1.3(2) | C1-C2-C11-C12 | 168.99(13) |
| C4-C5-C6-C7 | 0.7(2) | O2-C11-C12-F1 | 4.49(18) |
| C5-C6-C7-C8 | 0.1(2) | C2-C11-C12-F1 | −172.83(12) |
| C6-C7-C8-C9 | −0.3(2) | O2-C11-C12-F3 | 123.80(14) |
| C7-C8-C9-C10 | 179.23(15) | C2-C11-C12-F3 | −53.51(17) |
| C7-C8-C9-C4 | −0.3(2) | O2-C11-C12-F2 | −115.41(15) |
| C3-C4-C9-C10 | 1.6(2) | C2-C11-C12-F2 | 67.28(18) |
| C5-C4-C9-C10 | −178.48(14) |
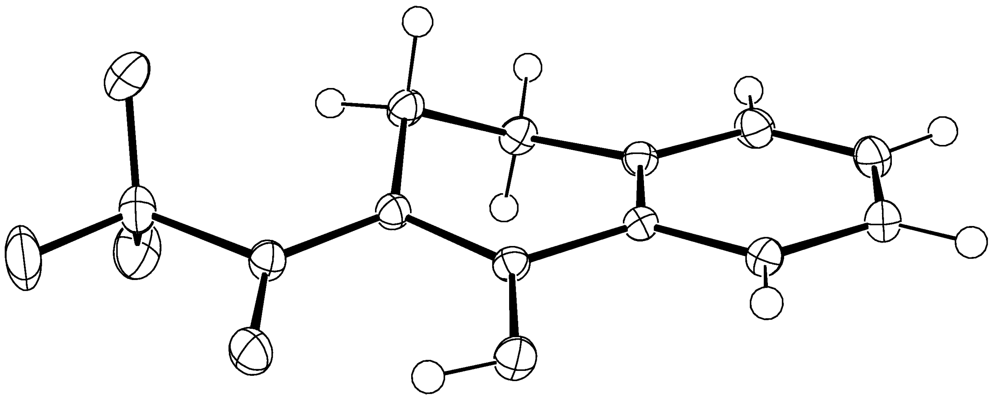
4. Compound 4c
Experimental for C12H9F3O2 (4c)

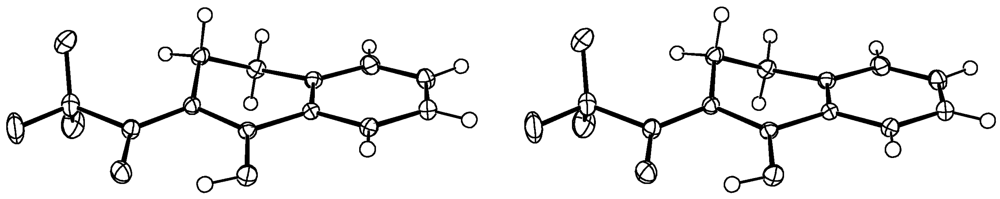
| Formula | C12H9F3O2 | |
| Formula Weight (g/mol) | 242.19 | |
| Crystal Dimensions (mm ) | 0.38 × 0.28 × 0.04 | |
| Crystal Color and Habit | colourless plate | |
| Crystal System | triclinic | |
| Space Group | P -1 | |
| Temperature, K | 110 | |
| a, Å | 7.3528(2) | |
| b, Å | 7.9165(2) | |
| c, Å | 9.7991(2) | |
| α,° | 73.0533(11) | |
| β,° | 85.3968(12) | |
| γ,° | 68.3581(11) | |
| V, Å3 | 506.92(2) | |
| Reflections to determine final unit cell | 6416 | |
| Min and Max 2θ for cell determination, ° | 5.78, 71.38 | |
| Z | 2 | |
| F(000) | 248 | |
| ρ (g/cm) | 1.587 | |
| λ, Å, (MoK ) | 0.71073 | |
| μ, (cm−1) | 0.143 | |
| Number of reflections measured | 20479 | |
| Unique reflections measured | 4691 | |
| Rmerge | 0.0265 | |
| Number of reflections included in refinement | 4691 | |
| Cut off Threshold Expression | >2sigma(I) | |
| Structure refined using | full matrix least-squares using F2 | |
| Weighting Scheme | w = 1/[sigma2(Fo2) + (0.0707P)2 + 0.0436P] where P = (Fo2 + 2Fc2)/3 | |
| R1 | 0.0382 | |
| wR2 | 0.1082 | |
| R1 (all data) | 0.0525 | |
| wR2 (all data) | 0.1220 | |
| GOF | 1.048 | |
| Atom | x | y | z | Uiso/equiv |
|---|---|---|---|---|
| O1 | 0.77448(8) | 0.28007(9) | 0.27345(6) | 0.01748(12) |
| O2 | 0.99446(9) | 0.28522(9) | 0.06442(6) | 0.02064(13) |
| C1 | 0.92164(10) | 0.26838(10) | 0.34926(7) | 0.01305(12) |
| C2 | 1.10003(10) | 0.26974(10) | 0.29126(8) | 0.01358(12) |
| C3 | 1.25154(11) | 0.27188(11) | 0.38557(8) | 0.01584(13) |
| C4 | 1.24786(11) | 0.15172(11) | 0.53827(8) | 0.01618(13) |
| C5 | 1.04451(11) | 0.19875(10) | 0.59451(8) | 0.01444(13) |
| C6 | 1.00916(12) | 0.18008(12) | 0.73914(8) | 0.01899(15) |
| C7 | 0.82002(13) | 0.21517(12) | 0.78875(9) | 0.02070(15) |
| C8 | 0.66361(12) | 0.26962(12) | 0.69526(9) | 0.01971(15) |
| C9 | 0.69574(11) | 0.28853(11) | 0.55100(8) | 0.01598(13) |
| C10 | 0.88579(10) | 0.25358(10) | 0.50052(7) | 0.01326(12) |
| C11 | 1.12232(11) | 0.27815(10) | 0.14448(8) | 0.01590(13) |
| C12 | 1.31898(12) | 0.27354(12) | 0.07494(9) | 0.02008(15) |
| F1 | 1.37562(8) | 0.40737(8) | 0.09309(6) | 0.02557(13) |
| F2 | 1.46211(8) | 0.10631(8) | 0.12997(6) | 0.03004(14) |
| F3 | 1.30731(9) | 0.29835(10) | −0.06471(6) | 0.03180(15) |
| H1 | 0.818(3) | 0.282(3) | 0.190(2) | 0.066(5) |
| H3A | 1.2210(17) | 0.4028(17) | 0.3866(12) | 0.018(3) |
| H3B | 1.386(2) | 0.2261(18) | 0.3516(13) | 0.026(3) |
| H4A | 1.3333(19) | 0.1700(17) | 0.5989(13) | 0.021(3) |
| H4B | 1.2933(18) | 0.0209(17) | 0.5436(13) | 0.020(3) |
| H6 | 1.120(2) | 0.1395(19) | 0.8020(14) | 0.030(3) |
| H7 | 0.806(2) | 0.204(2) | 0.8859(17) | 0.041(4) |
| H8 | 0.526(2) | 0.304(2) | 0.7231(14) | 0.033(3) |
| H9 | 0.5882(18) | 0.3326(17) | 0.4836(13) | 0.019(3) |
| Atom | u11 | u22 | u33 | u12 | u13 | u23 |
|---|---|---|---|---|---|---|
| O1 | 0.0149(2) | 0.0246(3) | 0.0147(2) | −0.0091(2) | −0.00183(19) | −0.0048(2) |
| O2 | 0.0229(3) | 0.0269(3) | 0.0145(2) | −0.0113(2) | −0.0001(2) | −0.0061(2) |
| C1 | 0.0127(3) | 0.0135(3) | 0.0133(3) | −0.0052(2) | −0.0005(2) | −0.0035(2) |
| C2 | 0.0129(3) | 0.0155(3) | 0.0131(3) | −0.0059(2) | 0.0007(2) | −0.0042(2) |
| C3 | 0.0137(3) | 0.0189(3) | 0.0166(3) | −0.0081(2) | 0.0007(2) | −0.0046(2) |
| C4 | 0.0134(3) | 0.0184(3) | 0.0162(3) | −0.0059(2) | −0.0019(2) | −0.0035(2) |
| C5 | 0.0151(3) | 0.0148(3) | 0.0143(3) | −0.0062(2) | 0.0003(2) | −0.0044(2) |
| C6 | 0.0226(4) | 0.0212(3) | 0.0144(3) | −0.0091(3) | −0.0007(3) | −0.0051(3) |
| C7 | 0.0270(4) | 0.0225(3) | 0.0156(3) | −0.0115(3) | 0.0055(3) | −0.0078(3) |
| C8 | 0.0210(4) | 0.0215(3) | 0.0203(3) | −0.0104(3) | 0.0077(3) | −0.0097(3) |
| C9 | 0.0142(3) | 0.0171(3) | 0.0185(3) | −0.0069(2) | 0.0030(2) | −0.0068(2) |
| C10 | 0.0133(3) | 0.0143(3) | 0.0136(3) | −0.0061(2) | 0.0012(2) | −0.0047(2) |
| C11 | 0.0174(3) | 0.0162(3) | 0.0140(3) | −0.0067(2) | 0.0021(2) | −0.0038(2) |
| C12 | 0.0204(3) | 0.0226(4) | 0.0166(3) | −0.0077(3) | 0.0049(3) | −0.0058(3) |
| F1 | 0.0244(3) | 0.0283(3) | 0.0279(3) | −0.0162(2) | 0.0068(2) | −0.0065(2) |
| F2 | 0.0215(3) | 0.0254(3) | 0.0345(3) | −0.0013(2) | 0.0080(2) | −0.0070(2) |
| F3 | 0.0341(3) | 0.0473(4) | 0.0169(2) | −0.0181(3) | 0.0108(2) | −0.0115(2) |
| O1-C1 | 1.3215(9) | C5-C10 | 1.4016(10) |
| O1-H1 | 0.855(19) | C6-C7 | 1.3903(12) |
| O2-C11 | 1.2476(10) | C6-H6 | 0.960(14) |
| C1-C2 | 1.3895(10) | C7-C8 | 1.3860(12) |
| C1-C10 | 1.4626(10) | C7-H7 | 0.931(15) |
| C2-C11 | 1.4193(10) | C8-C9 | 1.3877(11) |
| C2-C3 | 1.5112(10) | C8-H8 | 0.984(14) |
| C3-C4 | 1.5255(11) | C9-C10 | 1.3992(10) |
| C3-H3A | 0.979(12) | C9-H9 | 0.964(12) |
| C3-H3B | 0.984(13) | C11-C12 | 1.5408(11) |
| C4-C5 | 1.5016(10) | C12-F3 | 1.3298(10) |
| C4-H4A | 0.972(12) | C12-F1 | 1.3322(10) |
| C4-H4B | 0.950(12) | C12-F2 | 1.3410(10) |
| C5-C6 | 1.3946(10) |
| C1-O1-H1 | 105.2(13) | C7-C6-H6 | 122.1(8) |
| O1-C1-C2 | 123.08(7) | C5-C6-H6 | 117.4(8) |
| O1-C1-C10 | 115.65(6) | C8-C7-C6 | 120.54(7) |
| C2-C1-C10 | 121.28(6) | C8-C7-H7 | 123.4(9) |
| C1-C2-C11 | 116.88(7) | C6-C7-H7 | 116.1(9) |
| C1-C2-C3 | 118.29(6) | C7-C8-C9 | 119.86(7) |
| C11-C2-C3 | 124.76(6) | C7-C8-H8 | 124.2(8) |
| C2-C3-C4 | 111.06(6) | C9-C8-H8 | 115.9(8) |
| C2-C3-H3A | 108.3(7) | C8-C9-C10 | 119.82(7) |
| C4-C3-H3A | 108.5(7) | C8-C9-H9 | 121.1(7) |
| C2-C3-H3B | 113.2(7) | C10-C9-H9 | 119.0(7) |
| C4-C3-H3B | 108.2(8) | C9-C10-C5 | 120.59(7) |
| H3A-C3-H3B | 107.4(10) | C9-C10-C1 | 120.17(7) |
| C5-C4-C3 | 112.07(6) | C5-C10-C1 | 119.22(6) |
| C5-C4-H4A | 110.3(7) | O2-C11-C2 | 125.09(7) |
| C3-C4-H4A | 109.1(7) | O2-C11-C12 | 115.47(7) |
| C5-C4-H4B | 105.9(7) | C2-C11-C12 | 119.42(7) |
| C3-C4-H4B | 111.1(7) | F3-C12-F1 | 107.53(7) |
| H4A-C4-H4B | 108.2(10) | F3-C12-F2 | 107.30(7) |
| C6-C5-C10 | 118.70(7) | F1-C12-F2 | 107.24(7) |
| C6-C5-C4 | 121.98(7) | F3-C12-C11 | 110.83(7) |
| C10-C5-C4 | 119.24(6) | F1-C12-C11 | 112.56(6) |
| C7-C6-C5 | 120.50(8) | F2-C12-C11 | 111.13(6) |
| O1-C1-C2-C11 | −2.04(11) | C4-C5-C10-C9 | 176.58(7) |
| C10-C1-C2-C11 | 177.93(6) | C6-C5-C10-C1 | −178.26(7) |
| O1-C1-C2-C3 | 175.03(7) | C4-C5-C10-C1 | −1.57(10) |
| C10-C1-C2-C3 | −4.99(10) | O1-C1-C10-C9 | −12.24(10) |
| C1-C2-C3-C4 | 36.71(9) | C2-C1-C10-C9 | 167.79(7) |
| C11-C2-C3-C4 | −146.47(7) | O1-C1-C10-C5 | 165.92(6) |
| C2-C3-C4-C5 | −49.62(8) | C2-C1-C10-C5 | −14.06(10) |
| C3-C4-C5-C6 | −149.82(7) | C1-C2-C11-O2 | 0.34(11) |
| C3-C4-C5-C10 | 33.60(9) | C3-C2-C11-O2 | −176.52(7) |
| C10-C5-C6-C7 | 0.04(11) | C1-C2-C11-C12 | −178.03(6) |
| C4-C5-C6-C7 | −176.56(7) | C3-C2-C11-C12 | 5.11(11) |
| C5-C6-C7-C8 | −0.13(12) | O2-C11-C12-F3 | 6.60(10) |
| C6-C7-C8-C9 | 0.30(12) | C2-C11-C12-F3 | −174.87(7) |
| C7-C8-C9-C10 | −0.37(11) | O2-C11-C12-F1 | 127.07(8) |
| C8-C9-C10-C5 | 0.28(11) | C2-C11-C12-F1 | −54.40(10) |
| C8-C9-C10-C1 | 178.41(7) | O2-C11-C12-F2 | −112.62(8) |
| C6-C5-C10-C9 | −0.11(11) | C2-C11-C12-F2 | 65.91(9) |
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sloop, J.C.; Boyle, P.D.; Fountain, A.W.; Gomez, C.; Jackson, J.L.; Pearman, W.F.; Schmidt, R.D.; Weyand, J. Novel Fluorinated Indanone, Tetralone and Naphthone Derivatives: Synthesis and Unique Structural Features. Appl. Sci. 2012, 2, 61-99. https://doi.org/10.3390/app2010061
Sloop JC, Boyle PD, Fountain AW, Gomez C, Jackson JL, Pearman WF, Schmidt RD, Weyand J. Novel Fluorinated Indanone, Tetralone and Naphthone Derivatives: Synthesis and Unique Structural Features. Applied Sciences. 2012; 2(1):61-99. https://doi.org/10.3390/app2010061
Chicago/Turabian StyleSloop, Joseph C., Paul D. Boyle, Augustus W. Fountain, Cristina Gomez, James L. Jackson, William F. Pearman, Robert D. Schmidt, and Jonathan Weyand. 2012. "Novel Fluorinated Indanone, Tetralone and Naphthone Derivatives: Synthesis and Unique Structural Features" Applied Sciences 2, no. 1: 61-99. https://doi.org/10.3390/app2010061







