Overexpression of the QKI Gene Promotes Differentiation of Goat Myoblasts into Myotubes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Goat-Myoblast Isolation and Culture
2.2. Total-RNA Extraction and cDNA Synthesis
2.3. Primer Design and Overexpression Vector Construction
2.4. Cell Transfection and Induced Differentiation
2.5. Immunofluorescence
2.6. Western Blotting
2.7. RNA Extraction, Library Construction, and Transcriptome Sequencing
2.8. Quality Control and Sequence Alignment
2.9. Gene-Expression Quantification and Analysis of Differentially Expressed Genes (DEGs)
2.10. GO and KEGG Enrichment Analyses of DEGs
3. Results
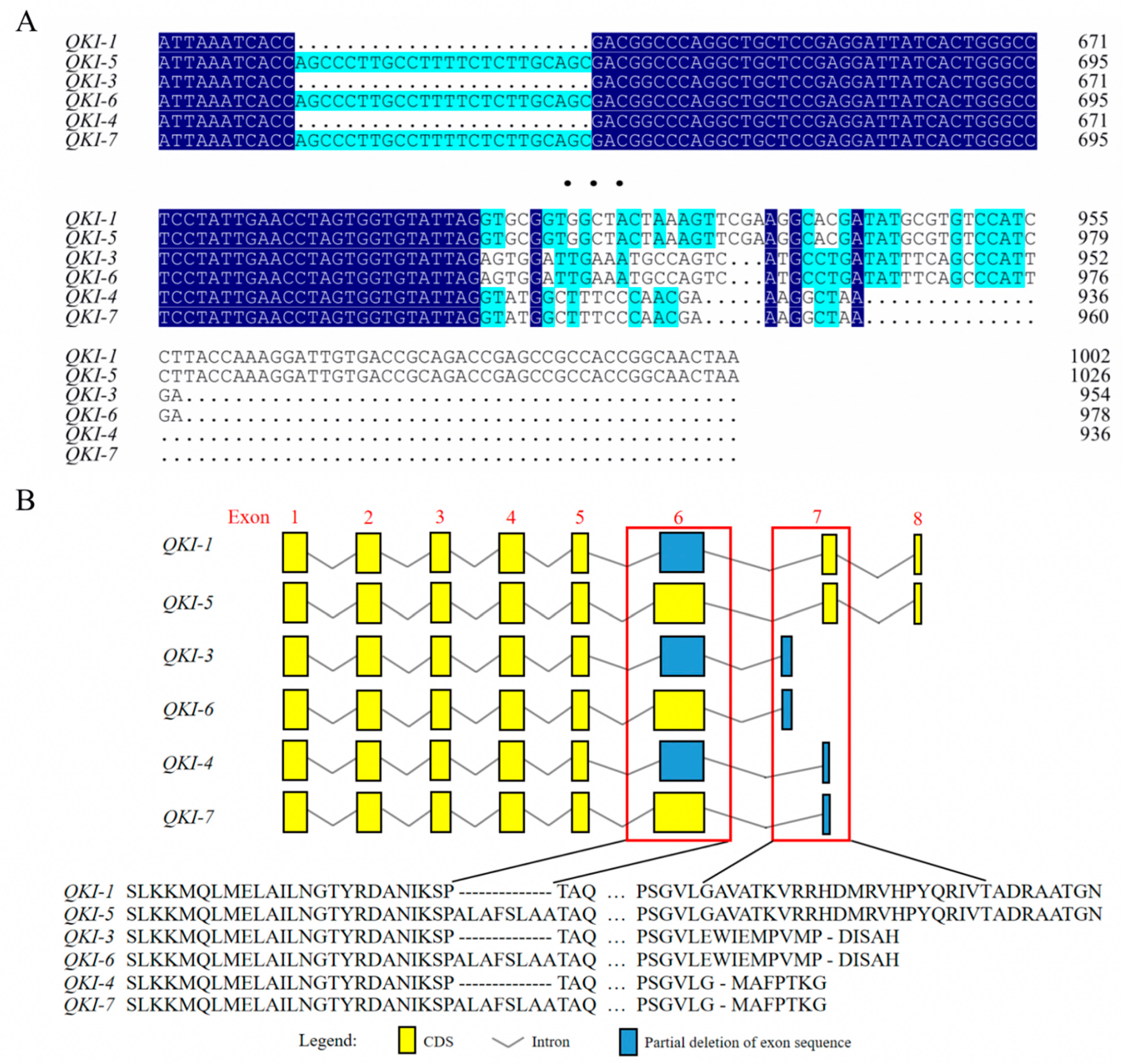
3.1. Sequencing Analyses of Various Goat QKI Isoforms
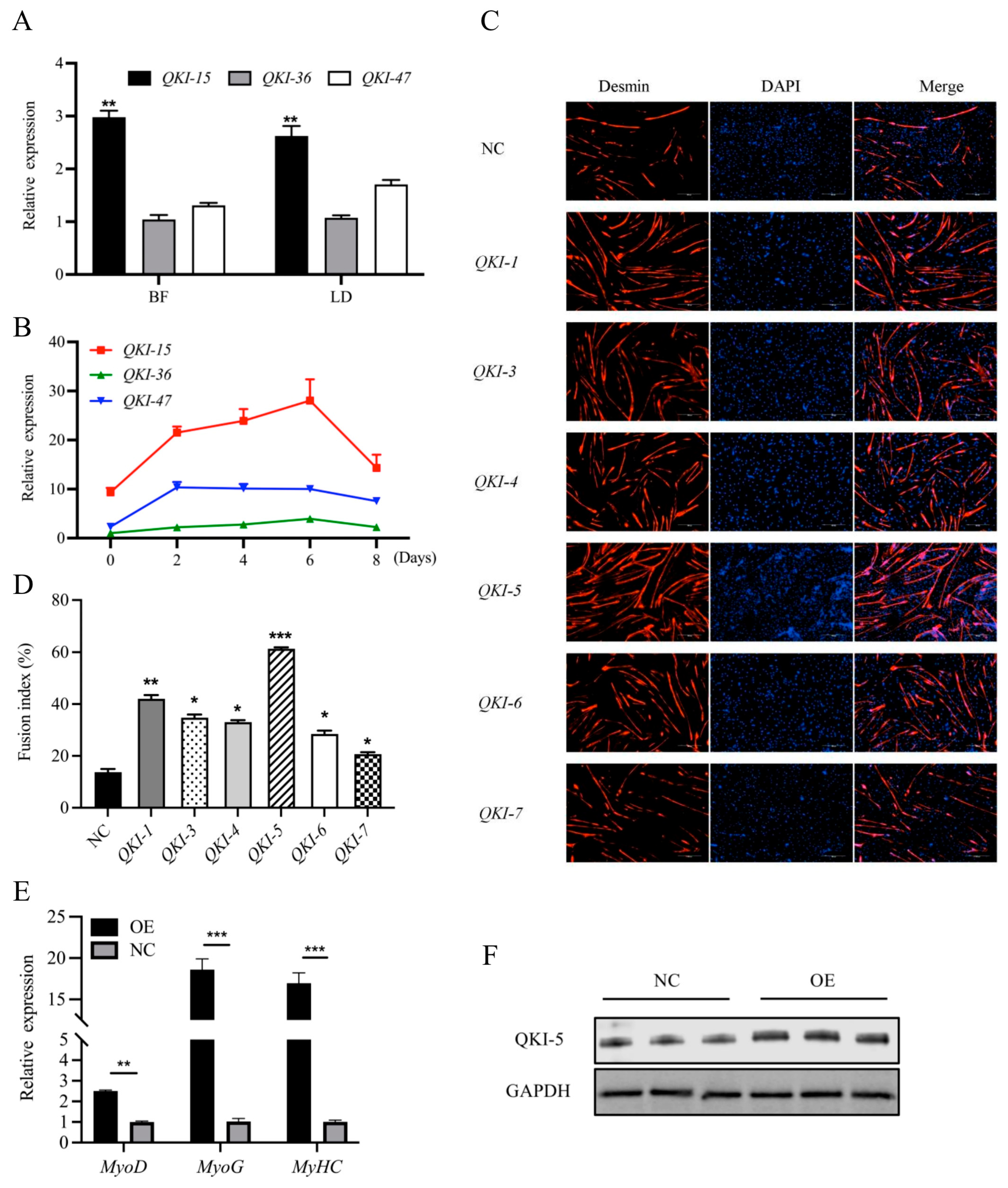
3.2. Expression Analysis of Different QKI Isoforms in Goats-Muscle Cells and Tissues
3.3. QKI Regulation of Myoblast Myogenesis and Identification of the Major QKI Isoform
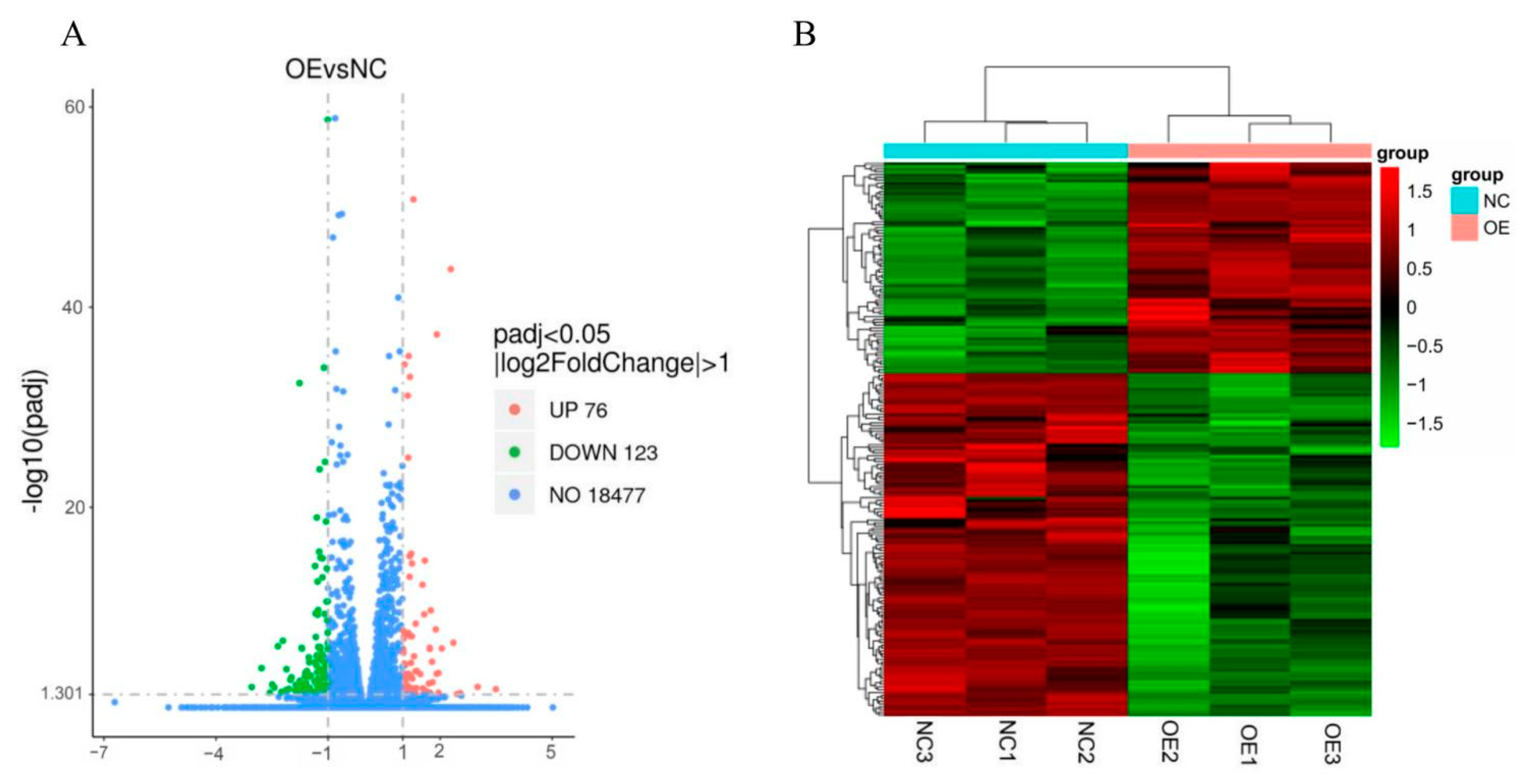
3.4. Summary of RNA-Seq Data
3.5. Differentially Expressed Gene (DEG) Analysis
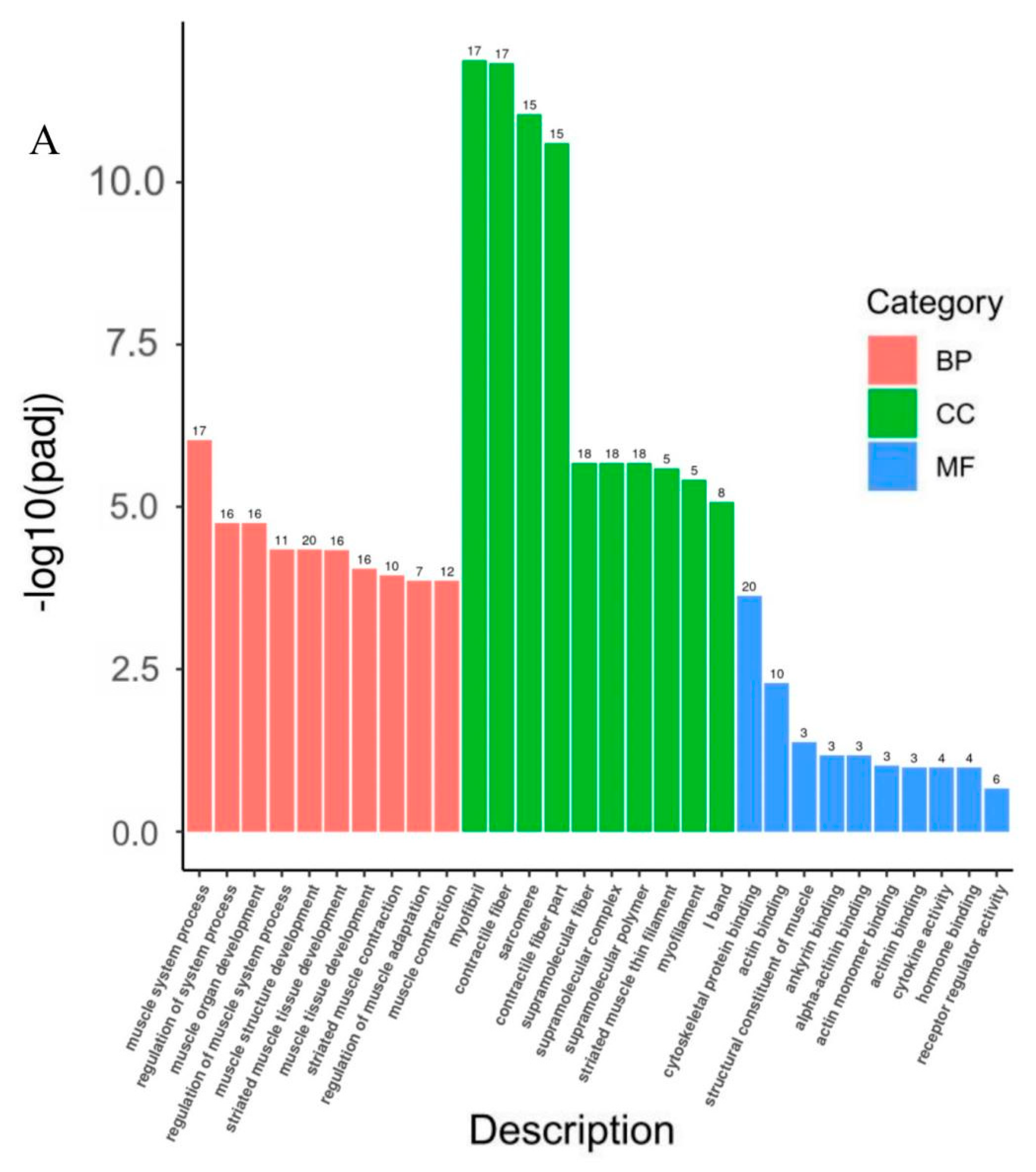
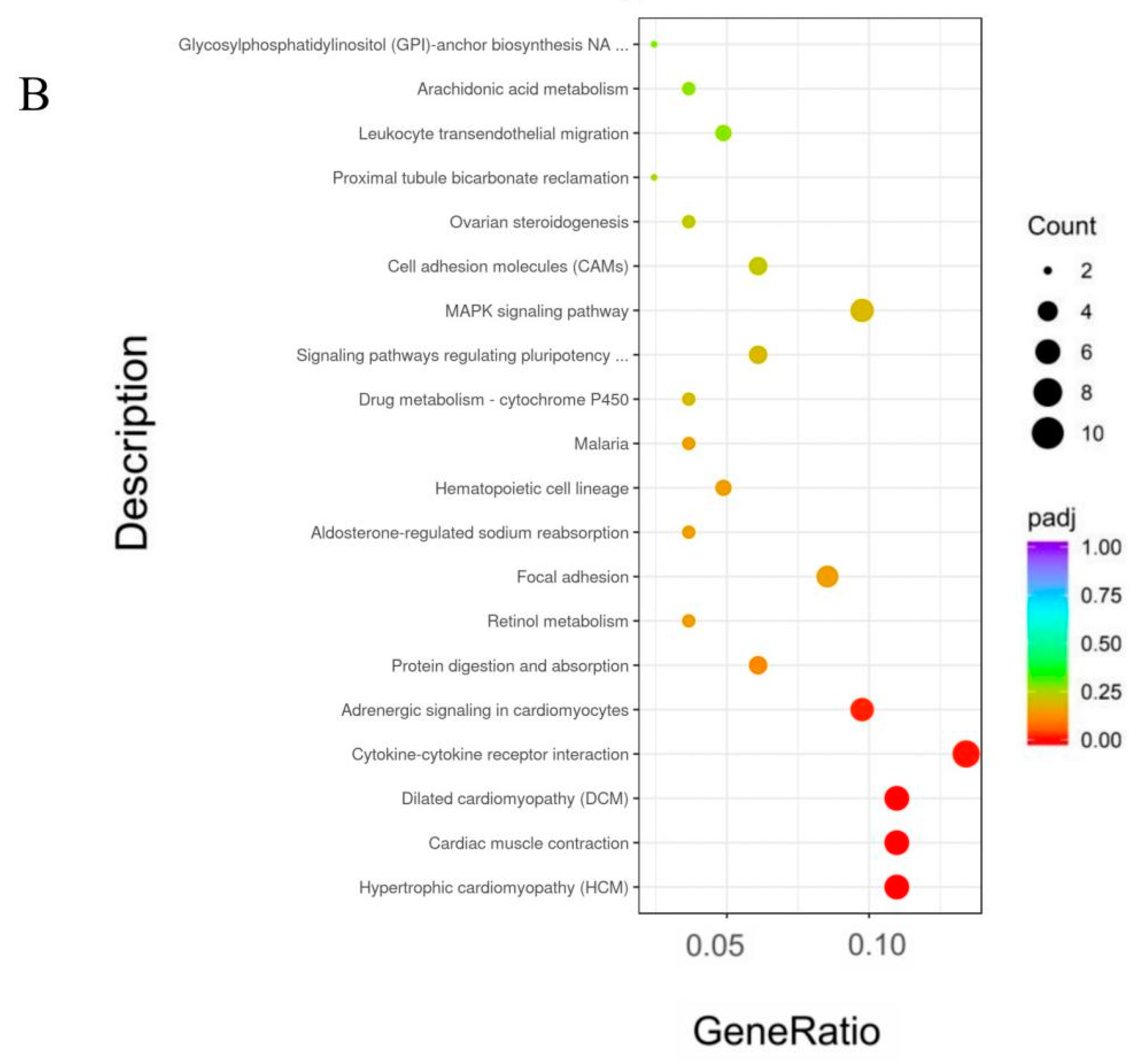
3.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analyses of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mukund, K.; Subramaniam, S. Skeletal muscle: A review of molecular structure and function, in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1462. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Dilworth, F.J. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. FEBS J. 2013, 280, 3991–4003. [Google Scholar] [CrossRef] [PubMed]
- Bismuth, K.; Relaix, F. Genetic regulation of skeletal muscle development. Exp. Cell Res. 2010, 316, 3081–3086. [Google Scholar] [CrossRef]
- Apponi, L.H.; Corbett, A.H.; Pavlath, G.K. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol. Sci. 2011, 32, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef]
- Blech-Hermoni, Y.; Ladd, A.N. RNA binding proteins in the regulation of heart development. Int. J. Biochem. Cell Biol. 2013, 45, 2467–2478. [Google Scholar] [CrossRef]
- Fredericks, A.M.; Cygan, K.J.; Brown, B.A.; Fairbrother, W.G. RNA-binding proteins: Splicing factors and disease. Biomolecules 2015, 5, 893–909. [Google Scholar] [CrossRef]
- Hinkle, E.R.; Wiedner, H.J.; Black, A.J.; Giudice, J. RNA processing in skeletal muscle biology and disease. Transcription 2019, 10, 1–20. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Li, K.; Du, Y.; Cui, K.; Yu, P.; Zhang, T.; Liu, H.; Ma, W. An interactive network of alternative splicing events with prognostic value in geriatric lung adenocarcinoma via the regulation of splicing factors. Biosci. Rep. 2020, 40, BSR20202338. [Google Scholar] [CrossRef]
- Pedrotti, S.; Giudice, J.; Dagnino-Acosta, A.; Knoblauch, M.; Singh, R.K.; Hanna, A.; Mo, Q.X.; Hicks, J.; Hamilton, S.; Cooper, T.A. The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function. Hum. Mol. Genet. 2015, 24, 2360–2374. [Google Scholar] [CrossRef]
- Biedermann, B.; Hotz, H.R.; Ciosk, R. The Quaking family of RNA-binding proteins: Coordinators of the cell cycle and differentiation. Cell Cycle 2010, 9, 1929–1933. [Google Scholar] [CrossRef] [PubMed]
- Galarneau, A.; Richard, S. Target RNA motif and target mRNAs of the quaking STAR protein. Nat. Struct Mol. Biol. 2005, 12, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Nagel, R.J.; Fagg, W.S.; Shiue, L.; Cline, M.S.; Perriman, R.J.; Donohue, J.P.; Ares, M. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA 2013, 19, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Furuta, T.; Mitsunaga, K.; Ebersole, T.A.; Shichiri, M.; Wu, J.; Artzt, K.; Yamamura, K.; Abe, K. Genomic organization and expression analysis of the mouse QKI locus. Mammal. Genome 1999, 10, 662–669. [Google Scholar]
- Darbelli, L.; Choquet, K.; Richard, S.; Kleinman, C.L. Transcriptome profiling of mouse brains with QKI-deficient oligodendrocytes reveals major alternative splicing defects including self-splicing. Sci. Rep. 2017, 7, 7554. [Google Scholar] [CrossRef]
- Fagg, W.S.; Liu, N.; Fair, J.H.; Shiue, L.; Katzman, S.; Donohue, J.P.; Ares, M. Autogenous cross-regulation of Quaking mRNA processing and translation balances Quaking functions in splicing and translation. Genes Dev. 2017, 31, 1894–1909. [Google Scholar] [CrossRef]
- Howe, D.G.; Bradford, Y.M.; Conlin, T.; Eagle, A.E.; Fashena, D.; Frazer, K.; Knight, J.; Mani, P.; Martin, R.; Moxon, S.A.; et al. ZFIN, the zebrafish model organism database: Increased support for mutants and transgenics. Nucl. Acid Res. 2013, 41, 854–860. [Google Scholar] [CrossRef]
- Bonnet, A.; Lambert, G.; Ernest, S.; Dutrieux, F.X.; Coulpier, F.; Lemoine, S.; Lobbardi, R.; Rosa, F.M. Quaking RNA-binding proteins control early myofibril formation by modulating tropomyosin. Develop. Cell 2017, 42, 527–541. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 2017, 66, 22–37. [Google Scholar] [CrossRef]
- Merkin, J.; Russell, C.; Chen, P.; Burge, C.B. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 2012, 338, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, K.; Su, R.; Liu, W.; Ren, Y.; Zhang, C.; Du, M.; Zhang, J. Effect of dietary Tartary buckwheat extract supplementation on growth performance, meat quality and antioxidant activity in ewe lambs. Meat Sci. 2017, 134, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhang, T.; Cao, Y.; Deng, B.; Zhang, J.; Zhao, J. Effects of dietary sea buckthorn pomace supplementation on skeletal muscle mass and meat quality in lambs. Meat Sci. 2020, 166, 108141. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Cote, J.; Carvajal, H.V.; Richard, S. Identification of Sam68 arginine glycine-rich sequences capable of conferring nonspecific RNA binding to the GSG domain. J. Biol. Chem. 2001, 276, 30803–30811. [Google Scholar] [CrossRef]
- Wu, J.I.; Reed, R.B.; Grabowski, P.J.; Artzt, K. Function of quaking in myelination: Regulation of alternative splicing. Proc. Natl. Acad. Sci. USA 2002, 99, 4233–4238. [Google Scholar] [CrossRef]
- Wang, J.; Fu, X.; Fang, Z.; Liu, H.; Zong, F.; Zhu, H.; Yu, Y.; Zhang, X.; Wang, S.; Huang, Y.; et al. QKI-5 regulates the alternative splicing of cytoskeletal gene ADD3 in lung cancer. J. Mol. Cell Biol. 2021, 13, 347–360. [Google Scholar] [CrossRef]
- Zong, F.; Fu, X.; Wei, W.; Luo, Y.; Heiner, M.; Cao, L.; Fang, Z.; Fang, R.; Lu, D.; Ji, H.B.; et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014, 10, e1004289. [Google Scholar] [CrossRef]
- Caines, R.; Cochrane, A.; Kelaini, S.; Vila-Gonzalez, M.; Yang, C.; Eleftheriadou, M.; Moze, A.; Stitt, A.W.; Zeng, L.F.; Grieve, D.J.; et al. The RNA-binding protein QKI controls alternative splicing in vascular cells, producing an effective model for therapy. J. Cell Sci. 2019, 132, jcs230276. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Xu, C.; Ba, L.; Liu, Z.; Li, X.; Huang, J.; Simpson, E.; Gao, H.; Cao, D.; et al. QKI is a critical pre-mRNA alternative splicing regulator of cardiac myofibrillogenesis and contractile function. Nat. Commun. 2021, 12, 89. [Google Scholar] [CrossRef]
- Barton, P.J.R.; Mullen, A.J.; Cullen, M.E.; Dhoot, G.K.; Simon-Chazottes, D.; Guénet, J.L. Genes encoding troponin I and troponin T are organized as three paralogous pairs in the mouse genome. Mammal. Genome 2000, 11, 926–929. [Google Scholar] [CrossRef]
- Gong, H.Y.; Hatch, V.; Ali, L.; Lehman, W.; Craig, R.; Tobacman, L.S. Mini-thin filaments regulated by troponin-tropomyosin. Proc. Natl. Acad. Sci. USA 2005, 102, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, J.; Yu, B.; Li, L.; Shang, Y. Molecular cloning, structural analysis, and tissue expression of the TNNT3 gene in Guizhou black goat. Gene 2015, 573, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Stefancsik, R.; Randall, J.D.; Mao, C.; Sarkar, S. Structure and sequence of the human fast skeletal troponin T (TNNT3) gene: Insight into the evolution of the gene and the origin of the developmentally regulated isoforms. Compar. Funct. Genom. 2003, 4, 609–625. [Google Scholar] [CrossRef]
- Sun, X.; Wang, W.; Dong, Y.; Wang, Y.; Zhang, M.; Wang, Z.; Yu, X.; Huang, J.; Cai, H. Relationship between calcium circulation-related factors and muscle strength in rat sciatic nerve injury model. Iran J. Basic Med. Sci. 2020, 23, 654–662. [Google Scholar] [PubMed]
- Yang, H.; Xu, Z.Y.; Lei, M.G.; Li, F.E.; Deng, C.Y.; Xiong, Y.Z.; Zuo, B. Real-time reverse transcription-PCR expression profiling of porcine troponin I family in three different types of muscles during development. Mol. Biol. Rep. 2011, 38, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Chothani, S.; Schäfer, S.; Adami, E.; Viswanathan, S.; Widjaja, A.A.; Langley, S.R.; Tan, J.; Wang, M.; Quaife, N.M.; Pua, C.J.; et al. Widespread translational control of fibrosis in the human heart by RNA-binding proteins. Circulation 2019, 140, 937–951. [Google Scholar] [CrossRef]
- Shelton, W.T.; Thomas, S.M.; Alexander, H.R.; Thomes, C.E.; Conway, D.E.; Dubash, A.D. Desmoglein-2 harnesses a PDZ-GEF2/Rap1 signaling axis to control cell spreading and focal adhesions independent of cell-cell adhesion. Sci. Rep. 2021, 11, 13295. [Google Scholar] [CrossRef] [PubMed]
- Leube, R.E.; Moch, M.; Windoffer, R. Intermediate filaments and the regulation of focal adhesion. Curr. Opin. Cell Biol. 2015, 32, 13–20. [Google Scholar] [CrossRef]
- Dalby, M.J.; Yarwood, S.J. Analysis of focal adhesions and cytoskeleton by custom microarray. Methods Mol. Biol. 2007, 370, 121–134. [Google Scholar]
- Le Lay, S.; Kurzchalia, T.V. Getting rid of caveolins: Phenotypes of caveolin-deficient animals. Biochim. Biophys. Acta 2005, 1746, 322–332. [Google Scholar] [CrossRef]
- Shin, D.H.; Kim, J.S.; Kwon, B.S.; Lee, K.S.; Kim, J.W.; Kim, M.H.; Cho, S.S.; Lee, W.J. Caveolin-3 expression during early chicken development. Brain Res. Dev. Brain Res. 2003, 141, 83–89. [Google Scholar] [CrossRef]
- Carlson, B.M.; Carlson, J.A.; Dedkov, E.I.; McLennan, I.S. Concentration of caveolin-3 at the neuro-muscular junction in young and old rat skeletal muscle fibers. J. Histochem. Cytochem. 2003, 51, 1113–1118. [Google Scholar] [CrossRef]
- Bennett, A.M.; Tonks, N.K. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science 1997, 278, 1288–1291. [Google Scholar] [CrossRef] [PubMed]
- Cuenda, A.; Cohen, P. Stress-activated protein kinase-2/p38 and a rapamycin-sensitive pathway are required for C2C12 myogenesis. J. Biol. Chem. 1999, 274, 4341–4346. [Google Scholar] [CrossRef] [PubMed]
- Horsley, V.; Friday, B.B.; Matteson, S.; Kegley, K.M.; Gephart, J.; Pavlath, G.K. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J. Cell Biol. 2001, 153, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Kineman, R.D.; Rio-Moreno, M.D.; Sarmento-Cabral, A. Understanding the tissue-specific roles of IGF1/IGF1R in regulating metabolism using the Cre/loxP system. J. Mol. Endocrinol. 2018, 61, T187–T198. [Google Scholar] [CrossRef] [PubMed]
- Philippou, A.; Maridaki, M.; Halapas, A.; Koutsilieris, M. The role of the insulin-like growth factor 1 (IGF-1) in skeletal muscle physiology. In Vivo 2007, 21, 45–54. [Google Scholar] [PubMed]
| Primer Name | Forward | Reverse |
|---|---|---|
| QKI-15 | ATGCCAAACGGAACTCCTCA | TAGCCACCGCACCTAATACAC |
| QKI-36 | GCTGTCATGCCAAACGGAAC | ATCAGGCATGACTGGCATTTC |
| QKI-47 | ATACCCCTACACGTTGGCAC | TCGTTGGGAAAGCCATACCT |
| GAPDH | GCAAGTTCCACGGCACAG | GGTTCACGCCCATCACAA |
| MyoD | GTGCAAACGCAAGACGACTA | GCTGGTTTGGGTTGCTAGAC |
| MyoG | GGACCCTACAGATGCCCACAA | TTGGTATGGTTTCATCTGGG |
| MyHC | CCACATCTTCTCCATCTCTG | GGTTCCTCCTTCTTCTTCTC |
| Gene | Description | log2 (FC) | p-Value | Enriched Terms |
|---|---|---|---|---|
| ACTA1 | actin, alpha 1, skeletal muscle | −1.002515693 | 0.001516326 | 1, 2, 3, 4, 6, 7, 8, 9, 10 |
| ATP1A2 | ATPase Na+/K+ transporting subunit alpha 2 | −1.121160481 | 0.000335005 | 5 |
| CACNG1 | calcium voltage-gated channel auxiliary subunit gamma 1 | −1.094915942 | 0.007555945 | 1, 4, 6, 7, 8, 10 |
| CAV3 | caveolin 3 | −1.120522417 | 0.016390643 | 1, 6, 8, 9, 10 |
| CHRNA1 | cholinergic receptor nicotinic alpha 1 subunit | −1.302552151 | 7.09 × 10−6 | 1, 4, 5, 7 |
| DES | desmin | −1.30009742 | 6.31 × 10−6 | 2, 3 |
| HRC | Histidine-rich calcium-binding protein | −1.12825787 | 0.000613572 | 2, 3 |
| IGF1 | insulin-like growth factor I | −1.079802091 | 2.91 × 10−25 | 1, 4, 6, 8 |
| JPH1 | junctophilin 1 | −1.013695004 | 0.018021031 | 1, 2, 3, 4 |
| LDB3 | LIM domain binding 3 | −1.09115513 | 4.54 × 10−10 | 1, 2, 3, 6, 8, 10 |
| NFATC2 | nuclear factor of activated T cells 2 | 3.004126141 | 0.008888678 | 1, 6, 8, 9 |
| SCX | scleraxis bHLH transcription factor | −1.682899293 | 0.025556357 | 1, 4, 7 |
| TNNC1 | troponin C1, slow skeletal and cardiac type | −1.251575045 | 1.19 × 10−5 | 1, 2, 3, 4, 5 |
| TNNC2 | capra hircus troponin C2, fast skeletal type (TNNC2), mRNA. | −1.333657493 | 4.12 × 10−10 | 2, 3, 5 |
| TNNI1 | troponin I1, slow skeletal type | −1.280560498 | 2.66 × 10−13 | 1, 4 |
| TNNT1 | troponin T1, slow skeletal type | −1.762686865 | 3.90 × 10−33 | 2, 3, 5 |
| TNNT3 | troponin T3, fast skeletal type | −1.60504666 | 0.000335191 | 2, 3, 5 |
| VGLL2 | vestigial like family member 2 | −1.006900776 | 0.003217405 | 1, 4, 7 |
| XIRP2 | xin actin binding repeat containing 2 | −1.210101158 | 0.000207475 | 1, 2, 3, 4 |
| CYP26B1 | cytochrome P450 family 26 subfamily B member 1 | 1.352173933 | 0.047638797 | 1, 4, 6, 7, 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Niu, S.; Wang, W.; Zhao, X.; Pan, Y.; Qiao, L.; Yang, K.; Liu, J.; Liu, W. Overexpression of the QKI Gene Promotes Differentiation of Goat Myoblasts into Myotubes. Animals 2023, 13, 725. https://doi.org/10.3390/ani13040725
Chen S, Niu S, Wang W, Zhao X, Pan Y, Qiao L, Yang K, Liu J, Liu W. Overexpression of the QKI Gene Promotes Differentiation of Goat Myoblasts into Myotubes. Animals. 2023; 13(4):725. https://doi.org/10.3390/ani13040725
Chicago/Turabian StyleChen, Sijia, Shu Niu, Wannian Wang, Xiang Zhao, Yangyang Pan, Liying Qiao, Kaijie Yang, Jianhua Liu, and Wenzhong Liu. 2023. "Overexpression of the QKI Gene Promotes Differentiation of Goat Myoblasts into Myotubes" Animals 13, no. 4: 725. https://doi.org/10.3390/ani13040725
APA StyleChen, S., Niu, S., Wang, W., Zhao, X., Pan, Y., Qiao, L., Yang, K., Liu, J., & Liu, W. (2023). Overexpression of the QKI Gene Promotes Differentiation of Goat Myoblasts into Myotubes. Animals, 13(4), 725. https://doi.org/10.3390/ani13040725





