Chicken Hypothalamic and Ovarian DNA Methylome Alteration in Response to Forced Molting
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Individual Selection and Tissue Collection
2.2. Animal Experimental Design
2.3. DNA Isolation and WGBS
2.4. Data Filtering and Identification of Methylated Cytosines
2.5. Identification and Distribution of Methylated C Sites
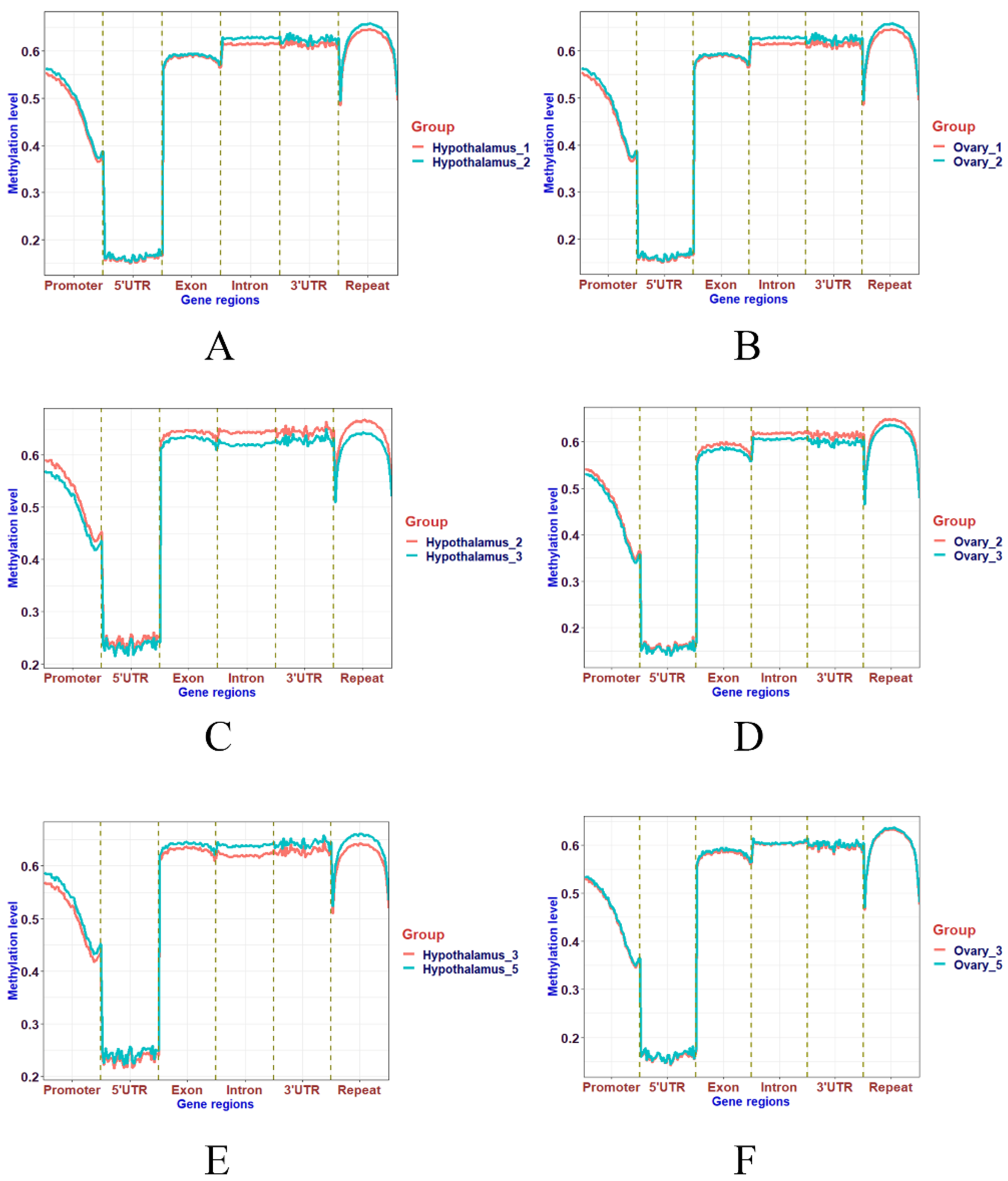
2.6. Horizontal Distribution of Methylation in Functional Areas
2.7. Identification of Differentially Methylated Regions (DMRs) and Functional Enrichment Analysis
3. Results
3.1. Sequencing Data Statistics
3.2. Identification of Methylated C Sites and Their Distribution
3.3. Cluster Analysis between Samples
3.4. Distribution of Methylation Levels in Functional Regions of the Genome
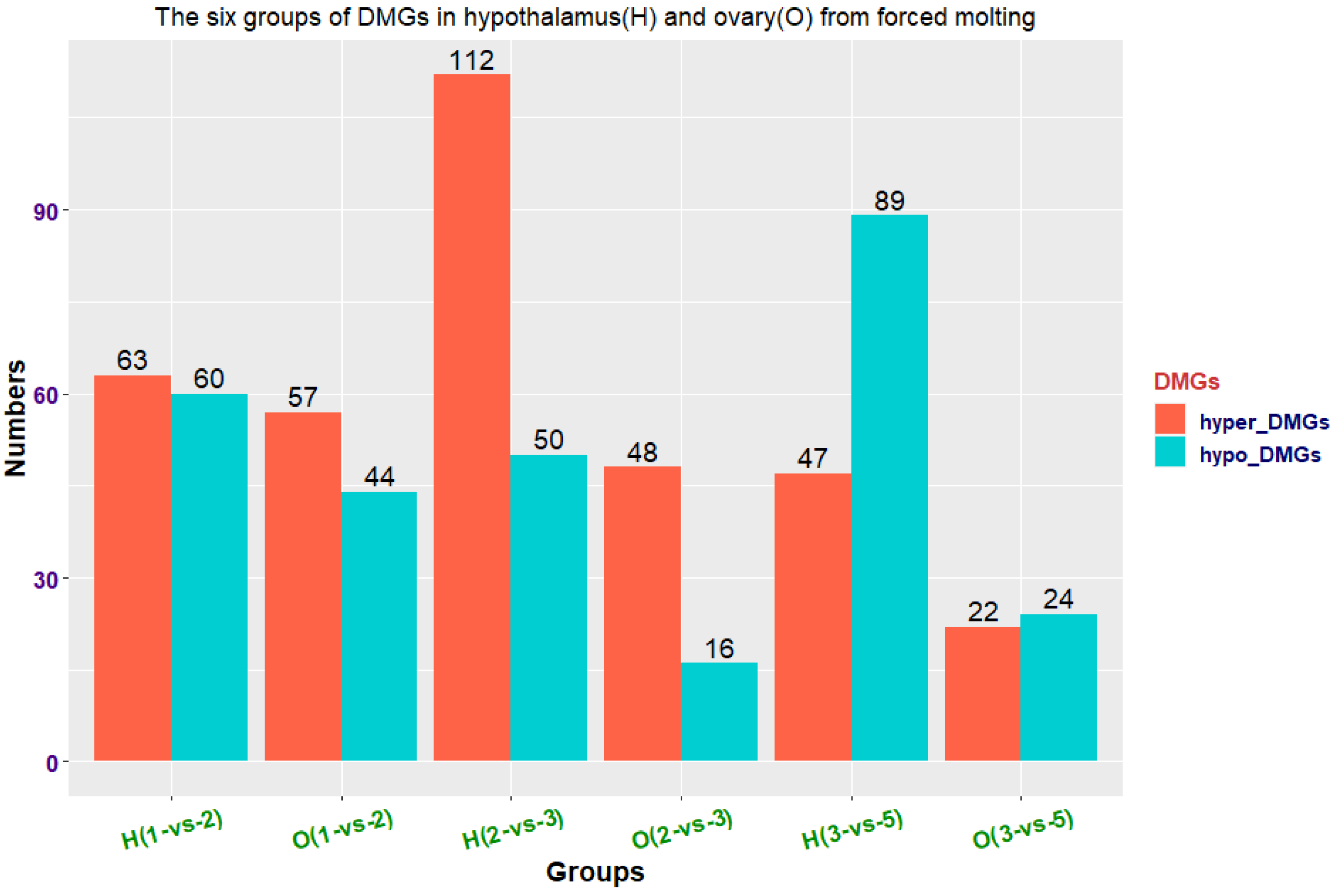
3.5. DMR Detection
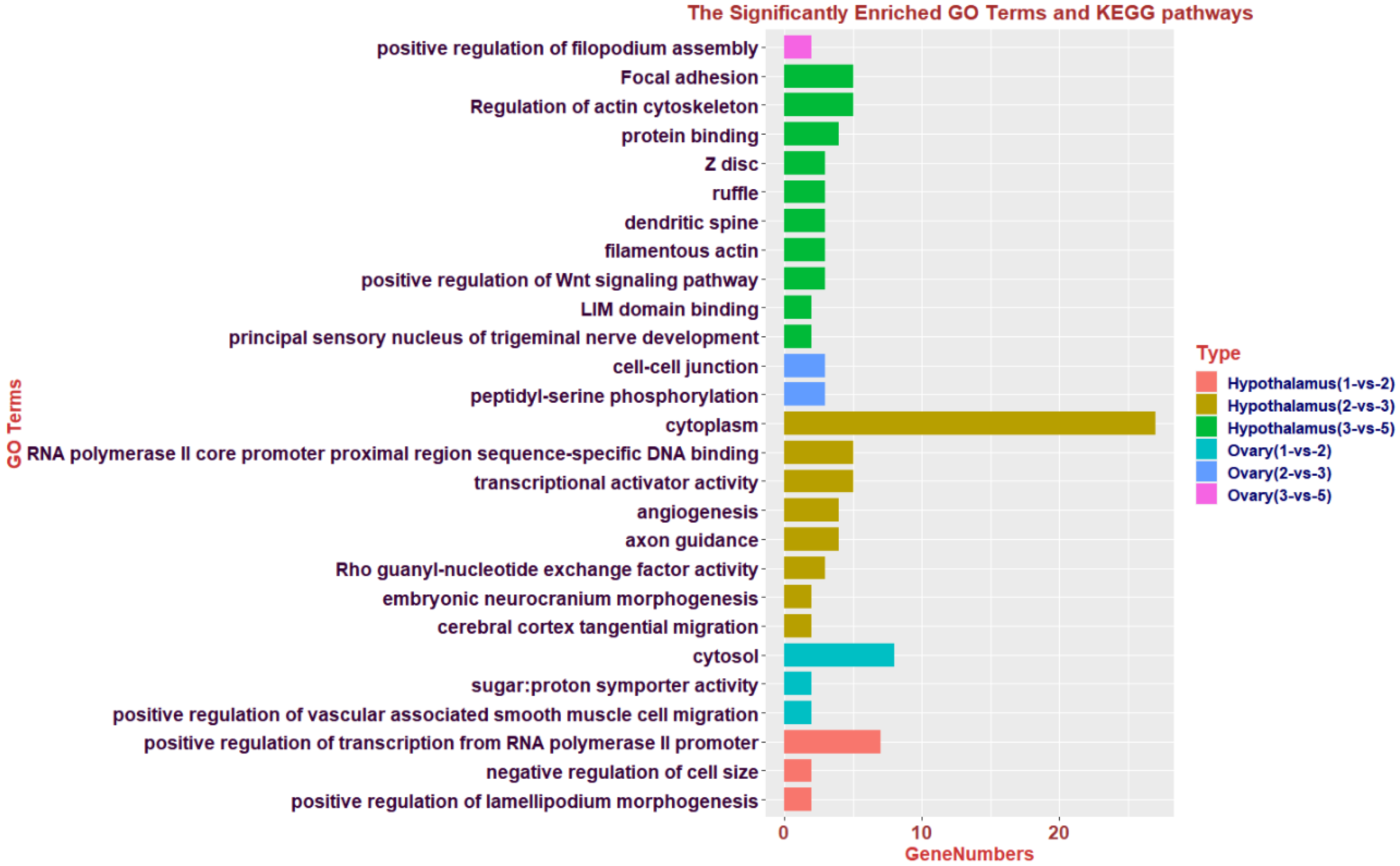
3.6. Identification of DMGs and Functional Enrichment Analysis
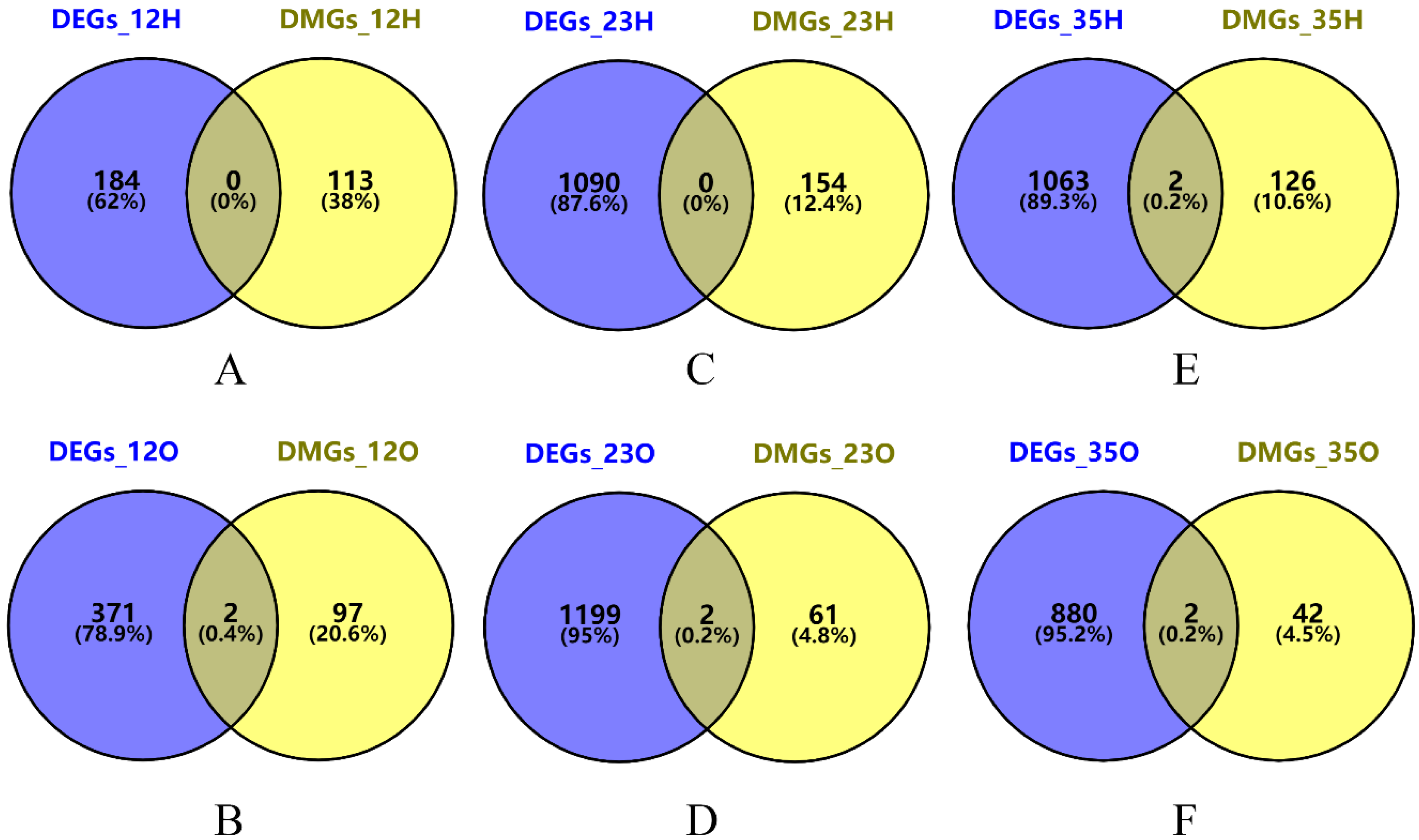
3.7. The Conjoint Analysis of Differential Expression Genes (DEGs) and DMGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andreatti Filho, R.L.; Milbradt, E.L.; Okamoto, A.S.; Silva, T.M.; Vellano, I.H.B.; Gross, L.S.; Oro, C.S.; Hataka, A. Salmonella Enteritidis infection, corticosterone levels, performance and egg quality in laying hens submitted to different methods of molting. Poult. Sci. 2009, 98, 4416–4425. [Google Scholar] [CrossRef]
- Nishiyama, T.; Nakagawa, K.; Imabayashi, T.; Iwatani, S.; Yamamoto, N.; Tsushima, N. Probiotic Bacillus subtilis C-3102 Improves Eggshell Quality after Forced Molting in Aged Laying Hens. J Poult. Sci. 2021, 58, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Pavlova, S.; Tena-Sempere, M.; Grossmann, R.; Jiménez, M.R.; Rodriguez, J.M.; Valenzuela, F. Food restriction, ghrelin, its antagonist and obestatin control expression of ghrelin and its receptor in chicken hypothalamus and ovary. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2013, 164, 141–153. [Google Scholar] [CrossRef]
- Mishra, S.K.; Chen, B.; Zhu, Q.; Xu, Z.; Ning, C.; Yin, H.; Wang, Y.; Zhao, X.; Fan, X.; Yang, M.; et al. Transcriptome analysis reveals differentially expressed genes associated with high rates of egg production in chicken hypothalamic-pituitary-ovarian axis. Sci. Rep. 2020, 10, 5976. [Google Scholar] [CrossRef] [PubMed]
- Abe, E.; Horikawa, H.; Masumura, T.; Sugahara, M.; Kubota, M.; Suda, T. Disorders of cholecalciferol metabolism in old egg-laying hens. J. Nutr. 1982, 112, 436–446. [Google Scholar] [CrossRef]
- Anderson, K.E. Comparison of fatty acid, cholesterol, vitamin A and E composition, and trans fats in eggs from brown and white egg strains that were molted or nonmolted. Poult. Sci. 2013, 92, 3259–3265. [Google Scholar] [CrossRef] [PubMed]
- Silva-Mendonça, M.C.; Fagundes, N.S.; Mendonça, G.A.; Gonçalves, F.C.; Fonseca, B.B.; Mundim, A.V.; Fernandes, E.A. Comparison of moulting methods for layers: High-zinc diet versus fasting. Br. Poult. Sci. 2015, 56, 598–604. [Google Scholar] [CrossRef]
- Kakhki, R.A.M.; Mousavi, Z.; Anderson, K.E. An appraisal of moulting on post-moult egg production and egg weight distribution in white layer hens; meta-analysis. Br. Poult. Sci. 2018, 59, 278–285. [Google Scholar] [CrossRef]
- Madekurozwa, M.N.; Mpango, M.M. Ultrastructure of the tubular glands in the isthmus region of the oviduct in laying and natural moulting commercial egg-type chickens. Anat. Histol. Embryol. 2018, 47, 493–497. [Google Scholar] [CrossRef]
- Madekurozwa, M.C.; Mpango, M.M. The shell gland in laying and natural moulting commercial egg-type chickens: A histomorphological and ultrastructural study. Anat. Histol. Embryol. 2020, 49, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Geng, X.; Zhang, Y.; Zhao, X.; Zhang, P.; Sun, G.; Li, W.; Li, D.; Han, R.; Li, G.; et al. Interaction Between Cecal Metabolites and Liver Lipid Metabolism Pathways During Induced Molting in Laying Hens. Front. Physiol. 2022, 13, 862721. [Google Scholar] [CrossRef] [PubMed]
- Bednarczyk, M.; Dunislawska, A.; Stadnicka, K.; Grochowska, E. Chicken embryo as a model in epigenetic research. Poult. Sci. 2021, 100, 101164. [Google Scholar] [CrossRef] [PubMed]
- Gryzinska, M.; Blaszczak, E.; Strachecka, A.; Jezewska-Witkowska, G. Analysis of age-related global DNA methylation in chicken. Biochem. Genet. 2013, 51, 554–563. [Google Scholar] [CrossRef]
- Li, Q.; Li, N.; Hu, X.; Li, J.; Du, Z.; Chen, L.; Yin, G.; Duan, J.; Zhang, H.; Zhao, Y.; et al. Genome-wide mapping of DNA methylation in chicken. PLoS ONE 2011, 6, e19428. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, H.; Li, Z.; Zheng, X.; Jia, X.; Nie, Q.; Zhang, X. Comparison of the genome-wide DNA methylation profiles between fast-growing and slow-growing broilers. PLoS ONE 2013, 8, e56411. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.N.; Gao, Y.; Qiao, S.P.; Wang, S.Z.; Duan, K.; Wang, Y.X.; Li, H.; Wang, N. Epigenetic DNA methylation in the promoters of peroxisome proliferator-activated receptor γ in chicken lines divergently selected for fatness. J. Anim. Sci. 2014, 92, 48–53. [Google Scholar] [CrossRef]
- Gao, Y.; Sun, Y.; Duan, K.; Shi, H.; Wang, S.; Li, H.; Wang, N. CpG site DNA methylation of the CCAAT/enhancer-binding protein, alpha promoter in chicken lines divergently selected for fatness. Anim. Genet. 2015, 46, 410–417. [Google Scholar] [CrossRef]
- Wu, C.; Wang, Y.; Gong, P.; Wang, L.; Liu, C.; Chen, C.; Jiang, X.; Dong, X.; Cheng, B.; Li, H. Promoter Methylation Regulates ApoA-I Gene Transcription in Chicken Abdominal Adipose Tissue. J. Agric. Food Chem. 2019, 67, 4535–4544. [Google Scholar] [CrossRef]
- Corbett, R.J.; Te Pas, M.F.W.; van den Brand, H.; Groenen, M.A.M.; Crooijmans, R.; Ernst, C.W.; Madsen, O. Genome-Wide Assessment of DNA Methylation in Chicken Cardiac Tissue Exposed to Different Incubation Temperatures and CO(2) Levels. Front. Genet. 2020, 11, 558189. [Google Scholar] [CrossRef]
- Zhang, Y.; Gou, W.; Ma, J.; Zhang, H.; Zhang, Y.; Zhang, H. Genome methylation and regulatory functions for hypoxic adaptation in Tibetan chicken embryos. PeerJ 2017, 5, e3891. [Google Scholar] [CrossRef]
- Zhang, Q.; Zheng, S.; Wang, S.; Jiang, Z.; Xu, S. The Effects of Low Selenium on DNA Methylation in the Tissues of Chickens. Biol. Trace Elem Res. 2019, 191, 474–484. [Google Scholar] [CrossRef]
- Dunislawska, A.; Slawinska, A.; Siwek, M. Hepatic DNA Methylation in Response to Early Stimulation of Microbiota with Lactobacillus Synbiotics in Broiler Chickens. Genes 2020, 11, 579. [Google Scholar] [CrossRef]
- Wang, F.; Li, J.; Li, Q.; Liu, R.; Zheng, M.; Wang, Q.; Wen, J.; Zhao, G. Changes of host DNA methylation in domestic chickens infected with Salmonella enterica. J. Genet. 2017, 96, 545–550. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, L.; Li, M.; Lin, L.; Su, P.; Tang, H.; Fan, X.; Li, X. Chicken cecal DNA methylome alteration in the response to Salmonella enterica serovar Enteritidis inoculation. BMC Genom. 2020, 21, 814. [Google Scholar] [CrossRef] [PubMed]
- Pejaković, S.; Mbouombouo Mfossa, A.C.; Wiggers, L.; Kheimar, A.; Coupeau, D.; Kaufer, B.B.; Muylkens, B. Role of DNA Methylation and CpG Sites in the Viral Telomerase RNA Promoter during Gallid Herpesvirus 2 Pathogenesis. J. Virol. 2020, 94, e01488-20. [Google Scholar] [CrossRef]
- Zhu, G.; Mao, Y.; Zhou, W.; Jiang, Y. Dynamic Changes in the Follicular Transcriptome and Promoter DNA Methylation Pattern of Steroidogenic Genes in Chicken Follicles throughout the Ovulation Cycle. PLoS ONE 2015, 10, e0146028. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Li, T.; Zhang, M.; Chen, C.; Gao, X.; Zhang, C.; Hu, C.; Zuo, Q.; Chen, G.; Li, B. Epigenetic modification cooperates with Zeb1 transcription factor to regulate Bmp4 to promote chicken PGCs formation. Gene 2021, 794, 145760. [Google Scholar] [CrossRef]
- Han, W.; Xue, Q.; Li, G.; Yin, J.; Zhang, H.; Zhu, Y.; Xing, W.; Cao, Y.; Su, Y.; Wang, K.; et al. Genome-wide analysis of the role of DNA methylation in inbreeding depression of reproduction in Langshan chicken. Genomics 2020, 112, 2677–2687. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, Y.; Wen, J.; Jia, Y.; Wang, L.; Lv, X.; Yang, W.; Qu, C.; Li, H.; Wang, H.; et al. Transcriptomic Analysis of Laying Hens Revealed the Role of Aging-Related Genes during Forced Molting. Genes 2021, 12, 1767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, D.; Zhai, Y.; Wang, Z.; Ma, X.; Zhang, D.; Li, G.; Han, R.; Jiang, R.; Li, Z.; et al. The Landscape of DNA Methylation Associated With the Transcriptomic Network of Intramuscular Adipocytes Generates Insight Into Intramuscular Fat Deposition in Chicken. Front. Cell Dev. Biol. 2020, 8, 206. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Zhang, T.; Ning, Z.; Chen, Y.; Wen, J.; Jia, Y.; Wang, L.; Lv, X.; Yang, W.; Qu, C.; Li, H.; et al. Understanding Transcriptomic and Serological Differences between Forced Molting and Natural Molting in Laying Hens. Genes 2021, 13, 89. [Google Scholar] [CrossRef]
- Sanna-Cherchi, S.; Sampogna, R.V.; Papeta, N.; Burgess, K.E.; Nees, S.N.; Perry, B.J.; Choi, M.; Bodria, M.; Liu, Y.; Weng, P.L.; et al. Mutations in DSTYK and dominant urinary tract malformations. N. Engl. J. Med. 2013, 369, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Maiti, M.; Hennessy, M.; Chang, T.; Kuo, P.; Addepalli, M.; Obalapur, P.; Sheibani, S.; Wilczek, J.; Pena, R.; et al. NKTR-255, a novel polymer-conjugated rhIL-15 with potent antitumor efficacy. J. Immunother. Cancer 2021, 9, e002024. [Google Scholar] [CrossRef] [PubMed]
- Pazin, D.E.; Albrecht, K.H. Developmental expression of Smoc1 and Smoc2 suggests potential roles in fetal gonad and reproductive tract differentiation. Dev. Dyn. 2009, 238, 2877–2890. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Z.; Lv, P.; Zhan, Y.; Zhong, Q. SCAMP3 Promotes Glioma Proliferation and Indicates Unfavorable Prognosis via Multiple Pathways. Onco Targets Ther. 2020, 13, 3677–3687. [Google Scholar] [CrossRef]
- Fragale, N.; Divvela, S.S.K.; Brand-Saberi, B. atoh8 expression pattern in early zebrafish embryonic development. Histochem. Cell Biol. 2021, 156, 209–226. [Google Scholar] [CrossRef]
- Samanta, D.; Arya, K. Electroclinical Findings of SYNJ1 Epileptic Encephalopathy. J. Pediatr. Neurosci. 2020, 15, 29–33. [Google Scholar]
- Vatsa, N.; Jana, N.R. UBE3A and Its Link With Autism. Front. Mol. Neurosci. 2018, 11, 448. [Google Scholar] [CrossRef]
- Elgersma, Y.; Sonzogni, M. UBE3A reinstatement as a disease-modifying therapy for Angelman syndrome. Dev. Med. Child. Neurol. 2021, 63, 802–807. [Google Scholar] [CrossRef]
- López Tobón, A.; Suresh, M.; Jin, J.; Vitriolo, A.; Pietralla, T.; Tedford, K.; Bossenz, M.; Mahnken, K.; Kiefer, F.; Testa, G.; et al. The guanine nucleotide exchange factor Arhgef7/βPix promotes axon formation upstream of TC10. Sci. Rep. 2018, 8, 8811. [Google Scholar] [CrossRef] [PubMed]
- Leachman, N.T.; Brellier, F.; Ferralli, J.; Chiquet-Ehrismann, R.; Tucker, R.P. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev. Growth Differ. 2010, 52, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Ngoh, A.; Bras, J.; Guerreiro, R.; Meyer, E.; McTague, A.; Dawson, E.; Mankad, K.; Gunny, R.; Clayton, P.; Mills, P.B.; et al. RARS2 mutations in a sibship with infantile spasms. Epilepsia 2016, 57, e97–e102. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Zhou, S.; Zhou, Z.; Liu, Y.; Yang, L.; Liu, J.; Zhang, Y.; Li, H.; Liu, Y.; Hou, F.F.; et al. Wnt9a Promotes Renal Fibrosis by Accelerating Cellular Senescence in Tubular Epithelial Cells. J. Am. Soc. Nephrol. 2018, 29, 1238–1256. [Google Scholar] [CrossRef]
- Larsson, C.; Ali, M.A.; Pandzic, T.; Lindroth, A.M.; He, L.; Sjöblom, T. Loss of DIP2C in RKO cells stimulates changes in DNA methylation and epithelial-mesenchymal transition. BMC Cancer 2017, 17, 487. [Google Scholar] [CrossRef]
- Le, P.T.; Bishop, K.A.; Maridas, D.E.; Motyl, K.J.; Brooks, D.J.; Nagano, K.; Baron, R.; Bouxsein, M.L.; Rosen, C.J. Spontaneous mutation of Dock7 results in lower trabecular bone mass and impaired periosteal expansion in aged female Misty mice. Bone 2017, 105, 103–114. [Google Scholar] [CrossRef]
- Kruta, M.; Sunshine, M.J.; Chua, B.A.; Fu, Y.; Chawla, A.; Dillingham, C.H.; Hidalgo San Jose, L.; De Jong, B.; Zhou, F.J.; Signer, R.A.J. Hsf1 promotes hematopoietic stem cell fitness and proteostasis in response to ex vivo culture stress and aging. Cell Stem Cell 2021, 28, 1950–1965. [Google Scholar] [CrossRef]
- Qin, Y.; Sun, M.; You, L.; Wei, D.; Sun, J.; Liang, X.; Zhang, B.; Jiang, H.; Xu, J.; Chen, Z.J. ESR1, HK3 and BRSK1 gene variants are associated with both age at natural menopause and premature ovarian failure. Orphanet. J. Rare Dis. 2012, 7, 5. [Google Scholar] [CrossRef]
- Innamaa, A.; Jackson, L.; Asher, V.; van Schalkwyk, G.; Warren, A.; Keightley, A.; Hay, D.; Bali, A.; Sowter, H.; Khan, R. Expression and effects of modulation of the K2P potassium channels TREK-1 (KCNK2) and TREK-2 (KCNK10) in the normal human ovary and epithelial ovarian cancer. Clin. Transl. Oncol. 2013, 15, 910–918. [Google Scholar] [CrossRef]
- Gibson, J.; Russ, T.C.; Clarke, T.K.; Howard, D.M.; Hillary, R.F.; Evans, K.L.; Walker, R.M.; Bermingham, M.L.; Morris, S.W.; Campbell, A.; et al. A meta-analysis of genome-wide association studies of epigenetic age acceleration. PLoS Genet. 2019, 15, e1008104. [Google Scholar] [CrossRef]
- Guay, S.P.; Légaré, C.; Houde, A.A.; Mathieu, P.; Bossé, Y.; Bouchard, L. Acetylsalicylic acid, aging and coronary artery disease are associated with ABCA1 DNA methylation in men. Clin. Epigenetics 2014, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Mockute, R.; Vilkeviciute, A.; Balciuniene, V.J.; Zemaitiene, R.; Liutkeviciene, R. ABCA1 rs1883025 and CYP4F2 rs2108622 Gene Polymorphism Association with Age-Related Macular Degeneration and Anti-VEGF Treatment. Medicina 2021, 57, 974. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Zhang, X.; Wang, L.; Li, Y. Curcumin induces ABCA1 expression and apolipoprotein A-I-mediated cholesterol transmembrane in the chronic cerebral hypoperfusion aging rats. Am. J. Chin. Med. 2013, 41, 1027–1042. [Google Scholar] [CrossRef] [PubMed]
- Zohar, K.; Lezmi, E.; Eliyahu, T.; Linial, M. Ladostigil Attenuates Induced Oxidative Stress in Human Neuroblast-like SH-SY5Y Cells. Biomedicines 2021, 9, 1251. [Google Scholar] [CrossRef]
- Kanazawa, Y.; Nagano, M.; Koinuma, S.; Sugiyo, S.; Shigeyoshi, Y. Effects of aging on basement membrane-related gene expression of the skeletal muscle in rat. Biomed. Res. 2021, 42, 115–119. [Google Scholar] [CrossRef]
- Diao, D.; Wang, H.; Li, T.; Shi, Z.; Jin, X.; Sperka, T.; Zhu, X.; Zhang, M.; Yang, F.; Cong, Y.; et al. Telomeric epigenetic response mediated by Gadd45a regulates stem cell aging and lifespan. EMBO Rep. 2018, 19, e45494. [Google Scholar] [CrossRef]
- Turner, D.C.; Gorski, P.P.; Maasar, M.F.; Seaborne, R.A.; Baumert, P.; Brown, A.D.; Kitchen, M.O.; Erskine, R.M.; Dos-Remedios, I.; Voisin, S.; et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: The role of HOX genes and physical activity. Sci. Rep. 2020, 10, 15360. [Google Scholar] [CrossRef]
- Chang-Panesso, M.; Kadyrov, F.F.; Machado, F.G.; Kumar, A.; Humphreys, B.D. Meis1 is specifically upregulated in kidney myofibroblasts during aging and injury but is not required for kidney homeostasis or fibrotic response. Am. J. Physiol.-Renal. 2018, 315, F275–F290. [Google Scholar] [CrossRef]
- Lindsey, J.C.; Lusher, M.E.; Strathdee, G.; Brown, R.; Gilbertson, R.J.; Bailey, S.; Ellison, D.W.; Clifford, S.C. Epigenetic inactivation of MCJ (DNAJD1) in malignant paediatric brain tumours. Int. J. Cancer 2006, 118, 346–352. [Google Scholar] [CrossRef]
- Moro, L.; Pagano, M. Epigenetic suppression of FBXL7 promotes metastasis. Mol. Cell Oncol. 2020, 7, 1833698. [Google Scholar] [CrossRef]
- Chiu, H.W.; Chang, J.S.; Lin, H.Y.; Lee, H.H.; Kuei, C.H.; Lin, C.H.; Huang, H.M.; Lin, Y.F. FBXL7 Upregulation Predicts a Poor Prognosis and Associates with a Possible Mechanism for Paclitaxel Resistance in Ovarian Cancer. J. Clin. Med. 2018, 7, 330. [Google Scholar] [CrossRef]
- Dathe, V.; Pröls, F.; Brand-Saberi, B. Expression of kinesin kif5c during chick development. Anat. Embryol. 2004, 207, 475–480. [Google Scholar]
- Li, J.; Li, C.; Li, Q.; Li, G.; Li, W.; Li, H.; Kang, X.; Tian, Y. Novel Regulatory Factors in the Hypothalamic-Pituitary-Ovarian Axis of Hens at Four Developmental Stages. Front. Genet. 2020, 11, 591672. [Google Scholar] [PubMed]
- Cheng, L.; Yang, Z.; Guo, W.; Wu, C.; Liang, S.; Tong, A.; Cao, Z.; Thorne, R.F.; Yang, S.Y.; Yu, Y.; et al. DCLK1 autoinhibition and activation in tumorigenesis. Innovation 2020, 3, 100191. [Google Scholar]
- Nevi, L.; Di Matteo, S.; Carpino, G.; Zizzari, I.G.; Samira, S.; Ambrosino, V.; Costantini, D.; Overi, D.; Giancotti, A.; Monti, M.; et al. DCLK1, a Putative Stem Cell Marker in Human Cholangiocarcinoma. Hepatology 2021, 73, 144–159. [Google Scholar] [PubMed]
- Liu, W.; Wang, S.; Sun, Q.; Yang, Z.; Liu, M.; Tang, H. DCLK1 promotes epithelial-mesenchymal transition via the PI3K/Akt/NF-κB pathway in colorectal cancer. Int. J. Cancer 2018, 142, 2068–2079. [Google Scholar] [CrossRef]
- Keder, A.; Tardieu, C.; Malong, L.; Filia, A.; Kashkenbayeva, A.; Newton, F.; Georgiades, M.; Gale, J.E.; Lovett, M.; Jarman, A.P.; et al. Homeostatic maintenance and age-related functional decline in the Drosophila ear. Sci. Rep. 2020, 10, 7431. [Google Scholar]
- De Gasperi, R.; Mo, C.; Azulai, D.; Wang, Z.; Harlow, L.M.; Du, Y.; Graham, Z.; Pan, J.; Liu, X.H.; Guo, L.; et al. Numb is required for optimal contraction of skeletal muscle. J. Cachexia Sarcopenia Muscle 2022, 13, 454–466. [Google Scholar] [CrossRef]
- Li, T.; Luo, Z.; Lin, S.; Li, C.; Dai, S.; Wang, H.; Huang, J.; Ma, W.; Songyang, Z.; Huang, Y. MiR-185 targets POT1 to induce telomere dysfunction and cellular senescence. Aging 2020, 12, 14791–14807. [Google Scholar] [CrossRef]
- Khairat, J.E.; Balasubramaniam, V.; Othman, I.; Omar, A.R.; Hassan, S.S. Interaction of Recombinant Gallus gallus SEPT5 and Brain Proteins of H5N1-Avian Influenza Virus-Infected Chickens. Proteomes 2017, 5, 23. [Google Scholar]
- Epifanova, E.; Babaev, A.; Newman, A.G.; Tarabykin, V. Role of Zeb2/Sip1 in neuronal development. Brain Res. 2019, 1705, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ferris, S.T.; Kim, S.; Choudhary, M.N.K.; Belk, J.A.; Fan, C.; Qi, Y.; Sudan, R.; Xia, Y.; Desai, P.; et al. Differential usage of transcriptional repressor Zeb2 enhancers distinguishes adult and embryonic hematopoiesis. Immunity 2021, 54, 1417–1432. [Google Scholar] [CrossRef]
- Hegarty, S.V.; Sullivan, A.M.; O’Keeffe, G.W. Zeb2: A multifunctional regulator of nervous system development. Prog Neurobiol. 2015, 132, 81–95. [Google Scholar] [CrossRef]
- Chen, X.; Li, Y.; Luo, J.; Hou, N. Molecular Mechanism of Hippo-YAP1/TAZ Pathway in Heart Development, Disease, and Regeneration. Front. Physiol. 2020, 11, 389. [Google Scholar] [CrossRef] [PubMed]
- Molina, L.; Nejak-Bowen, K.; Monga, S.P. Role of YAP1 Signaling in Biliary Development, Repair, and Disease. Semin. Liver Dis. 2022, 42, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Warnier, M.; Raynard, C.; Ferrand, M.; Kirsh, O.; Defossez, P.A.; Martin, N.; Bernard, D. The nuclear receptor RXRA controls cellular senescence by regulating calcium signaling. Aging Cell 2018, 17, e12831. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yang, Y.E.; Yin, Y.H.; Zhang, M.Y.; Li, H.; Qu, Y.Q. Methylation and transcriptome analysis reveal lung adenocarcinoma-specific diagnostic biomarkers. J. Transl. Med. 2019, 17, 324. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhou, X.; Zheng, R.; Jiang, Y.; Yao, Z.; Wang, X.; Zhang, Z.; Zhang, H.; Li, J.; Yuan, X. The Association of an SNP in the EXOC4 Gene and Reproductive Traits Suggests Its Use as a Breeding Marker in Pigs. Animals 2021, 11, 521. [Google Scholar] [CrossRef]
- Shen, L.F.; Chen, Y.J.; Liu, K.M.; Haddad, A.N.S.; Song, I.W.; Roan, H.Y.; Chen, L.Y.; Yen, J.J.Y.; Chen, Y.J.; Wu, J.Y.; et al. Role of S-Palmitoylation by ZDHHC13 in Mitochondrial function and Metabolism in Liver. Sci. Rep. 2017, 7, 2182. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, Q.; Kang, X.; Wang, K.; Wang, J.; Feng, D.; Bai, Y.; Fang, M. Dynamic changes of genomic methylation profiles at different growth stages in Chinese Tan sheep. J. Anim. Sci. Biotechnol. 2021, 12, 118. [Google Scholar] [CrossRef]
- Cao, Y.; Jin, H.G.; Ma, H.H.; Zhao, Z.H. Comparative analysis on genome-wide DNA methylation in longissimus dorsi muscle between Small Tailed Han and Dorper × Small Tailed Han crossbred sheep. Asian-Australas. J. Anim. Sci. 2017, 30, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, A.; Duhadaway, J.B.; Sutanto-Ward, E.; Wang, Y.; Dinchuk, J.; Huang, M.; Donover, P.S.; Boulden, J.; McNally, L.M.; Soler, A.P.; et al. Bin3 deletion causes cataracts and increased susceptibility to lymphoma during aging. Cancer Res 2008, 68, 1683–1690. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Long, J.; Chelariu-Raicu, A.; Mullikin, H.; Vilsmaier, T.; Vattai, A.; Heidegger, H.H.; Batz, F.; Keckstein, S.; Jeschke, U.; et al. Identification of a Novel Tumor Microenvironment Prognostic Signature for Advanced-Stage Serous Ovarian Cancer. Cancers 2021, 13, 3343. [Google Scholar] [CrossRef] [PubMed]
- Losi-Guembarovski, R.; Kuasne, H.; Guembarovski, A.L.; Rainho, C.A.; Cólus, I.M. DNA methylation patterns of the CDH1, RARB, and SFN genes in choroid plexus tumors. Cancer Genet Cytogenet. 2007, 179, 140–145. [Google Scholar] [CrossRef]
- Udhane, S.S.; Flück, C.E. Regulation of human (adrenal) androgen biosynthesis-New insights from novel throughput technology studies. Biochem. Pharmacol. 2016, 102, 20–33. [Google Scholar] [CrossRef]
- Wu, S.; Xu, S.; Li, R.; Li, K.; Zhong, X.; Li, Y.; Zhou, Z.; Liu, Y.; Feng, R.; Zheng, J.; et al. mTORC1-Rps15 Axis Contributes to the Mechanisms Underlying Global Translation Reduction During Senescence of Mouse Embryonic Fibroblasts. Front. Cell Dev. Biol. 2019, 7, 337. [Google Scholar] [CrossRef]

| Tissue | DMGs | Trends in Methylation Level |
|---|---|---|
| hypothalamus | CD2AP, CERK, FBXW11, ROS1, UNC13B, YAP1 |  |
| hypothalamus | ABLIM2, ENSGALG00000033770, FSD1, HOXA1, NUMB, SLC7A11 |  |
| ovary | CD2AP, CPXM1, KCNK13 |  |
| ovary | ENSGALG00000053295 |  |
| Tissues | Groups | Genes | Transcriptional Level | Methylation State | Gene Function |
|---|---|---|---|---|---|
| Ovary | 1-vs.-2 | PRIMA1 | −1.518 | hypo | proline-rich membrane anchor 1 |
| Ovary | 1-vs.-2 | DSTYK | −1.001 | hyper | dual serine/threonine and tyrosine protein kinase |
| Ovary | 2-vs.-3 | NKTR | −1.083 | hyper | natural killer cell triggering receptor |
| Ovary | 2-vs.-3 | DOCK4 | −1.196 | hypo | dedicator of cytokinesis 4 |
| Ovary | 3-vs.-5 | RPS15 | −1.680 | hypo | ribosomal protein S15 |
| Ovary | 3-vs.-5 | SMOC1 | −1.709 | hyper | SPARC-related modular calcium binding 1 |
| Hypothalamus | 3-vs.-5 | SCAMP3 | −1.062 | hyper | secretory carrier membrane protein 3 |
| Hypothalamus | 3-vs.-5 | ATOH8 | −1.050 | hyper | atonal bHLH transcription factor 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Li, C.; Deng, J.; Jia, Y.; Qu, L.; Ning, Z. Chicken Hypothalamic and Ovarian DNA Methylome Alteration in Response to Forced Molting. Animals 2023, 13, 1012. https://doi.org/10.3390/ani13061012
Zhang T, Li C, Deng J, Jia Y, Qu L, Ning Z. Chicken Hypothalamic and Ovarian DNA Methylome Alteration in Response to Forced Molting. Animals. 2023; 13(6):1012. https://doi.org/10.3390/ani13061012
Chicago/Turabian StyleZhang, Tongyu, Chengfeng Li, Jianwen Deng, Yaxiong Jia, Lujiang Qu, and Zhonghua Ning. 2023. "Chicken Hypothalamic and Ovarian DNA Methylome Alteration in Response to Forced Molting" Animals 13, no. 6: 1012. https://doi.org/10.3390/ani13061012





