Molecular Typing of Staphylococcus aureus Isolated from Patients with Autosomal Dominant Hyper IgE Syndrome
Abstract
:1. Introduction
2. Results
2.1. Patients
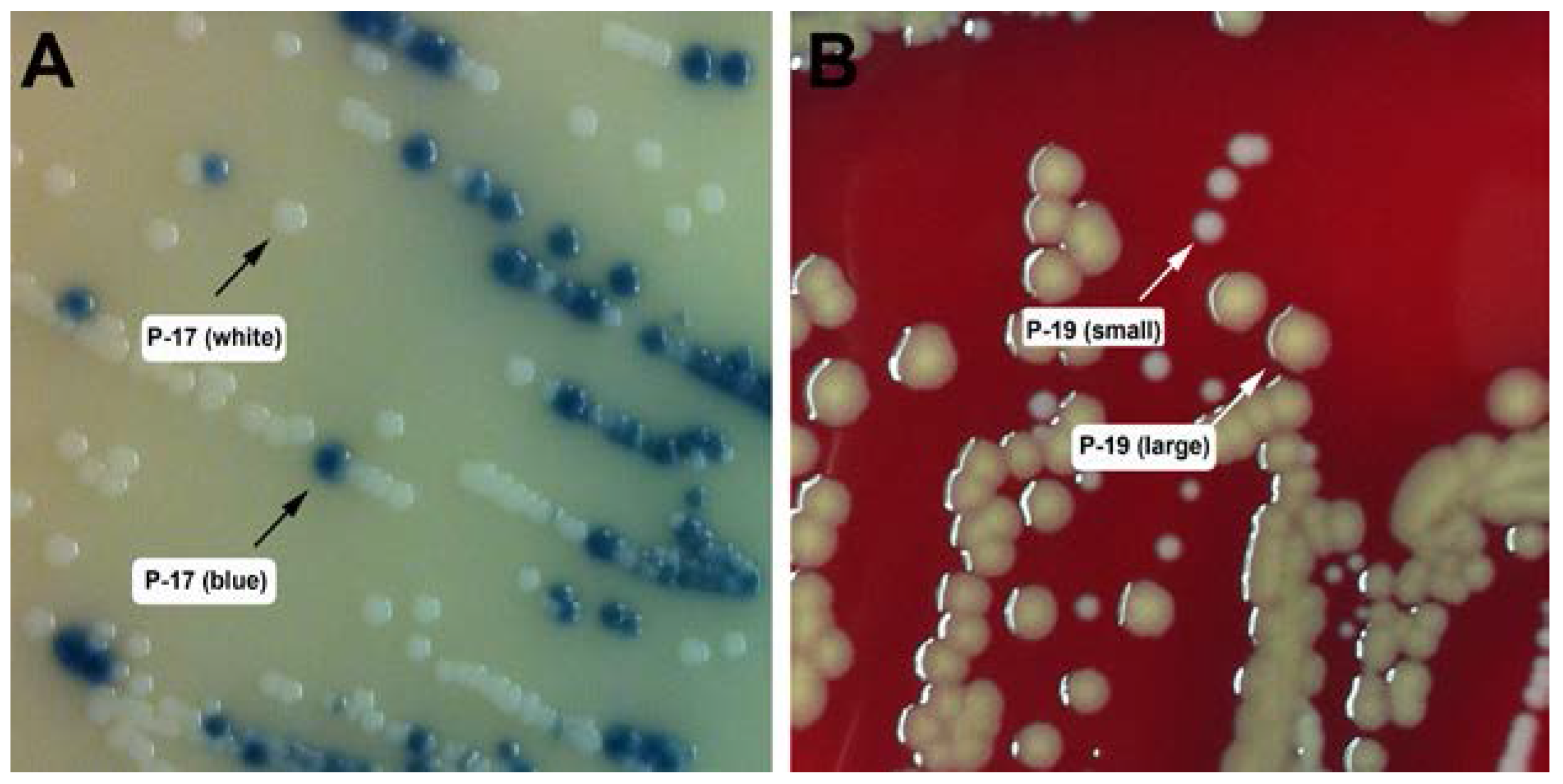
2.2. Strain Isolation and Identification
2.3. Typing of S. aureus Isolates
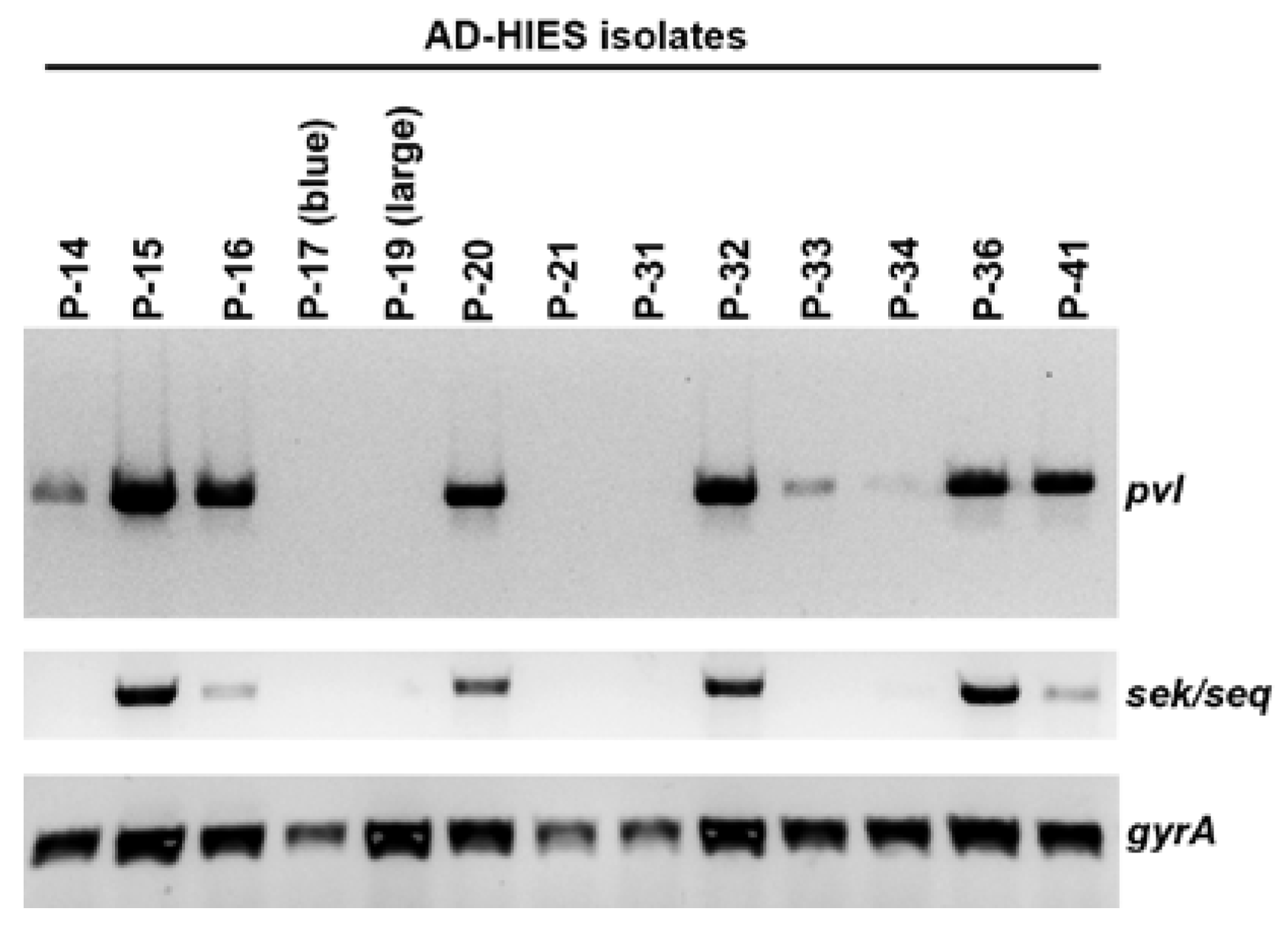
2.4. Presence of the Phage-Encoded Toxins Panton-Valentine Leukocidin and Enterotoxins SEK, SEQ
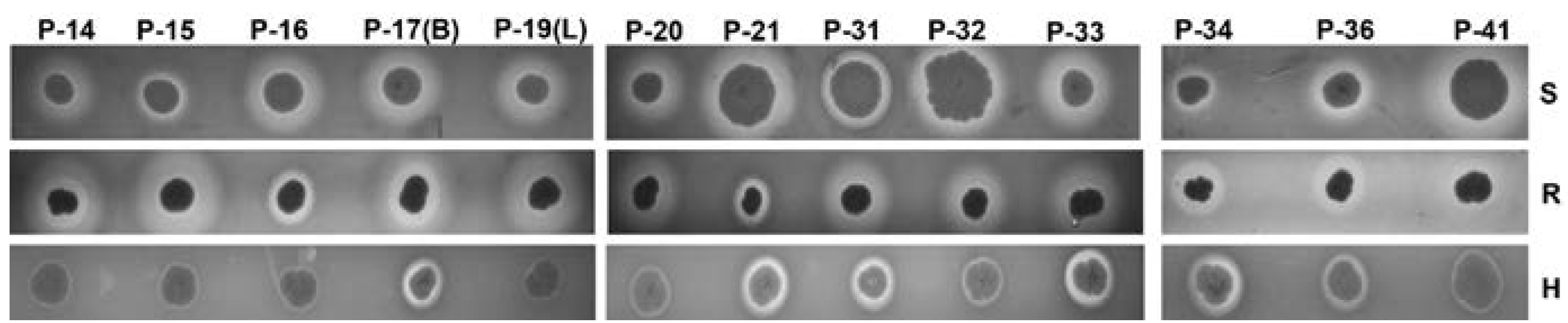
2.5. Hemolysis on Sheep, Rabbit, and Human Agar Plates
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Collection and Isolation of Staphylococci
4.3. DNA Isolation from Staphylococci and 16S rRNA Sequencing
4.4. Identification of MRSA Isolates
4.5. MLST and Spa Typing
4.6. Screening for pvl, sek, and seq
4.7. Blood Agar Hemolysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Ferry, T.; Perpoint, T.; Vandenesch, F.; Etienne, J. Virulence determinants in Staphylococcus aureus and their involvement in clinical syndromes. Curr. Infect. Dis. Rep. 2005, 7, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 2001, 7, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Moreno, F.; Crisp, C.; Jorgensen, J.H.; Patterson, J.E. Methicillin-resistant Staphylococcus aureus as a community organism. Clin. Infect. Dis. 1995, 21, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Carrel, M.; Perencevich, E.N.; David, M.Z. USA300 Methicillin-Resistant Staphylococcus aureus, United States, 2000–2013. Emerg. Infect. Dis. 2015, 21, 1973–1980. [Google Scholar] [CrossRef] [PubMed]
- Welte, K.; Zeidler, C. Severe congenital neutropenia. Hematol. Oncol. Clin. N. Am. 2009, 23, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Quie, P.G.; White, J.G.; Holmes, B.; Good, R.A. In vitro bactericidal capacity of human polymorphonuclear leukocytes: Diminished activity in chronic granulomatous disease of childhood. J. Clin. Investig. 1967, 46, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Guide, S.V.; Stock, F.; Gill, V.J.; Anderson, V.L.; Malech, H.L.; Gallin, J.I.; Holland, S.M. Reinfection, rather than persistent infection, in patients with chronic granulomatous disease. J. Infect. Dis. 2003, 187, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Holland, S.M.; DeLeo, F.R.; Elloumi, H.Z.; Hsu, A.P.; Uzel, G.; Brodsky, N.; Freeman, A.F.; Demidowich, A.; Davis, J.; Turner, M.L.; et al. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 2007, 357, 1608–1619. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, Y.; Saito, M.; Tsuchiya, S.; Tsuge, I.; Takada, H.; Hara, T.; Kawamura, N.; Ariga, T.; Pasic, A.; Stojkovic, O.; et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 2007, 448, 1058–1062. [Google Scholar] [CrossRef] [PubMed]
- Kane, A.; Deenick, E.K.; Ma, C.S.; Cook, M.C.; Uzel, G.; Tangye, S.G. STAT3 is a central regulator of lymphocyte differentiation and function. Curr. Opin. Immunol. 2014, 28, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Milner, J.D.; Brenchley, J.M.; Laurence, A.; Freeman, A.F.; Hill, B.J.; Elias, K.M.; Kanno, Y.; Spalding, C.; Elloumi, H.Z.; Paolson, M.L.; et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 2008, 452, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, Y.; Saito, M.; Nagasawa, M.; Takada, H.; Hara, T.; Tsuchiya, S.; Agematsu, K.; Yamada, M.; Kawamura, N.; Ariga, T.; et al. Molecular explanation for the contradiction between systemic Th17 defect and localized bacterial infection in hyper-IgE syndrome. J. Exp. Med. 2009, 206, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Freeman, A.F.; Program, N.C.S.; Park, M.; Sokolic, R.; Candotti, F.; Holland, S.M.; Segre, J.A.; Kong, H.H. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 2013, 23, 2103–2114. [Google Scholar] [CrossRef] [PubMed]
- Smeekens, S.P.; Huttenhower, C.; Riza, A.; van de Veerdonk, F.L.; Zeeuwen, P.L.J.M.; Schalkwijk, J.; van der Meer, J.W.M.; Xavier, R.J.; Netea, M.G.; Gevers, D. Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. J. Innate Immun. 2014, 6, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Severity scoring of atopic dermatitis: the SCORAD index. Consensus Report of the European Task Force on Atopic Dermatitis. Dermatology 1993, 186, 23–31. [Google Scholar]
- Hallin, M.; Friedrich, A.W.; Struelens, M.J. spa typing for epidemiological surveillance of Staphylococcus aureus. Methods Mol. Biol. 2009, 551, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Panton, P.N.; Camb, M.B.; Valentine, F.C.O. Staphylococcal Toxin. Lancet 1932, 219, 506–508. [Google Scholar] [CrossRef]
- Wiseman, G.M. The hemolysins of Staphylococcus aureus. Bacteriol. Rev. 1975, 39, 317–344. [Google Scholar] [PubMed]
- Vandenesch, F.; Lina, G.; Henry, T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: A redundant arsenal of membrane-damaging virulence factors? Front. Cell Infect. Microbiol. 2012, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Schaumburg, F.; Alabi, A.S.; Peters, G.; Becker, K. New epidemiology of Staphylococcus aureus infection in Africa. Clin. Microbiol. Infect. 2014, 20, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Borjesson, S.; Matussek, A.; Melin, S.; Lofgren, S.; Lindgren, P.E. Methicillin-resistant Staphylococcus aureus (MRSA) in municipal wastewater: an uncharted threat? J. Appl. Microbiol. 2010, 108, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.A.; Holden, M.T. Staphylococcus aureus: Superbug, super genome? Trends Microbiol. 2004, 12, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Gillet, Y.; Issartel, B.; Vanhems, P.; Fournet, J.C.; Lina, G.; Bes, M.; Vandenesch, F.; Piémont, Y.; Brousse, N.; Floret, D.; et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 2002, 359, 753–759. [Google Scholar] [CrossRef]
- Parker, D.; Ryan, C.L.; Alonzo, F., 3rd; Torres, V.J.; Planet, P.J.; Prince, A.S. CD4+ T cells promote the pathogenesis of Staphylococcus aureus pneumonia. J. Infect. Dis. 2015, 211, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Yoong, P.; Torres, V.J. Counter inhibition between leukotoxins attenuates Staphylococcus aureus virulence. Nat. Commun. 2015, 6, 8125. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.G.; Eells, S.J.; Taylor, A.R.; David, M.Z.; Ortiz, N; Zychowski, D; Kumar, N.; Cruz, D.; Boyle-Vavra, S.; Daum, R.S. Staphylocccus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clin. Infect. Dis. 2012, 54, 1523–1535. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, V.S.; Limbago, B.M.; Moran, G.J.; Krishnadasan, A.; Gorwitz, R.J.; McDougal, L.K.; Talan, D.A. Staphylococcus aureus Colonization and Strain Type at Various Body Sites among Patients with a Closed Abscess and Uninfected Controls at U.S. Emergency Departments. J. Clin. Microbiol. 2015, 53, 3478–3484. [Google Scholar] [CrossRef] [PubMed]
- Krausz, K.L.; Bose, J.L. Rapid Isolation of DNA from Staphylococcus. Methods Mol. Biol. 2016, 1373, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.X.; Galiana, A.; Pedreira, W.; Mowszowicz, M.; Christophersen, I.; Machiavello, S.; Lope, L.; Benaderet, S.; Buela, F.; Vicentino, W.; et al. Community-acquired methicillin-resistant Staphylococcus aureus, Uruguay. Emerg. Infect. Dis. 2005, 11, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Diep, B.A.; Gill, S.R.; Chang, R.F.; Phan, T.H.; Chen, J.H.; Davidson, M.G.; Lin, F.; Lin, J.; Carleton, H.A.; Mongodin, E.F.; et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 2006, 367, 731–739. [Google Scholar] [CrossRef]
| Patient ID | Gender | Age + | Race | STAT3 Mutation (Nucleotide; Amino Acid) | STAT3 Mutation Domain | History of: | On Antibiotics | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Eczema | Skin Abscess | PNA | CMC | |||||||
| P-9 | F | 45 | W | G1909A; V637M | SH2 | Y | Y | Y | Y | N |
| P-10 | M | 27 | B | C1144T; R382W | DNA | Y | Y | Y | Y | Y |
| P-14 | F | 45 | W | T1151C; F384S | DNA | Y | Y | Y | Y | Y |
| P-15 | F | 18 | W | C1144T; R382W | DNA | Y | Y | Y | Y | N |
| P-16 | M | 23 | W | T1997G; L666R | SH2 | Y | Y | Y | Y | Y |
| P-17 | F | 39 | W | C2003T; S668F | SH2 | Y | Y | Y | Y | Y |
| P-18 | F | 43 | W | T1861G; F621V | SH2 | Y | Y | Y | Y | Y |
| P-19 | F | 1 | W | A1939G; N647D | SH2 | Y | N | N | N | Y |
| P-20 | M | 7 | B | C1144T; R382G | DNA | Y | Y | Y | Y | Y |
| P-21 | F | 13 | W | T2117C; L706P | Trans-activation | Y | Y | Y | N | Y |
| P-25 | M | 44 | W | 1387delGTG; V463del | DNA | Y | Y | Y | Y | Y |
| P-26 | M | 9 | B | A1831G; S611G | SH2 | Y | Y | Y | N | Y |
| P-31 | F | 17 | A | G1268A; R423Q | DNA | Y | N | N | N | Y |
| P-32 | M | 17 | W | G1145A; R382Q | DNA | Y | Y | Y | Y | Y |
| P-33 | M | 6 | A | G1909A; V637M | SH2 | Y | Y | Y | N | Y |
| P-34 | M | 20 | W | G1909A; V637M | SH2 | Y | Y | Y | Y | Y |
| P-36 | F | 6 | B | G1909A; V637M | SH2 | Y | Y | Y | Y | Y |
| P-39 | F | 1 | W | G1268A; R423Q | DNA | Y | N | N | N | Y |
| P-40 | F | 36 | W | G1268A; R423Q | DNA | Y | Y | Y | Y | N |
| P-41 | F | 30 | H | G1909A; V637M; G1381C; V461L | SH2 | Y | Y | Y | Y | Y |
| P-53 | M | 53 | W | G1145A; R382Q | DNA | Y | Y | Y | Y | Y |
| S. aureus Isolates from Patients with AD-HIES | |||||
|---|---|---|---|---|---|
| Strain ID | MLST ST | Spa Type | MRSA | PVL | SEK/SEQ |
| P-14 | 88 | t4013 | N | Y | N |
| P-15 | 2148 | t4013 | N | Y | Y |
| P-16 | 88 | t4013 | Y | Y | Y |
| P-17 (blue) | 8 | t064 | Y | N | N |
| P-19 (large) | 88 | t4013 | N | N | N |
| P-20 | 8 | t008 | Y | Y | Y |
| P-21 | 8 | t064 | N | N | N |
| P-31 | N.D. | t189 | N | N | N |
| P-32 | 8 | t008 | Y | Y | Y |
| P-33 | 8 | t118 | N | Y | N |
| P-34 | 8 | t064 | Y | Y | N |
| P-36 | 8 | t451 | Y | Y | Y |
| P-41 | 8 | t008 | Y | Y | Y |
| Non-Aureus Staphylococcal Isolates from Patients with AD-HIES | |
|---|---|
| P-9 | S. warneri |
| P-10 a | S. haemolyticus |
| P-10 b | S. hominis |
| P-17 (white) | S. haemolyticus |
| P-18 | S. haemolyticus |
| P-19 (small) | S. hominis |
| P-25 a | S. haemolyticus |
| P-25 b | S. haemolyticus |
| P-26 | S. simulans |
| P-39 | S. hominis |
| P-40 | S. hominis |
| P-53 | S. haemolyticus |
| Name | 5′–3′ Sequence | Purpose | Reference |
|---|---|---|---|
| 16S-2F | GCRKGCYTAAYACATGCAAGTCGA | 16S typing | This study |
| 16S-1088R | CACGACACGAGCTGACGACAGCCA | ||
| arcC-up | TTGATTCACCAGCGCGTATTGTC | MLST typing | saureus.mlst.net |
| arcC-dn | AGGTATCTGCTTCAATCAGCG | ||
| aroE-up | ATCGGAAATCCTATTTCACATTC | ||
| aroE-dn | GGTGTTGTATTAATAACGATATC | ||
| glpf-up | CTAGGAACTGCAATCTTAATC | ||
| glpf-dn | TGGTAAAATCGCATGTCCAATTC | ||
| gmk-up | ATCGTTTTATCGGGACCATC | ||
| gmk-dn | TCATTAACTACAACGTAATCGTA | ||
| pta-up | GTTAAAATCGTATTACCTGAAGG | ||
| pta-n | GACCCTTTTCTTCAAAAGCTTAA | ||
| tpi-up | TCGTTCATTCTGAACGTCGTGAA | ||
| tpi-dn | TTTGCACCTTCTAACAATTGTAC | ||
| yqiL-up | CAGATACAGGACACCTATTGGC | ||
| yqiL-dn | CGTTGAGGAATCGATACTGGAAC | ||
| spa-1113f | TAAAGACGATCCTTCGGTGAGC | spa typing | www.ridom.de |
| spa-1514r | CAGCAGTAGTGCCGTTTGCTT | ||
| PVL-1 | ATGTCTGGACATGATCCA | LukSF-PV amplification | [30] |
| PVL-2 | AACTATCTCTGCCATATGGT | ||
| SEQ/SEK1 | GTATGGCGGAATTACGTTGG | sek/seq amplification | This study |
| SEQ/SEK2 | TTGGTAACCCATCATCTCCTG |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sastalla, I.; Williams, K.W.; Anderson, E.D.; Myles, I.A.; Reckhow, J.D.; Espinoza-Moraga, M.; Freeman, A.F.; Datta, S.K. Molecular Typing of Staphylococcus aureus Isolated from Patients with Autosomal Dominant Hyper IgE Syndrome. Pathogens 2017, 6, 23. https://doi.org/10.3390/pathogens6020023
Sastalla I, Williams KW, Anderson ED, Myles IA, Reckhow JD, Espinoza-Moraga M, Freeman AF, Datta SK. Molecular Typing of Staphylococcus aureus Isolated from Patients with Autosomal Dominant Hyper IgE Syndrome. Pathogens. 2017; 6(2):23. https://doi.org/10.3390/pathogens6020023
Chicago/Turabian StyleSastalla, Inka, Kelli W. Williams, Erik D. Anderson, Ian A. Myles, Jensen D. Reckhow, Marlene Espinoza-Moraga, Alexandra F. Freeman, and Sandip K. Datta. 2017. "Molecular Typing of Staphylococcus aureus Isolated from Patients with Autosomal Dominant Hyper IgE Syndrome" Pathogens 6, no. 2: 23. https://doi.org/10.3390/pathogens6020023






