Quantitative Analysis of the KSHV Transcriptome Following Primary Infection of Blood and Lymphatic Endothelial Cells
Abstract
:1. Introduction
2. Results
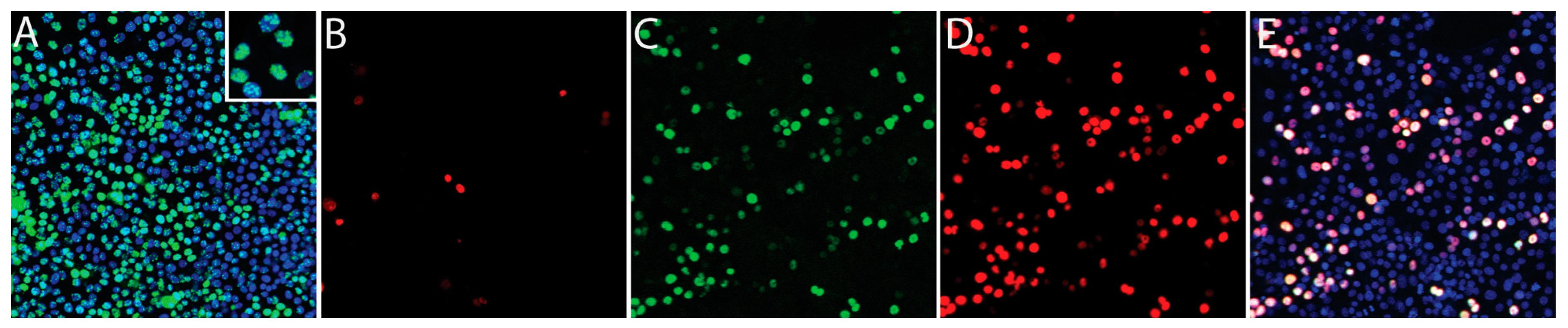
2.1. Comparison of Conventional Latency in KSHV-Infected Cell Lines
2.2. RNA-seq Analysis of KSHV Latent Transcriptomes
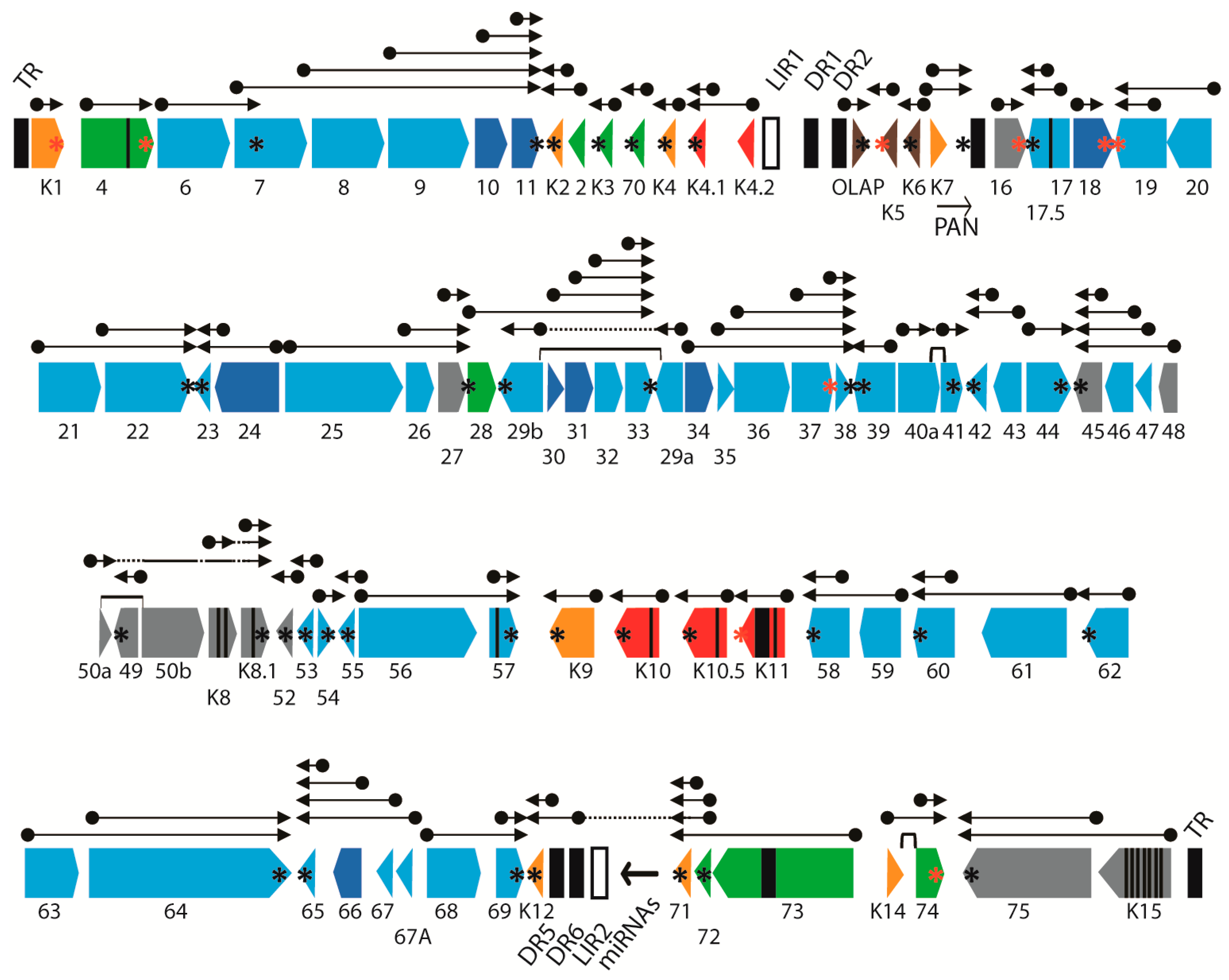
2.3. RNA-seq Analysis of KSHV Transcripts
2.4. Quantitation of RNA Transcripts Using Unique UCDS Features
2.5. Quantitation of RNA Transcripts Is Confounded by Overlapping Transcripts
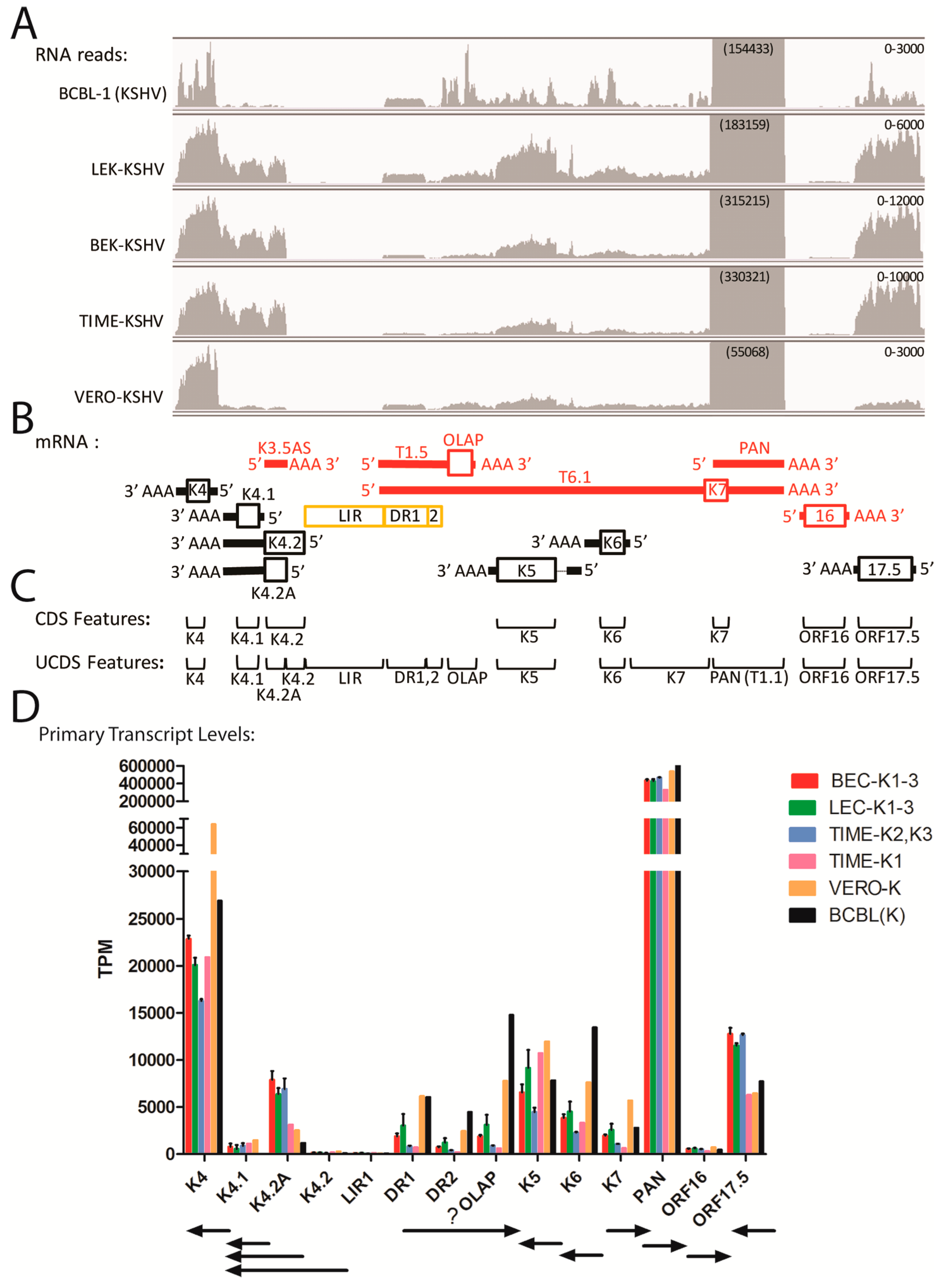
2.6. Quantitation of Overlapping Transcripts Terminating at Common Poly(A) Sites (ORF7-K3)
2.7. Gene Expression in the Divergent Locus B and PAN Region
2.8. Quantitation of Overlapping Transcripts for ORFs 17 and 17.5
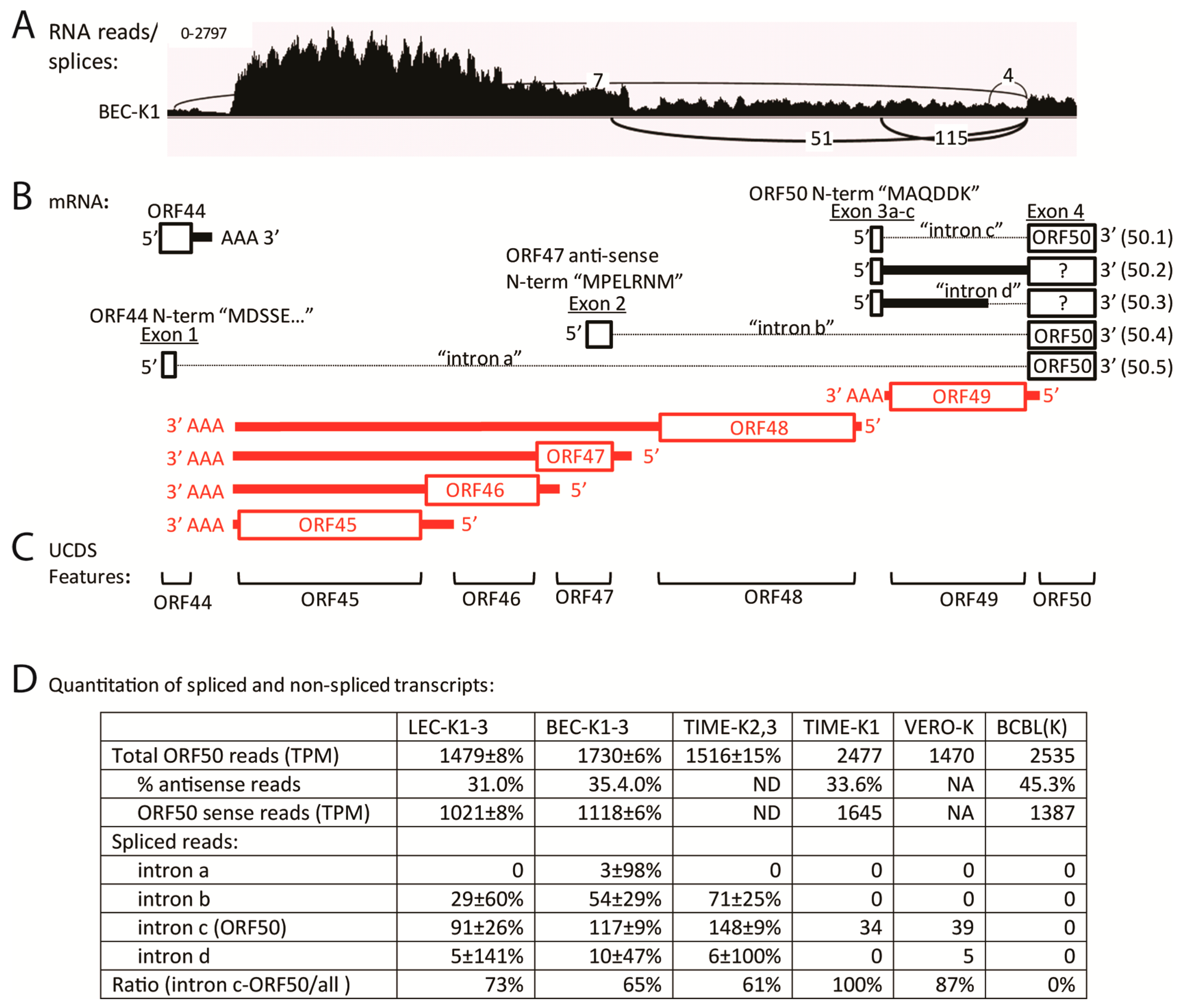
2.9. Quantitation of Spliced Transcripts, ORFs 50/K8/K8.1
2.10. Characterization ORF57 Alternate Splicing
2.11. Characterization of vIRF Alternate Splicing
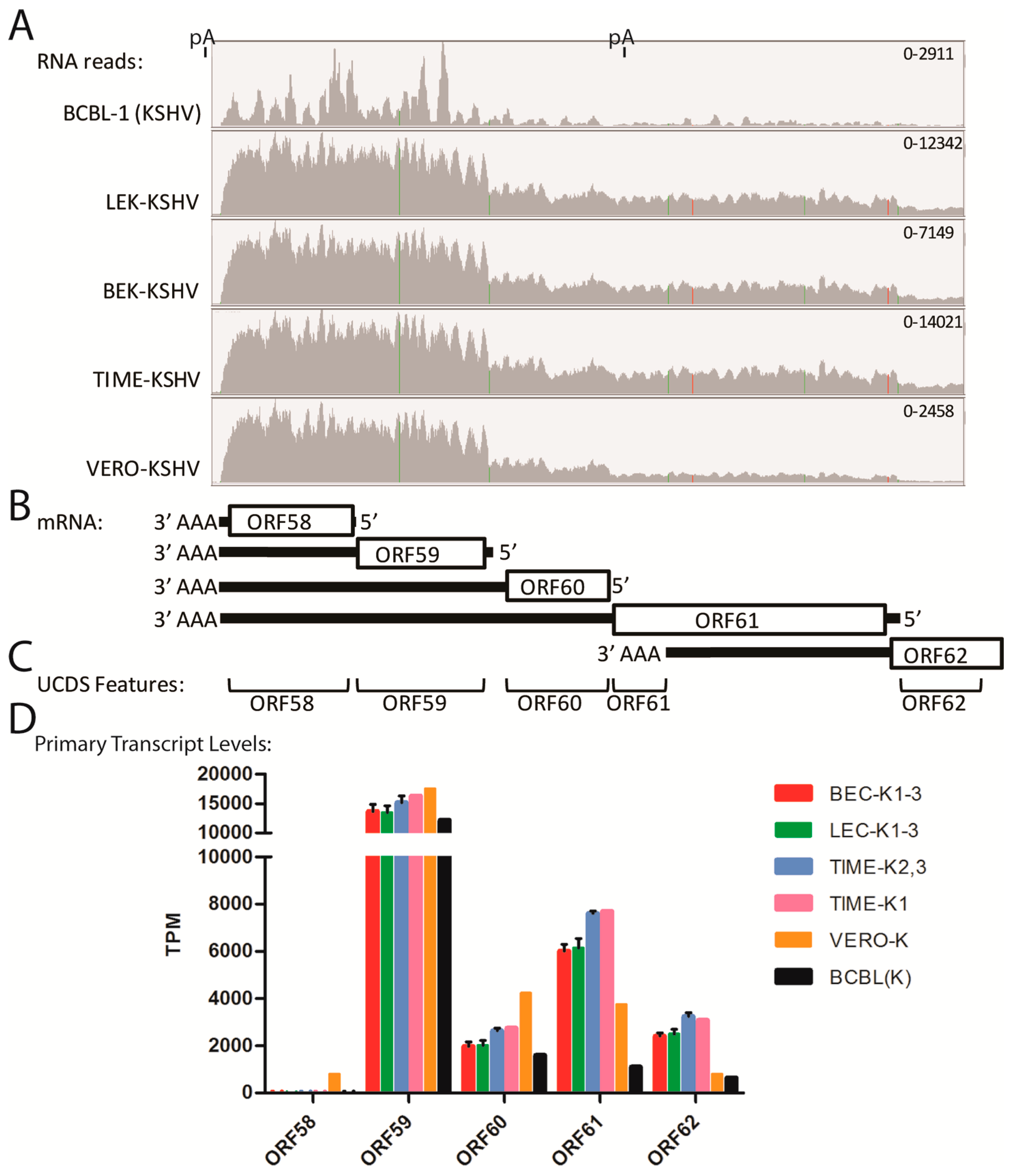
2.12. Quantitation of Bicistronic Transcripts, ORFs 58–62
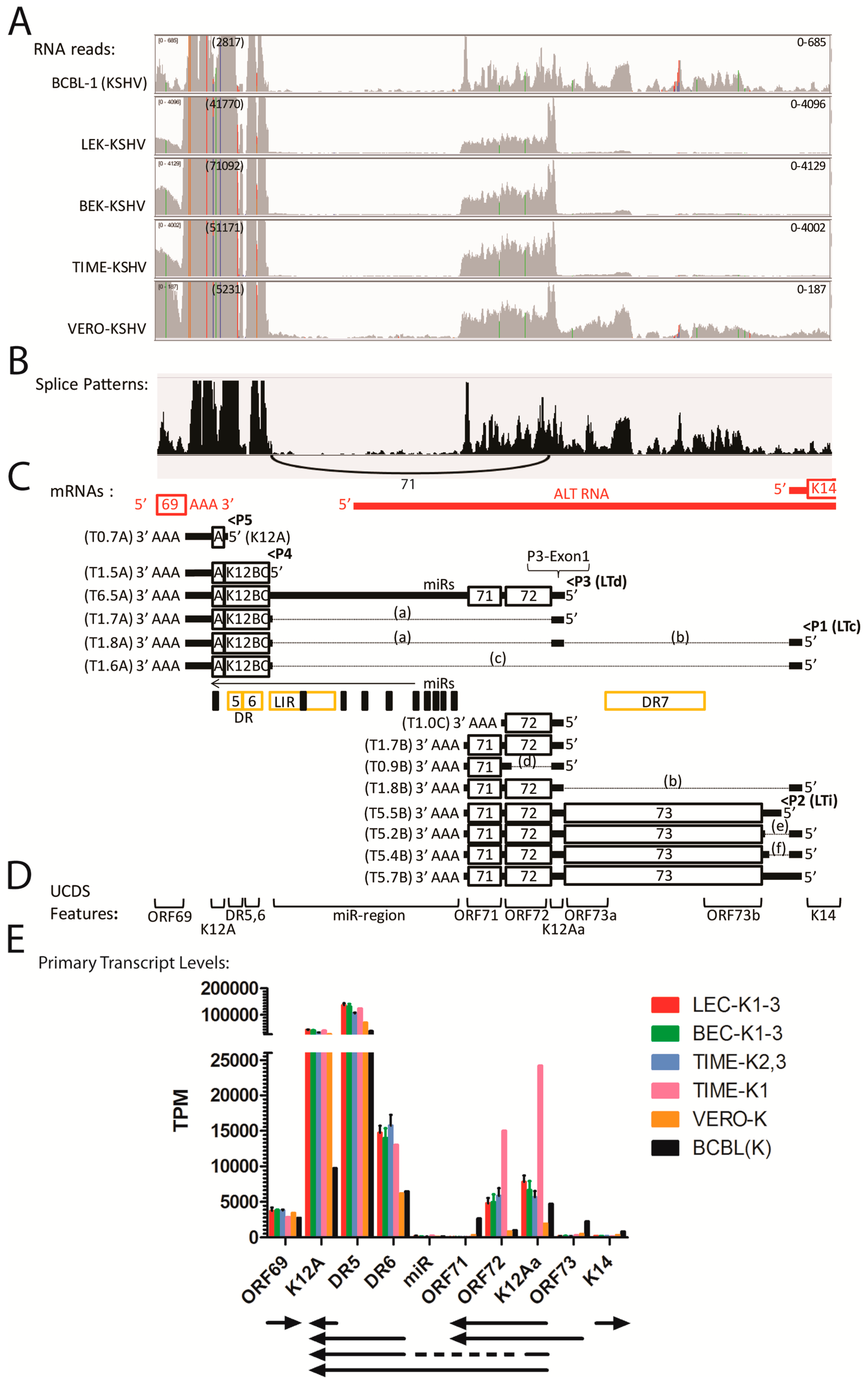
2.13. Quantitation of Transcripts in the ORFK12-73 Latency Region
2.14. Antisense Transcripts
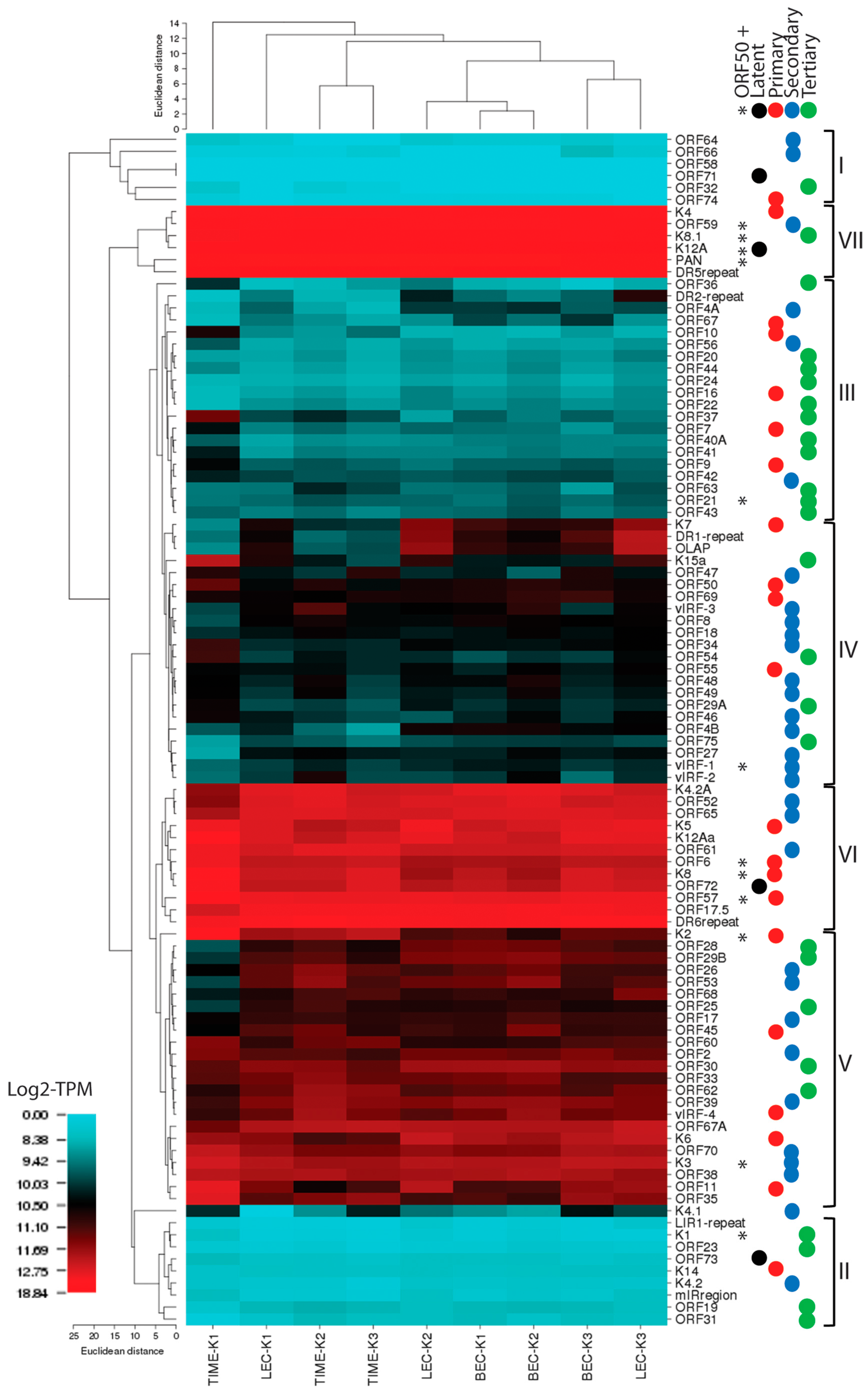
2.15. Hierarchical Clustering of RNA Reads
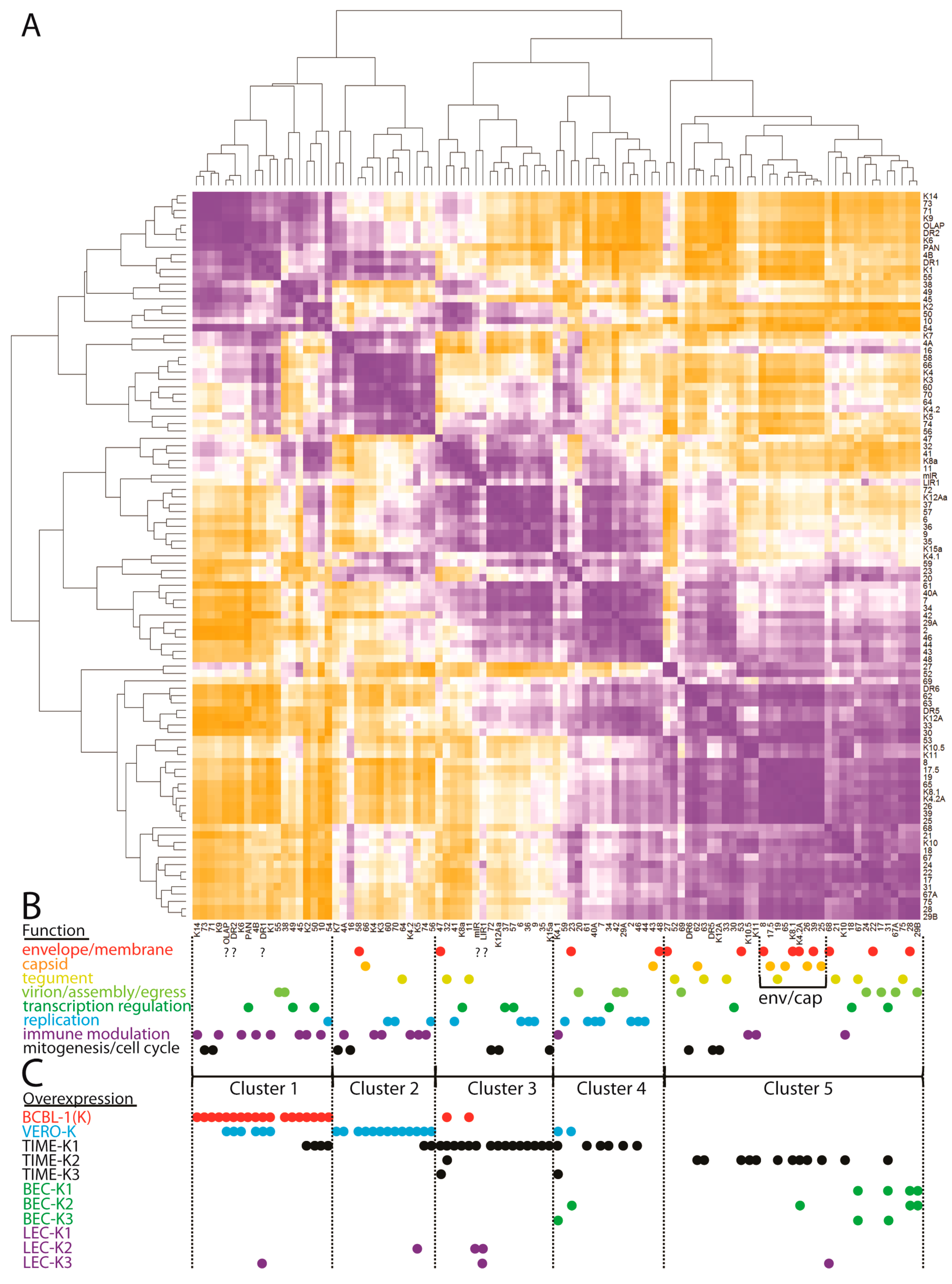
2.16. Hierarchical Clustering of the Matrix of Gene-Gene Expression Correlations
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Immunological Reagents
4.3. KSHV Infection
4.4. Immunofluorescence Assays
4.5. Confocal Microscopy
4.6. RNA-Sequencing
4.7. Data Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Han, Y.; Gao, S.; Muegge, K.; Zhang, W.; Zhou, B. Advanced applications of RNA sequencing and challenges. Bioinform. Biol. Insights 2015, 9, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Gatherer, D.; Seirafian, S.; Cunningham, C.; Holton, M.; Dargan, D.J.; Baluchova, K.; Hector, R.D.; Galbraith, J.; Herzyk, P.; Wilkinson, G.W.; et al. High-resolution human cytomegalovirus transcriptome. Proc. Natl. Acad. Sci. USA 2011, 108, 19755–19760. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Tarrant-Elorza, M.; Pari, G.S. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013, 9, e1003366. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Xu, G.; Deng, N.; Taylor, C.; Zhu, D.; Flemington, E.K. Quantitative and qualitative RNA-seq-based evaluation of Epstein-Barr virus transcription in type I latency Burkitt’s lymphoma cells. J. Virol. 2010, 84, 13053–13058. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.; Weisburd, B.; Stern-Ginossar, N.; Mercier, A.; Madrid, A.S.; Bellare, P.; Holdorf, M.; Weissman, J.S.; Ganem, D. KSHVKSHV 2.0: A comprehensive annotation of the KKaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014, 10, e1003847. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Strong, M.J.; Wang, X.; Moss, W.N.; Concha, M.; Lin, Z.; O’Grady, T.; Baddoo, M.; Fewell, C.; Renne, R.; et al. High-throughput RNA sequencing-based virome analysis of 50 lymphoma cell lines from the cancer cell line encyclopedia project. J. Virol. 2015, 89, 713–729. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, P.; Thakker, S.; Verma, S.C. Transcriptome analysis of Kaposi’s sarcoma-associated herpesvirus during de novo primary infection of human b and endothelial cells. J. Virol. 2015, 89, 3093–3111. [Google Scholar] [CrossRef] [PubMed]
- Juranic Lisnic, V.; Babic Cac, M.; Lisnic, B.; Trsan, T.; Mefferd, A.; Das Mukhopadhyay, C.; Cook, C.H.; Jonjic, S.; Trgovcich, J. Dual analysis of the murine cytomegalovirus and host cell transcriptomes reveal new aspects of the virus-host cell interface. PLoS Pathog. 2013, 9, e1003611. [Google Scholar] [CrossRef] [PubMed]
- Sijmons, S.; Van Ranst, M.; Maes, P. Genomic and functional characteristics of human cytomegalovirus revealed by next-generation sequencing. Viruses 2014, 6, 1049–1072. [Google Scholar] [CrossRef] [PubMed]
- Krug, L.T. Complexities of gammaherpesvirus transcription revealed by microarrays and RNA-seq. Curr. Opin. Virol. 2013, 3, 276–284. [Google Scholar] [CrossRef] [PubMed]
- O’Grady, T.; Wang, X.; Honer Zu Bentrup, K.; Baddoo, M.; Concha, M.; Flemington, E.K. Global transcript structure resolution of high gene density genomes through multi-platform data integration. Nucleic Acids Res. 2016, 44, e45. [Google Scholar] [CrossRef] [PubMed]
- Ganem, D. KSHVand the pathogenesis of KKaposi sarcoma: Listening to human biology and medicine. J. Clin. Investig. 2010, 120, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the KKaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, J.; Zong, J.C.; Alcendor, D.J.; Ciufo, D.M.; Poole, L.J.; Sarisky, R.T.; Chiou, C.J.; Zhang, X.; Wan, X.; Guo, H.G.; et al. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHVKSHV/HHVHHV8. J. Natl. Cancer Inst. Monogr. 1998, 23, 79–88. [Google Scholar] [CrossRef]
- Boshoff, C.; Schulz, T.F.; Kennedy, M.M.; Graham, A.K.; Fisher, C.; Thomas, A.; McGee, J.O.; Weiss, R.A.; O’Leary, J.J. Kaposi’s sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1995, 1, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Ensoli, B.; Sgadari, C.; Barillari, G.; Sirianni, M.C.; Sturzl, M.; Monini, P. Biology of Kaposi’s sarcoma. Eur. J. Cancer 2001, 37, 1251–1269. [Google Scholar] [CrossRef]
- Cesarman, E.; Chang, Y.; Moore, P.S.; Said, J.W.; Knowles, D.M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas [see comments]. N. Engl. J. Med. 1995, 332, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Soulier, J.; Grollet, L.; Oksenhendler, E.; Cacoub, P.; Cazals-Hatem, D.; Babinet, P.; d’Agay, M.F.; Clauvel, J.P.; Raphael, M.; Degos, L.; et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease [see comments]. Blood 1995, 86, 1276–1280. [Google Scholar] [PubMed]
- Kedes, D.H.; Lagunoff, M.; Renne, R.; Ganem, D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J. Clin. Investig. 1997, 100, 2606–2610. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, D.; Lagunoff, M.; Renne, R.; Staskus, K.; Haase, A.; Ganem, D. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J. Virol. 1998, 72, 8309–8315. [Google Scholar] [PubMed]
- Staskus, K.A.; Zhong, W.; Gebhard, K.; Herndier, B.; Wang, H.; Renne, R.; Beneke, J.; Pudney, J.; Anderson, D.J.; Ganem, D.; et al. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 1997, 71, 715–719. [Google Scholar] [PubMed]
- Zhong, W.; Wang, H.; Herndier, B.; Ganem, D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 1996, 93, 6641–6646. [Google Scholar] [CrossRef] [PubMed]
- Parravicini, C.; Chandran, B.; Corbellino, M.; Berti, E.; Paulli, M.; Moore, P.S.; Chang, Y. Differential viral protein expression in Kaposi’s sarcoma-associated herpesvirus-infected diseases: Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Am. J. Pathol. 2000, 156, 743–749. [Google Scholar] [CrossRef]
- Du, M.Q.; Liu, H.; Diss, T.C.; Ye, H.; Hamoudi, R.A.; Dupin, N.; Meignin, V.; Oksenhendler, E.; Boshoff, C.; Isaacson, P.G. Kaposi sarcoma-associated herpesvirus infects monotypic (IGM lambda) but polyclonal naive B cells in Castleman disease and associated lymphoproliferative disorders. Blood 2001, 97, 2130–2136. [Google Scholar] [CrossRef] [PubMed]
- Bechtel, J.T.; Liang, Y.; Hvidding, J.; Ganem, D. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J. Virol. 2003, 77, 6474–6481. [Google Scholar] [CrossRef] [PubMed]
- Lagunoff, M.; Bechtel, J.; Venetsanakos, E.; Roy, A.M.; Abbey, N.; Herndier, B.; McMahon, M.; Ganem, D. De novo infection and serial transmission of Kaposi’s sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol. 2002, 76, 2440–2448. [Google Scholar] [CrossRef] [PubMed]
- Naranatt, P.P.; Krishnan, H.H.; Svojanovsky, S.R.; Bloomer, C.; Mathur, S.; Chandran, B. Host gene induction and transcriptional reprogramming in Kaposi’s sarcoma-associated herpesvirus (KSHVKSHV/HHV-8)-infected endothelial, fibroblast, and B cells: Insights into modulation events early during infection. Cancer Res. 2004, 64, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Greene, W.; Kuhne, K.; Ye, F.; Chen, J.; Zhou, F.; Lei, X.; Gao, S.J. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Cancer Treat. Res. 2007, 133, 69–127. [Google Scholar] [PubMed]
- Lu, F.; Zhou, J.; Wiedmer, A.; Madden, K.; Yuan, Y.; Lieberman, P.M. Chromatin remodeling of the Kaposi’s sarcoma-associated herpesvirus orf50 promoter correlates with reactivation from latency. J. Virol. 2003, 77, 11425–11435. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Shedd, D.; Miller, G. An sp1 response element in the Kaposi’s sarcoma-associated herpesvirus open reading frame 50 promoter mediates lytic cycle induction by butyrate. J. Virol. 2005, 79, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- Lukac, D.M.; Renne, R.; Kirshner, J.R.; Ganem, D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 1998, 252, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Lin, S.F.; Gradoville, L.; Yuan, Y.; Zhu, F.; Miller, G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 1998, 95, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- DeMaster, L.K.; Rose, T.M. A critical SP1 element in the rhesus rhadinovirus (RRV) RTA promoter confers high-level activity that correlates with cellular permissivity for viral replication. Virology 2014, 448, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Lan, K.; Kuppers, D.A.; Verma, S.C.; Sharma, N.; Murakami, M.; Robertson, E.S. Induction of Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: A novel mechanism for establishment of latency. J. Virol. 2005, 79, 7453–7465. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.; Ganem, D. A unique herpesviral transcriptional program in KSHV-infected lymphatic endothelial cells leads to mtorc1 activation and rapamycin sensitivity. Cell Host Microbe 2013, 13, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, H.J.; DeMaster, L.K.; Rubinchikova, Y.E.; Rose, T.M. KSHV attachment and entry are dependent on alphavbeta3 integrin localized to specific cell surface microdomains and do not correlate with the presence of heparan sulfate. Virology 2014, 464–465, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Chandran, B. Early events in Kaposi’s sarcoma-associated herpesvirus infection of target cells. J. Virol. 2010, 84, 2188–2199. [Google Scholar] [CrossRef] [PubMed]
- McClure, L.V.; Kincaid, R.P.; Burke, J.M.; Grundhoff, A.; Sullivan, C.S. Comprehensive mapping and analysis of Kaposi’s sarcoma-associated herpesvirus 3’ UTRs identify differential posttranscriptional control of gene expression in lytic versus latent infection. J. Virol. 2013, 87, 12838–12849. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Ni, T.; Yang, W.; Meng, B.; Zhu, J.; Zheng, Z.M. A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog. 2013, 9, e1003749. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Tarrant-Elorza, M.; Verma, S.; Purushothaman, P.; Pari, G.S. Regulation of viral and cellular gene expression by Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 2013, 87, 5540–5553. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.L.; Bennett, H.N.; Snyder, B.A.; Moore, P.S.; Chang, Y. Transcriptional analysis of latent and inducible Kaposi’s sarcoma-associated herpesvirus transcripts in the k4 to k7 region. J. Virol. 2005, 79, 15099–15106. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H.; Chan, M.Y.; Zhu, F.X.; Lukac, D.M.; Yuan, Y. Kaposi’s sarcoma-associated herpesvirus ori-lyt-dependent DNA replication: Cis-acting requirements for replication and ori-lyt-associated rna transcription. J. Virol. 2004, 78, 8615–8629. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Okruzhnov, Y.; Li, H.; Nicholas, J. Human herpesvirus 8 (HHV-8)-encoded cytokines induce expression of and autocrine signaling by vascular endothelial growth factor (VEGF) in HHV-8-infected primary-effusion lymphoma cell lines and mediate VEGF-independent antiapoptotic effects. J. Virol. 2001, 75, 10933–10940. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.X.; Cusano, T.; Yuan, Y. Identification of the immediate-early transcripts of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 1999, 73, 5556–5567. [Google Scholar] [PubMed]
- Saveliev, A.; Zhu, F.; Yuan, Y. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi’s sarcoma-associated herpesvirus. Virology 2002, 299, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Wakeman, B.S.; Izumiya, Y.; Speck, S.H. Identification of novel Kaposi’s sarcoma-associated herpesvirus ORF50 transcripts: Discovery of new RTA isoforms with variable transactivation potential. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Zheng, Z.M. KSHV ORF57, a protein of many faces. Viruses 2015, 7, 604–633. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.R.; Damania, B. The viral interferon regulatory factors of KSHV: Immunosuppressors or oncogenes? Front. Immunol. 2011, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Jenner, R.G.; Alba, M.M.; Boshoff, C.; Kellam, P. Kaposi’s sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 2001, 75, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Thandapani, P.; O'Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Yamanegi, K.; Zheng, Z.M. Gene structure and expression of Kaposi’s sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 2006, 80, 11968–11981. [Google Scholar] [CrossRef] [PubMed]
- Sarid, R.; Wiezorek, J.S.; Moore, P.S.; Chang, Y. Characterization and cell cycle regulation of the major Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 1999, 73, 1438–1446. [Google Scholar] [PubMed]
- Grundhoff, A.; Ganem, D. Mechanisms governing expression of the v-flip gene of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2001, 75, 1857–1863. [Google Scholar] [CrossRef] [PubMed]
- Talbot, S.J.; Weiss, R.A.; Kellam, P.; Boshoff, C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, k14, and 74 in a primary effusion lymphoma cell line. Virology 1999, 257, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Sadler, R.; Wu, L.; Forghani, B.; Renne, R.; Zhong, W.; Herndier, B.; Ganem, D. A complex translational program generates multiple novel proteins from the latently expressed Kaposin (k12) locus of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 1999, 73, 5722–5730. [Google Scholar] [PubMed]
- Li, H.; Komatsu, T.; Dezube, B.J.; Kaye, K.M. The Kaposi’s sarcoma-associated herpesvirus k12 transcript from a primary effusion lymphoma contains complex repeat elements, is spliced, and initiates from a novel promoter. J. Virol. 2002, 76, 11880–11888. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Cullen, B.R. Transcriptional origin of Kaposi’s sarcoma-associated herpesvirus micrornas. J. Virol. 2006, 80, 2234–2242. [Google Scholar] [CrossRef] [PubMed]
- Chandriani, S.; Xu, Y.; Ganem, D. The lytic transcriptome of Kaposi’s sarcoma-associated herpesvirus reveals extensive transcription of noncoding regions, including regions antisense to important genes. J. Virol. 2010, 84, 7934–7942. [Google Scholar] [CrossRef] [PubMed]
- Chandriani, S.; Ganem, D. Array-based transcript profiling and limiting-dilution reverse transcription-PCR analysis identify additional latent genes in Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2010, 84, 5565–5573. [Google Scholar] [CrossRef] [PubMed]
- Dresang, L.R.; Teuton, J.R.; Feng, H.; Jacobs, J.M.; Camp, D.G., 2nd; Purvine, S.O.; Gritsenko, M.A.; Li, Z.; Smith, R.D.; Sugden, B.; et al. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: Insights on the detection and discovery of viral genes. BMC Genom. 2011, 12, 625. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Myers, T.; Buolamwini, J.; Raghavan, K.; van Osdol, W.; Licht, J.; Viswanadhan, V.N.; Kohn, K.W.; Rubinstein, L.V.; Koutsoukos, A.D.; et al. Predictive statistics and artificial intelligence in the U.S. National cancer institute’s drug discovery program for cancer and AIDS. Stem Cells 1994, 12, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Serio, T.R.; Cahill, N.; Prout, M.E.; Miller, G. A functionally distinct tata box required for late progression through the Epstein-Barr virus life cycle. J. Virol. 1998, 72, 8338–8343. [Google Scholar] [PubMed]
- Chang, J.; Ganem, D. On the control of late gene expression in Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 2000, 81 Pt 8, 2039–2047. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Persson, L.M.; O’Brien, M.W.; Mohr, I.; Wilson, A.C. Cooperation between viral interferon regulatory factor 4 and RTA to activate a subset of Kaposi’s sarcoma-associated herpesvirus lytic promoters. J. Virol. 2012, 86, 1021–1033. [Google Scholar] [CrossRef] [PubMed]
- Kliche, S.; Nagel, W.; Kremmer, E.; Atzler, C.; Ege, A.; Knorr, T.; Koszinowski, U.; Kolanus, W.; Haas, J. Signaling by human herpesvirus 8 Kaposin a through direct membrane recruitment of cytohesin-1. Mol. Cell 2001, 7, 833–843. [Google Scholar] [CrossRef]
- Gong, D.; Wu, N.C.; Xie, Y.; Feng, J.; Tong, L.; Brulois, K.F.; Luan, H.; Du, Y.; Jung, J.U.; Wang, C.Y.; et al. Kaposi’s sarcoma-associated herpesvirus ORF18 and ORF30 are essential for late gene expression during lytic replication. J. Virol. 2014, 88, 11369–11382. [Google Scholar] [CrossRef] [PubMed]
- Brulois, K.; Wong, L.Y.; Lee, H.R.; Sivadas, P.; Ensser, A.; Feng, P.; Gao, S.J.; Toth, Z.; Jung, J.U. Association of Kaposi’s sarcoma-associated herpesvirus ORF31 with ORF34 and ORF24 is critical for late gene expression. J. Virol. 2015, 89, 6148–6154. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.R.; Bloomer, C.; Chandran, B. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology 1998, 240, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Dai, C.Y.; Ricciardi, R.P. Cloning and functional analysis of Kaposi’s sarcoma-associated herpesvirus DNA polymerase and its processivity factor. J. Virol. 1998, 72, 6228–6232. [Google Scholar] [PubMed]
- Gao, S.J.; Deng, J.H.; Zhou, F.C. Productive lytic replication of a recombinant Kaposi's sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J. Virol. 2003, 77, 9738–9749. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.; Matsumura, S.; Wilson, A.C. Transcripts encoding K12, v-flip, v-cyclin, and the microrna cluster of Kaposi’s sarcoma-associated herpesvirus originate from a common promoter. J. Virol. 2005, 79, 14457–14464. [Google Scholar] [CrossRef] [PubMed]
- Izumiya, Y.; Izumiya, C.; Van Geelen, A.; Wang, D.H.; Lam, K.S.; Luciw, P.A.; Kung, H.J. Kaposi’s sarcoma-associated herpesvirus-encoded protein kinase and its interaction with k-bzip. J. Virol. 2007, 81, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Glaunsinger, B.; Chavez, L.; Ganem, D. The exonuclease and host shutoff functions of the SOX protein of Kaposi’s sarcoma-associated herpesvirus are genetically separable. J. Virol. 2005, 79, 7396–7401. [Google Scholar] [CrossRef] [PubMed]
- Venetsanakos, E.; Mirza, A.; Fanton, C.; Romanov, S.R.; Tlsty, T.; McMahon, M. Induction of tubulogenesis in telomerase-immortalized human microvascular endothelial cells by glioblastoma cells. Exp. Cell Res. 2002, 273, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Renne, R.; Zhong, W.; Herndier, B.; McGrath, M.; Abbey, N.; Kedes, D.; Ganem, D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 1996, 2, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, H.J.; Rubinchikova, Y.E.; Dipersio, C.M.; Rose, T.M. Integrin alphavbeta3 binds to the RGD motif of glycoprotein b of Kaposi’s sarcoma-associated herpesvirus and functions as an RGD-dependent entry receptor. J. Virol. 2008, 82, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed]









| Infected Cell | IFA 1 | RNA Library | Total Reads | KSHV | ||
|---|---|---|---|---|---|---|
| ORF73+ | ORF59+ | Reads | % Total | |||
| TIME-K1 2 | 89% | 3.4% | Paired-end | 1.41 × 108 | 6.25 × 108 | 0.4% |
| Stranded | 4.06 × 107 | 1.75 × 108 | 0.4% | |||
| TIME-K2 2 | 89% | 1.0% | Paired-end | 1.34 × 108 | 8.19 × 106 | 6.1% |
| TIME-K3 2 | 94% | 2.7% | Paired-end | 1.22 × 108 | 8.83 × 106 | 7.2% |
| LEC-K1 3 | 91% | 8.6% | Paired-end | 1.37 × 108 | 7.25 × 106 | 5.3% |
| Stranded | 4.17 × 107 | 2.15 × 106 | 5.1% | |||
| LEC-K2 3 | 97% | 6.0% | Paired-end | 1.27 × 108 | 4.61 × 106 | 3.6% |
| LEC-K3 3 | 84% | 2.9% | Paired-end | 1.37 × 108 | 3.50 × 106 | 2.6% |
| BEC-K1 4 | 91% | 12.3% | Paired-end | 1.35 × 108 | 6.29 × 106 | 4.7% |
| Stranded | 4.09 × 107 | 1.88 × 106 | 4.6% | |||
| BEC-K2 4 | 84% | <1% | Paired-end | 1.47 × 108 | 7.85 × 106 | 5.4% |
| BEC-K3 4 | 79% | 2.6% | Paired-end | 1.64 × 108 | 5.51 × 106 | 3.4% |
| Vero-K 5 | 95% | <2% | Paired-end | 1.92 × 108 | 1.21 × 106 | 0.6% |
| BCBL1(K) 6 | 100% | <2% | Stranded | 4.99 × 107 | 1.01 × 106 | 2.0% |
| Gene | TATA-Like Signal 1 | Start (bp) 2 | Stop (bp) 2 | PolyA-Signal | Start (bp) 2 | Stop (bp) 2 | Strand | Verified 3 |
|---|---|---|---|---|---|---|---|---|
| K1 | TAATTTT | 4 | 9 | AUUAAA | 1053 | 1058 | + | ? |
| ORF4 | TTATTAAAA | 1051 | 1059 | AAUACA | 2922 | 2927 | + | mapped |
| ORF6 | TATATAAA | 3090 | 3097 | AAUAAA | 6974 | 6979 | + | mapped |
| ORF7 | Nd 4 | - | - | AAUAAA | 17,021 | 17,026 | + | mapped |
| ORF8 | TATTTAAA | 8611 | 8618 | AAUAAA | 17,021 | 17,026 | + | mapped |
| ORF9 | TAAATTTA | 11,230 | 11,237 | AAUAAA | 17,021 | 17,026 | + | mapped |
| ORF10 | TATAA | 14,326 | 14,330 | AAUAAA | 17,021 | 17,026 | + | mapped |
| ORF11 | TATAT | 15,568 | 15,572 | AAUAAA | 17,021 | 17,026 | + | mapped |
| K2 | TATTTTTAA | 18,122 | 18,114 | AAUAAA | 17,171 | 17,166 | - | mapped |
| ORF2 | TATATAA | 18,570 | 18,564 | AAUAAA | 17,171 | 17,166 | - | mapped |
| K3 | TTACAATAAA | 19,793 | 19,784 | AUUAAA | 18,578 | 18,573 | - | mapped |
| ORF70 | TATA | 21,116 | 21,113 | AAUAAA | 19,799 | 19,794 | - | ? |
| K4 | TAATAAAA | 21,912 | 21,906 | AAUAAA | 21,280 | 21,275 | - | mapped |
| K4.1 | TAATTTATAA | 22,532 | 22,523 | AAUAAA | 21,912 | 21,907 | - | ? |
| K4.2A | TATTAAA | 22,875 | 22,871 | AAUAAA | 21,912 | 21,907 | - | ? |
| K4.2 | TAATTTAT | 23,106 | 23,099 | AAUAAA | 21,912 | 21,907 | - | ? |
| OLAP (T1.5) | TATAA | 24,137 | 24,141 | AAUAAA | 26,222 | 26,227 | + | mapped |
| K5 | TTATTT | 27,017 | 27,022 | AUUAAA | 25,712 | 25,717 | - | mapped |
| K6 | TATAA | 27,663 | 27,703 | AAUAAA | 27,067 | 27,072 | - | mapped |
| K7 | TATAT | 28,639 | 28,643 | AAUAAA | 29,871 | 29,876 | + | mapped |
| PAN | GATAAAA | 28,788 | 28,794 | AAUAAA | 29,871 | 29,876 | + | mapped |
| ORF16 | TATATAAAA | 30,176 | 30,184 | AUUAAA | 30,829 | 30,834 | + | mapped |
| ORF17.5 | TATTTAAA | 31,838 | 31,846 | AAUAAA | 30,857 | 30,862 | - | mapped |
| ORF17 | TATTTAAA | 32,606 | 32,613 | AAUAAA | 30,857 | 30,862 | - | mapped |
| ORF18 | TTATT | 32,398 | 32,402 | AUUAAA | 33,530 | 33,535 | + | mapped |
| ORF19 | TATTTAA | 35,050 | 35,056 | AUUAAA | 32,967 | 32,972 | - | ? |
| ORF20 | CATTAAAT | 35,711 | 35,718 | AUUAAA | 32,967 | 32,972 | - | ? |
| ORF21 | AATAAT | 35,296 | 35,291 | AAUAAA | 39,411 | 39,416 | + | mapped |
| ORF22 | TATTAAA | 37,140 | 37,146 | AAUAAA | 39,411 | 39,416 | + | mapped |
| ORF23 | Nd | - | - | AAUAAA | 39,345 | 39,350 | - | mapped |
| ORF24 | TATATA | 43,042 | 43,047 | AAUAAA | 39,345 | 39,350 | - | mapped |
| ORF25 | TATAAAA | 42,629 | 42,635 | AAUAAA | 48,841 | 48,846 | + | mapped |
| ORF26 | TATTAAA | 46,975 | 46,981 | AAUAAA | 48,841 | 48,846 | + | mapped |
| ORF27 | TATATTTAAT | 47,865 | 47,874 | AAUAAA | 48,841 | 48,846 | + | mapped |
| ORF28 | TATATATAA | 48,999 | 49,006 | AAUAAA | 54,171 | 54,176 | + | mapped |
| ORF29B | TTATTAAAAA | 50,546 | 50,555 | AAUAAA | 49,461 | 49,466 | - | mapped |
| ORF30 | Nd | - | - | AAUAAA | 54,171 | 54,176 | + | mapped |
| ORF31 | Nd | - | - | AAUAAA | 54,171 | 54,176 | + | mapped |
| ORF32 | TATTTAAA | 51,446 | 51,453 | AAUAAA | 54,171 | 54,176 | + | mapped |
| ORF33 | TATTAAA | 52,805 | 52,811 | AAUAAA | 54,171 | 54,176 | + | mapped |
| ORF29A | TTAAATT | 54,880 | 54,886 | AAUAAA | 49,461 | 49,466 | - | mapped |
| ORF34 | AATAAAA | 54,635 | 54,641 | AUUAAA | 58,933 | 58,938 | + | mapped |
| ORF35 | TATATAA | 55,635 | 55,641 | AUUAAA | 58,933 | 58,938 | + | mapped |
| ORF36 | Nd | - | - | AUUAAA | 58,933 | 58,938 | + | mapped |
| ORF37 | TATTTATATA | 57,257 | 57,266 | AUUAAA | 58,933 | 58,938 | + | mapped |
| ORF38 | TATTAAA | 58,439 | 58,974 | AAUAAA | 58,952 | 58,957 | + | mapped |
| ORF39 | TATTTAAAA | 60,355 | 60,363 | AAUAAA | 59,000 | 60,363 | - | mapped |
| ORF40A | TTAAATA | 60,357 | 60,363 | AAUAAA | 62,639 | 62,644 | + | mapped |
| ORF41 | Nd | - | - | AAUAAA | 62,639 | 62,644 | + | mapped |
| ORF42 | TATTTATAT | 63,445 | 63,453 | AAUAAA | 62,534 | 62,639 | - | mapped |
| ORF43 | ATATTT | 65,104 | 65,109 | AAUAAA | 62,534 | 62,639 | - | mapped |
| ORF44 | TATAAATA | 63,446 | 63,453 | AAUAAA | 67,394 | 67,399 | + | mapped |
| ORF45 | TAAATTT | 68,707 | 68,713 | AAUAAA | 67,444 | 67,449 | - | mapped |
| ORF46 | TTATAATAA | 69,612 | 69,620 | AAUAAA | 67,444 | 67,449 | - | mapped |
| ORF47 | TATATAAA | 70,088 | 70,096 | AAUAAA | 67,444 | 67,449 | - | mapped |
| ORF48 | TATTTA | 71,551 | 71,556 | AAUAAA | 67,444 | 67,449 | - | mapped |
| ORF49 | ATTTATAA | 72,856 | 72,863 | AAUAAA | 71,728 | 71,733 | - | mapped |
| ORF50 | TTTAAAAA | 71,627 | 71,634 | AAUAAA | 76,813 | 76,818 | + | mapped |
| ORF50AS(T1.2) | TATA | 74,618 | 74,621 | AAUAAA | 73,621 | 73,616 | - | mapped |
| ORF50AS(T3.0) | TATA | 74,618 | 74,621 | AAUAAA | 71,728 | 71,733 | - | mapped |
| K8 | ATATA | 74,592 | 74,596 | AAUAAA | 76,813 | 76,818 | + | mapped |
| K8.1 | TATTAAA | 75,966 | 75,972 | AAUAAA | 76,813 | 76,818 | + | mapped |
| ORF52 | TATTTAAA | 77,368 | 77,375 | AAUAAA | 76,823 | 76,828 | - | mapped |
| ORF53 | TATATAA | 77,819 | 77,825 | AAUAAA | 77,377 | 77,382 | - | ? |
| ORF54 | TTATATA | 77,819 | 77,825 | AAUAAA | 78,787 | 78,792 | + | mapped |
| ORF55 | TATATTTAAT | 79,832 | 79,841 | AAUAAA | 78,818 | 78,823 | - | mapped |
| ORF56 | TTTTATA | 79,507 | 79,512 | AAUAAA | 83,078 | 83,713 | + | mapped |
| ORF57 | TATATAA | 82,073 | 82,079 | AAUAAA | 83,078 | 83,713 | + | mapped |
| vIRF-1 | TATATA | 85,410 | 85,415 | AAUAAA | 83,903 | 83,908 | - | mapped |
| vIRF-4 | TATAA | 89,066 | 89,071 | AAUAAA | 86,125 | 86,130 | - | mapped |
| vIRF-3 | Nd | - | - | AAUAAA | 89,487 | 89,492 | - | mapped |
| vIRF_2 | ATATATT | 94,348 | 94,353 | AUUAAA | 91,881 | 91,886 | - | mapped |
| ORF58 | Nd | - | - | AAUAAA | 94,584 | 94,589 | - | mapped |
| ORF59 | TATTTAA | 96,923 | 96,929 | AAUAAA | 94,584 | 94,589 | - | mapped |
| ORF60 | Nd | - | - | AAUAAA | 94,584 | 94,589 | - | mapped |
| ORF61 | TATATAAA | 100,459 | 100,466 | AAUAAA | 94,584 | 94,589 | - | mapped |
| ORF62 | TATTTAAAAA | 101,458 | 101,467 | AAUAAA | 98,399 | 98,404 | - | mapped |
| ORF63 | CATATTTA | 101,165 | 101,172 | AAUAAA | 111,992 | 111,997 | + | mapped |
| ORF64 | TATTAAA | 103,950 | 103,956 | AAUAAA | 111,992 | 111,997 | + | mapped |
| ORF65 | TATTAAA | 112,652 | 112,658 | AAUAAA | 111,927 | 111,932 | - | mapped |
| ORF66 | TATATT | 113,951 | 113,956 | AAUAAA | 111,927 | 111,932 | - | mapped |
| ORF67 | TATTTAAAAA | 114,809 | 114,818 | AAUAAA | 111,927 | 111,932 | - | mapped |
| ORF67A | AAATATTT | 114,989 | 114,996 | AAUAAA | 111,927 | 111,932 | - | mapped |
| ORF68 | AAATATTT | 114,989 | 114,996 | AAUAAA | 117,509 | 117,514 | + | mapped |
| ORF69 | TATAAAAA | 116,287 | 116,294 | AAUAAA | 117,509 | 117,514 | + | mapped |
| K12A | Nd | - | - | AAUAAA | 117,548 | 117,553 | - | mapped |
| miR-region | Nd | - | - | AAUAAA | 117,548 | 117,553 | - | mapped |
| DR5-repeat | Nd | - | - | AAUAAA | 117,548 | 117,553 | - | mapped |
| DR6-repeat | Nd | - | - | AAUAAA | 117,548 | 117,553 | - | mapped |
| Alt RNA | TATAT | 120,457 | 120,461 | AUUAAA | 130,666 | 130,671 | + | mapped |
| ORF71 | Nd | - | - | AAUAAA | 122,337 | 122,342 | - | mapped |
| ORF72 | ACATAAAA | 124,055 | 124,062 | AAUAAA | 123,010 | 123,015 | - | ? |
| ORF73 | TATAA | 128,185 | 128,189 | AAUAAA | 123,010 | 123,015 | - | ? |
| K14 | TATAA | 127,966 | 127,970 | AUUAAA | 130,666 | 130,671 | + | mapped |
| ORF74 | TTTATTA | 129,380 | 129,386 | AUUAAA | 130,666 | 130,671 | + | mapped |
| ORF75 | TATAGA | 134,891 | 134,896 | AAUAAA | 130,655 | 130,660 | - | mapped |
| K15 | TATTTAT | 137,077 | 137,083 | AAUAAA | 130,655 | 130,660 | - | mapped |
| UCDS | Description 1 | Start (bp) | Stop (bp) | Size (bp) | Strand | Primary Transcript (Subtract) 2 |
|---|---|---|---|---|---|---|
| K1 | N-term CDS domain | 105 | 747 | 643 | + | |
| ORF4a | N-term CDS domain | 1096 | 1985 | 890 | + | |
| ORF4b | C-term CDS domain | 2595 | 2764 | 170 | + | |
| ORF6 | Normal CDS | 3179 | 6577 | 3399 | + | |
| ORF7 | N-term trunc/ORF6(3′NC), C-term trunc/ORF8(5′NC) | 6980 | 8618 | 1639 | + | |
| ORF8 | N-term trunc/ORF7(CDS,3′NC) | 8681 | 11,202 | 2522 | + | ORF7 UCDS |
| ORF9 | Normal CDS | 11,329 | 14,367 | 3039 | + | ORF8 UCDS |
| ORF10 | C-term trunc/ORF11(5′NC,CDS) | 14,485 | 15,522 | 1257 | + | ORF9 UCDS |
| ORF11 | N-term trunc/ORF10(CDS) | 15,791 | 16,979 | 1223 | + | ORF10 UCDS |
| K2 | Normal CDS | 17,227 | 17,841 | 615 | - | ORF2 UCDS |
| ORF2 | CDS C-term trunc/K2(5′NC) | 18,114 | 18,519 | 406 | - | |
| K3 | Normal CDS | 18,574 | 19,542 | 969 | - | |
| ORF70 | Normal CDS | 20,023 | 21,036 | 1014 | - | |
| K4 | Normal CDS | 21,480 | 21,764 | 285 | - | |
| K4.1 | Normal CDS | 22,117 | 22,461 | 345 | - | K4.2A UCDS |
| K4.2A | C-term domain K4.2 | 22,530 | 22,835 | 306 | - | K4.2 UCDS |
| K4.2 | N-term domain K4.2 | 22,885 | 23,078 | 549 | - | |
| LIR1 | 5′ trunc/K4.2(CDS), 3′ trunc/DR1-repeat | 23,128 | 24,180 | 1147 | + | |
| DR1 | Normal domain | 24,230 | 24,808 | 579 | + | |
| DR2 | 5′ trunc/DR1-repeat | 24,858 | 25,045 | 223 | + | |
| OLAP | N-term trunc/DR2-repeat | 25,095 | 25,569 | 524 | + | |
| K5 | Normal CDS | 25,865 | 26,635 | 771 | - | |
| K6 | Normal CDS | 27,289 | 27,576 | 288 | - | |
| K7 | 3′ trunc/PAN RNA | 27,693 | 28,818 | 1126 | + | |
| PAN | 5′ trunc/K7(CDS) | 28,868 | 29,895 | 1077 | + | K7 UCDS |
| ORF16 | Normal CDS | 30,242 | 30,769 | 528 | + | |
| ORF17.5 | Normal CDS | 30,920 | 31,786 | 867 | - | ORF17 UCDS |
| ORF17 | C-term trunc/ORF17.5 CDS | 31,838 | 32,524 | 687 | - | |
| ORF18 | N-term trunc/ORF17(5′NC), C-term trunc/ORF19(CDS,3′NC) | 32,605 | 32,967 | 363 | + | |
| ORF19 | N-term trunc/ORF20(CDS), C-term trunc/ORF18(3′NC) | 33,535 | 34,709 | 1175 | - | ORF20 UCDS |
| ORF20 | N-term trunc/ORF19(CDS), C-term trunc/ORF21(CDS) | 35,049 | 35,401 | 353 | - | |
| ORF21 | N-term trunc/ORF20(CDS), C-term trunc/ORF22(5′NC) | 35,672 | 37,147 | 1476 | + | |
| ORF22 | C-term trunc/ORF23(CDS) | 37,212 | 39,351 | 2193 | + | ORF21 UCDS |
| ORF23 | Normal CDS | 39,401 | 40,615 | 1215 | - | ORF24 UCDS |
| ORF24 | N-term trunc/ORF25(3′NC), C-term trunc/ORF23(CDS) | 40,665 | 42,636 | 2018 | - | |
| ORF25 | N-term trunc/ORF24(5′NC), C-term trunc/ORF26(5′NC) | 43,041 | 46,982 | 3942 | + | |
| ORF26 | C-term trunc/ORF27(5′NC) | 47,032 | 47,873 | 842 | + | ORF25 UCDS |
| ORF27 | Normal CDS | 47,973 | 48,845 | 873 | + | ORF26 UCDS |
| ORF28 | Normal CDS | 49,091 | 49,399 | 309 | + | |
| ORF29B | Normal CDS | 49,462 | 50,517 | 1056 | - | |
| ORF30 | Normal CDS | 50,723 | 50,952 | 230 | + | |
| ORF31 | N-term trunc/ORF30(CDS), C-term trunc/ORF32(5′NC,CDS) | 51,002 | 51,452 | 500 | + | ORF30 UCDS |
| ORF32 | N-term trunc/ORF31(CDS) | 51,537 | 52,860 | 1324 | + | ORF31 UCDS |
| ORF33 | N-term trunc/ORF32(CDS) | 52,910 | 53,865 | 1005 | + | ORF32 UCDS |
| ORF29A | N-term trunc/ORF34(5′NC), C-term trunc/ORF33(3′NC) | 54,176 | 54,665 | 490 | - | |
| ORF34 | C-term trunc/ORF35(5′NC,CDS) | 54,774 | 55,688 | 984 | + | |
| ORF35 | C-term trunc/ORF36(CDS) | 55,738 | 56,075 | 338 | + | ORF34 UCDS |
| ORF36 | N-term trunc/ORF35(CDS), C-term trunc/ORF37(5′NC,CDS) | 56,190 | 57,269 | 1080 | + | ORF35 UCDS |
| ORF37 | N-term trunc/ORF36(CDS), C-term trunc/ORF38(5′NC,CDS) | 57,409 | 58,396 | 988 | + | ORF36 UCDS |
| ORF38 | N-term trunc/ORF37(5′NC,CDS) | 58,446 | 58,972 | 527 | + | ORF37 UCDS |
| ORF39 | Normal CDS | 59,072 | 60,274 | 1203 | - | |
| ORF40A | Normal CDS | 60,407 | 61,780 | 1374 | + | |
| ORF41 | C-term trunc/ORF42(CDS) | 61,926 | 62,543 | 618 | + | Variable 3 |
| ORF42 | C-term trunc/ORF41(3'NC), N-term trunc/ORF43(CDS) | 62,644 | 63,235 | 592 | - | ORF43 UCDS |
| ORF43 | N-term trunc/ORF44(CDS), C-term trunc/ORF42(5′NC,CDS) | 63,444 | 64,991 | 1548 | - | |
| ORF44 | N-term trunc/ORF43(CDS) | 65,052 | 67,357 | 2306 | + | |
| ORF45 | Normal CDS | 67,452 | 68,675 | 1224 | - | ORF46 UCDS |
| ORF46 | C-term trunc/ORF45(5′NC) | 68,963 | 69,503 | 541 | - | ORF47 UCDS |
| ORF47 | C-term trunc/ORF46(5′NC) | 69,611 | 70,014 | 404 | - | ORF48 UCDS |
| ORF48 | Normal CDS | 70,272 | 71,480 | 1209 | - | |
| ORF49 | Normal CDS | 71,729 | 72,637 | 909 | - | |
| ORF50 | N-term trunc/ORF49(5′NC,CDS), C-term trunc/K8(5′NC) | 72,856 | 74,688 | 1833 | + | |
| K8a | N-term domain | 74,949 | 75,422 | 474 | + | ORF50 UCDS |
| K8.1 | N-term domain | 76,014 | 76,254 | 241 | + | |
| ORF52 | Normal CDS | 76,901 | 77,296 | 396 | - | |
| ORF53 | Normal CDS | 77,432 | 77,764 | 333 | - | |
| ORF54 | Normal CDS | 77,835 | 78,722 | 888 | + | |
| ORF55 | N-term trunc/ORF56(5′NC) | 78,864 | 79,513 | 650 | - | |
| ORF56 | N-term trunc/ORF55(5′NC) | 79,831 | 82,066 | 2236 | + | |
| ORF57 | Normal CDS | 82,326 | 83,644 | 1319 | + | ORF56 UCDS |
| vIRF-1 | Normal CDS | 83,960 | 85,309 | 1350 | - | |
| vIRF-4 | C-terminal domain | 86,174 | 88,442 | 2269 | - | |
| vIRF-3 | C-terminal domain | 89,700 | 90,945 | 1246 | - | |
| vIRF_2 | C-terminal domain | 92,066 | 93,620 | 1555 | - | |
| ORF58 | Normal CDS | 94,577 | 95,650 | 1074 | - | ORF59 UCDS |
| ORF59 | N-term trunc/ORF60(3′NC), C-term trunc/ORF58(CDS) | 95,700 | 96,658 | 1004 | - | |
| ORF60 | Normal CDS | 96,976 | 97,893 | 918 | - | ORF61 UCDS |
| ORF61 | N-term trunc/ORF62(3′NC) | 97,922 | 98,399 | 478 | - | |
| ORF62 | N-term trunc/ORF63(5′NC), C-term trunc/ORF61(5′NC) | 100,458 | 101,171 | 714 | - | |
| ORF63 | N-term trunc/ORF62(5′NC) | 101,495 | 104,100 | 2606 | + | |
| ORF64 | N-term trunc/ORF63(CDS), C-term trunc/ORF65(5′NC) | 104,150 | 111,927 | 7822 | + | ORF63 UCDS |
| ORF65 | Normal CDS | 112,037 | 112,549 | 513 | - | ORF66 UCDS |
| ORF66 | C-term trunc/ORF65(5′NC), N-term trunc/ORF67(CDS) | 112,651 | 113,799 | 1149 | - | ORF67 UCDS |
| ORF67 | C-term trunc/ORF66(5′NC,CDS) | 113,957 | 114,614 | 658 | - | ORF67A UCDS |
| ORF67A | C-term trunc/ORF67(5′NC) | 114,808 | 114,911 | 104 | - | |
| ORF68 | C-term trunc/ORF69(5′NC) | 115,108 | 116,295 | 1188 | + | |
| ORF69 | Normal CDS | 116,544 | 117,452 | 909 | + | ORF68 UCDS |
| K12A | Normal CDS | 118,025 | 118,207 | 183 | - | |
| miR | Normal domain | 119,048 | 122,159 | 3132 | - | |
| DR5 | 5′ trunc/K12A(CDS) | 118,257 | 118,490 | 262 | + | |
| DR6 | 5′ trunc/DR5repeat | 118,540 | 118,845 | 355 | + | |
| ORF71 | Normal CDS | 122,393 | 122,959 | 567 | - | ORF72 UCDS 3 |
| ORF72 | C-term trunc/ORF71(5′NC) | 123,042 | 123,815 | 774 | - | Variable 3 |
| K12Aa | Small exon upstream of ORF72 | 123,843 | 124,054 | 212 | - | |
| ORF73a | C-term domain | 124,104 | 124,783 | 680 | - | |
| ORF73b | N-term domain | 126,456 | 127,446 | 991 | - | |
| K14 | Normal CDS | 128,264 | 129,079 | 816 | + | |
| ORF74 | Normal CDS | 129,520 | 130,548 | 1029 | + | K14 UCDS |
| ORF75 | Normal CDS | 130,699 | 134,589 | 3891 | - | K15a UCDS |
| K15a | Exon 8 | 134,824 | 135,287 | 464 | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruce, A.G.; Barcy, S.; DiMaio, T.; Gan, E.; Garrigues, H.J.; Lagunoff, M.; Rose, T.M. Quantitative Analysis of the KSHV Transcriptome Following Primary Infection of Blood and Lymphatic Endothelial Cells. Pathogens 2017, 6, 11. https://doi.org/10.3390/pathogens6010011
Bruce AG, Barcy S, DiMaio T, Gan E, Garrigues HJ, Lagunoff M, Rose TM. Quantitative Analysis of the KSHV Transcriptome Following Primary Infection of Blood and Lymphatic Endothelial Cells. Pathogens. 2017; 6(1):11. https://doi.org/10.3390/pathogens6010011
Chicago/Turabian StyleBruce, A. Gregory, Serge Barcy, Terri DiMaio, Emilia Gan, H. Jacques Garrigues, Michael Lagunoff, and Timothy M. Rose. 2017. "Quantitative Analysis of the KSHV Transcriptome Following Primary Infection of Blood and Lymphatic Endothelial Cells" Pathogens 6, no. 1: 11. https://doi.org/10.3390/pathogens6010011





