Gut Transcription in Helicoverpa zea is Dynamically Altered in Response to Baculovirus Infection
Abstract
:1. Introduction
2. Experimental Section
2.1. Insect Rearing, Infection, and Gut Isolation
2.2. Total RNA Isolation
2.3. Amplification and Labeling of mRNA
2.4. Hybridization and Scanning of the Microarray Chip
2.5. Microarray Experimental Design and Analysis
2.6. Primer Design and qRT-PCR
| Gene Identifier 1 | Gene Name | Forward Primer | Reverse Primer |
|---|---|---|---|
| AAL34109.1 | Alpha-amylase | 5'-accgttggcgtcaaatctac-3' | 5'-ggagctgctgtagtggtcgt-3' |
| AAT08964.1 | Cytochrome P450 (CYP) | 5'-gttggatcccattcagtgct-3' | 5'-ctggctcttgagatgctggt-3' |
| EEB16964.1 | Glucose Dehydrogenase (GDH) | 5'-tgactgtccaagactggtg-3' | 5'-atcgctccgctctttctccta-3' |
| AAZ52554.1 | Prophenoloxidase Subunit 2 (PROPO) | 5'-gcctaccagttgttcgttat-3' | 5'-agggtactttctgtccttcag-3' |
| AAO16241.1 | Effector Caspase (ECAS) | 5'-caagccagagaacctgtggt-3' | 5'-gaagcagaaggtgagccatc-3' |
| ACL51928.1 | Lysozyme (LYZ) | 5'-atgagttgaggaggcaagga-3' | 5'-gcccactttgtctgtcttcc-3' |
| ACD74811.1 | Eukaryotic Initiation Factor 5c | 5'-agaggttcgtcgagtggcta-3' | 5'-agcctcactggctgctctta-3' |
3. Results and Discussion
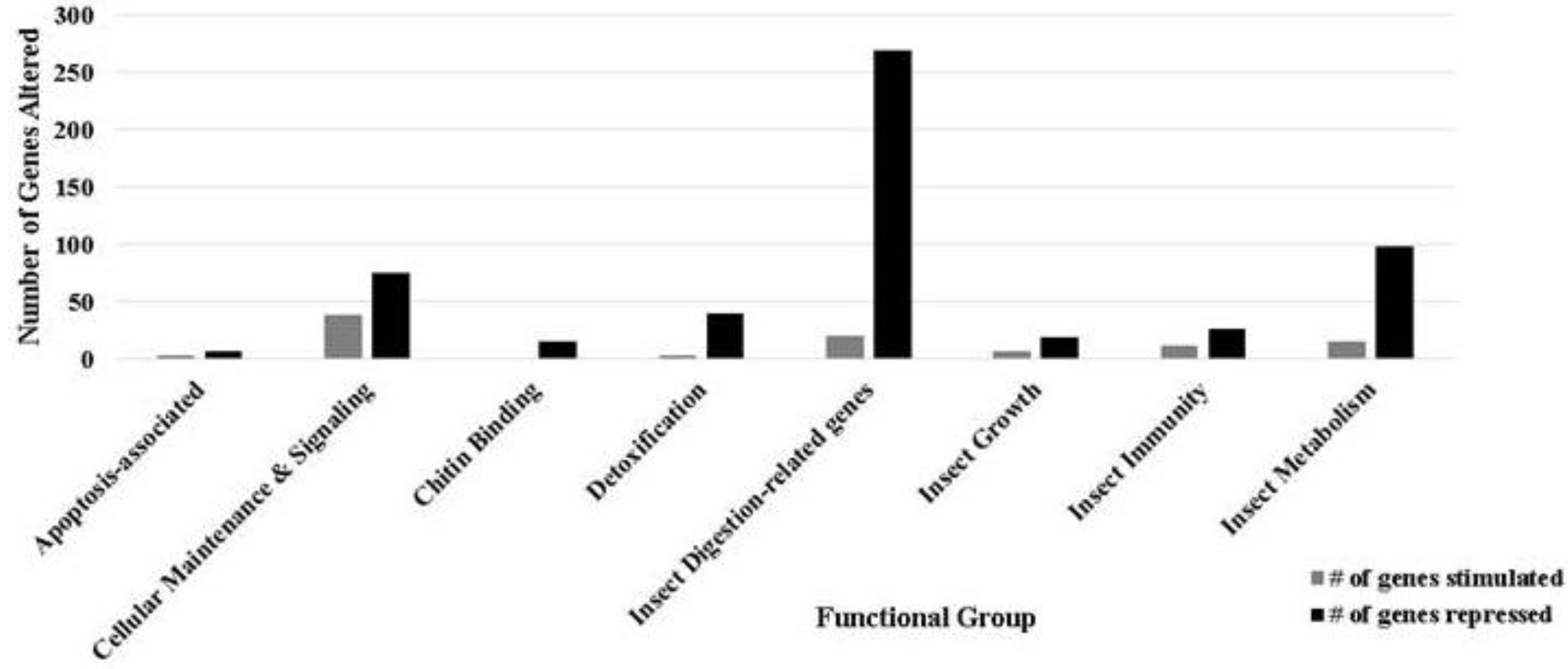
3.1. Ontology of Differentially-Regulated Genes in Response to Baculovirus Infection

| Gene classification/name | Fold change | Gene Identifier |
|---|---|---|
| Apoptosis | ||
| Caspase short class | −1.97 | XP_320581.3 |
| CG10641-PA | 2.41 | XP_624640.1 |
| Daxx-like protein | 2.67 | XP_973237.1 |
| Death related ced-3/Nedd2-like protein; Initiator caspase | −1.92 | NP_001108337.1 |
| Death related ced-3/Nedd2-like protein; Initiator caspase | −1.65 | NP_001108337.1 |
| Death related ced-3/Nedd2-like protein; Initiator caspase | −1.35 | NP_001108337.1 |
| Death related ced-3/Nedd2-like protein; Initiator caspase | −1.24 | NP_001108337.1 |
| Effector caspase; SI-caspase-1 | −2.57 | AAO16241.1 |
| Survivin | −1.86 | XP_001662572.1 |
| Detoxification | ||
| Cytochrome P450 | −8.26 | XP_0010604810.1 |
| Cytochrome P450 | −6.82 | XP_0010604810.1 |
| Cytochrome P450 | −3.59 | XP_0010604810.1 |
| Cytochrome P450 | −16.09 | NP_001104007.1 |
| Cytochrome P450 | −11.02 | NP_001104007.1 |
| Cytochrome P450 CYP4S1 | −6.88 | ABU88427.1 |
| Cytochrome P450 monooxygenase | −5.41 | NP_001104007.1 |
| GA10313-PA | 3.22 | XP_001601512.1 |
| Glutathione S-transferase | 3.39 | AAL23839.1 |
| Glutathione S-transferase | −9.38 | ABU88426.1 |
| Glutathione S-transferase GSTX01 | −11.14 | ABK29516.1 |
| Glutathione S-transferase 1 | −8.69 | P46430.1 |
| Insect Immunity | ||
| Adhesion-like transmembrane protein | 2.61 | CAB65413.1 |
| H. armigera cecropin-D (HacD) | 2.49 | AAX51193.1 |
| Hemolymph proteinase 18 | 4.41 | AAZ52554.1 |
| M-like protein | 5.07 | YP_313517.1 |
| Prophenoloxidase subunit 2 | 5.46 | ABF18489.1 |
| Toll-interacting protein | 1.66 | XP_975168.1 |
3.2. Apoptosis
3.3. Cellular Maintenance and Signaling
3.4. Chitin Binding
3.5. Detoxification
3.6. Digestive Function
3.7. Immunity
3.9. Quantitative Real Time-Polymerase Chain Reaction

4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Conflicts of Interest
References
- Kozlov, E.A.; Levitina, T.L.; Gusak, N.M. The primary structure of baculovirus inclusion body proteins. Evolution and structure-function aspects. Curr. Top. Microbiol. Immunol. 1986, 131, 135–164. [Google Scholar] [CrossRef]
- Rohrmann, G.F.; Pearson, M.N.; Bailey, T.J.; Becker, R.R.; Beaudreau, G.S. N-Terminal polyhedrin sequences and occluded Baculovirus evolution. J. Mol. Evol. 1981, 17, 329–333. [Google Scholar] [CrossRef]
- Wang, Y.; Jehle, J.A. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: New insights on an old topic. J. Invertebr. Pathol. 2009, 101, 187–193. [Google Scholar] [CrossRef]
- Shim, H.J.; Choi, J.Y.; Wang, Y.; Tao, X.Y.; Liu, Q.; Roh, J.Y.; Kim, J.S.; Kim, W.J.; Woo, S.D.; Jin, B.R.; Je, Y.H. NeuroBactrus, a novel, highly effective, and environmentally friendly recombinant baculovirus insecticide. Appl. Environ. Microbiol. 2013, 79, 141–149. [Google Scholar] [CrossRef]
- Gramkow, A.W.; Perecmanis, S.; Sousa, R.L.; Noronha, E.F.; Felix, C.R.; Nagata, T.; Ribeiro, B.M. Insecticidal activity of two proteases against Spodoptera frugiperda larvae infected with recombinant baculoviruses. Virology J. 2010, 7. [Google Scholar] [CrossRef]
- Jakubowska, A.K.; Vogel, H.; Herrero, S. Increase in gut microbiota after immune suppression in baculovirus-infected larvae. PLoS Pathog. 2013, 9, e1003379. [Google Scholar] [CrossRef]
- Jayachandran, B.; Hussain, M.; Asgari, S. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 2012, 86, 13729–13734. [Google Scholar] [CrossRef]
- Mitchell, J.K.; Friesen, P.D. Baculoviruses modulate a proapoptotic DNA damage response to promote virus multiplication. J. Virol. 2012, 86, 13542–13553. [Google Scholar] [CrossRef]
- Hammock, B.D.; McCutchen, B.F.; Beetham, J.; Choudary, P.V.; Fowler, E.; Ichinose, R.; Ward, V.K.; Vickers, J.M.; Bonning, B.C.; Harshman, L.G.; et al. Development of recombinant viral insecticides by expression of an insect-specific toxin and insect-specific enzyme in nuclear polyhedrosis viruses. Arch. Insect Biochem. Physiol. 1993, 22, 315–344. [Google Scholar] [CrossRef]
- Menn, J.J. Current trends and new directions in crop protection. Am. J. Ind. Med. 1990, 18, 499–504. [Google Scholar] [CrossRef]
- Jehle, J.A.; Blissard, G.W.; Bonning, B.C.; Cory, J.S.; Herniou, E.A.; Rohrmann, G.F.; Theilmann, D.A.; Thiem, S.M.; Vlak, J.M. On the classification and nomenclature of baculoviruses: A proposal for revision. Arch. Virol. 2006, 151, 1257–1266. [Google Scholar] [CrossRef]
- Braunagel, S.C.; Summers, M.D. Molecular biology of the baculovirus occlusion-derived virus envelope. Curr. Drug Targets 2007, 8, 1084–1095. [Google Scholar] [CrossRef]
- Slack, J.; Arif, B.M. The baculoviruses occlusion-derived virus: Virion structure and function. Adv. Virus Res. 2007, 69, 99–165. [Google Scholar] [CrossRef]
- Volkman, L.E. Nucleopolyhedrovirus interactions with their insect hosts. Adv. Virus Res. 1997, 48, 313–348. [Google Scholar] [CrossRef]
- Cory, J.S.; Hails, R.S. The ecology and biosafety of baculoviruses. Curr. Opin. Biotechnol. 1997, 8, 323–327. [Google Scholar] [CrossRef]
- Blissard, G.W.; Rohrmann, G.F. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 1990, 35, 127–155. [Google Scholar] [CrossRef]
- Clem, R.J. The role of apoptosis in defense against baculovirus infection in insects. Curr. Top. Microbiol. Immunol. 2005, 289, 113–129. [Google Scholar] [CrossRef]
- Clarke, T.E.; Clem, R.J. Insect defenses against virus infection: The role of apoptosis. Int. Rev. Immunol. 2003, 22, 401–424. [Google Scholar] [CrossRef]
- Clem, R.J. Baculoviruses and apoptosis: The good, the bad, and the ugly. Cell Death Differ. 2001, 8, 137–143. [Google Scholar] [CrossRef]
- Breitenbach, J.E.; Shelby, K.S.; Popham, H.J.R. Baculovirus induced transcripts in hemocytes from the larvae of Heliothis virescens. Viruses 2011, 3, 2047–2064. [Google Scholar] [CrossRef]
- Huang, L.; Cheng, T.; Xu, P.; Fang, T.; Xia, Q. Bombyx mori transcription factors: Genome-wide identification, expression profiles and response to pathogens by microarray analysis. J. Insect Sci. 2012. [Google Scholar] [CrossRef]
- Yamagishi, J.; Burnett, E.D.; Harwood, S.H.; Blissard, G.W. The AcMNPV pp31 gene is not essential for productive AcMNPV replication or late gene transcription but appears to increase levels of most viral transcripts. Virology 2007, 365, 34–47. [Google Scholar] [CrossRef]
- Fujita, R.; Matsuyama, T.; Yamagishi, J.; Sahara, K.; Asano, S.; Bando, H. Expression of Autographa californica multiple nucleopolyhedrovirus genes in mammalian cells and upregulation of the host beta-actin gene. J. Virol. 2006, 80, 2390–2395. [Google Scholar] [CrossRef]
- Ote, M.; Mita, K.; Kawasaki, H.; Daimon, T.; Kobayashi, M.; Shimada, T. Identification of molting fluid carboxypeptidase A (MF-CPA) in Bombyx mori. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 141, 314–322. [Google Scholar] [CrossRef]
- Chen, Y.R.; Zhong, S.; Fei, Z.; Hashimoto, Y.; Xiang, J.Z.; Zhang, S.; Blissard, G.W. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J. Virol. 2013, 87, 6391–6405. [Google Scholar] [CrossRef]
- Choi, J.Y.; Roh, J.Y.; Wang, Y.; Zhen, Z.; Tao, X.Y.; Lee, J.H.; Liu, Q.; Kim, J.S.; Shin, S.W.; Je, Y.H. Analysis of genes expression of Spodoptera exigua larvae upon AcMNPV infection. PLoS One 2012, 7, e42462. [Google Scholar] [CrossRef]
- Nguyen, Q.; Palfreyman, R.W.; Chan, L.C.; Reid, S.; Nielsen, L.K. Transcriptome sequencing of and microarray development for a Helicoverpa zea cell line to investigate in vitro insect cell-baculovirus interactions. PLoS One 2012, 7, e36324. [Google Scholar]
- Celorio-Mancera, M.P.; Ahn, S.-J.; Vogel, H.; Heckel, D.G. Transcriptional responses underlying the hormetic and detrimental effects of the plant secondary metabolite gossypol on the generalist herbivore Helicovepa armigera. BMC Genomics 2011, 12. [Google Scholar] [CrossRef]
- Musser, R.O.; Hum-Musser, S.M.; Lee, H.K.; Des Rochers, B.L.; Williams, S.A.; Vogel, H. Caterpillar labial saliva alters tomato plant gene expression. J. Chem. Ecol. 2012, 38, 1381–1401. [Google Scholar]
- PerkinElmer, Inc. “geospiza”. Available online: http://www.geospiza.com/index.shtml/ (accessed on 1 January 2012).
- Klipper-Aurbach, Y.; Wasserman, M.; Braunspiegel-Weintrob, N.; Borstein, D.; Peleg, S.; Assa, S.; Karp, M.; Benjamini, Y.; Hochberg, Y.; Laron, Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses 1995, 45, 486–490. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; Vandesompele, J.; Wittwer, C.T. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Hughes, A.L.; Friedman, R. Genome-wide survey for genes horizontally transferred from cellular organisms to baculoviruses. Mol. Biol. Evol. 2003, 20, 979–987. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Hasson, A.; Spira, M.E.; Gray, W.R.; Li, W.; Marsh, M.; Hillyard, D.R.; Olivera, B.M. A new family of conotoxins that blocks voltage-gated sodium channels. J. Biol. Chem. 1995, 270, 16796–16802. [Google Scholar] [CrossRef]
- Nie, Z.M.; Zhang, Z.F.; Wang, D.; He, P.A.; Jiang, C.Y.; Song, L.; Chen, F.; Xu, J.; Yang, L.; Yu, L.L.; et al. Complete sequence and organization of Antheraea pernyi nucleopolyhedrovirus, a dr-rich baculovirus. BMC Genomics 2007. [Google Scholar] [CrossRef]
- Faulkner, P.; Kuzio, J.; Williams, G.V.; Wilson, J.A. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J. Gen. Virol. 1997, 78, 3091–3100. [Google Scholar]
- Zeng, R.S.; Wen, Z.; Niu, G.; Schuler, M.A.; Berenbaum, M.R. Enhanced toxicity and induction of cytochrome P450s suggest a cost of “eavesdropping” in a multitrophic interaction. J. Chem. Ecol. 2009, 35, 526–532. [Google Scholar] [CrossRef]
- Kostaropoulos, I.; Kalmanti, D.; Theodoropoulou, B.; Loumbourdis, N.S. Effects of exposure to a mixture of cadmium and chromium on detoxification enzyme (GST, P450-MO) activities in the frog Rana ridibunda. Ecotoxicology 2005, 14, 439–447. [Google Scholar] [CrossRef]
- Schmid-Hempel, P. Evolutionary ecology of insect immune defenses. Annu. Rev. Entomol. 2005, 50, 529–551. [Google Scholar] [CrossRef]
- Shelby, K.S.; Popham, H.J.R. Cloning and characterization of the secreted hemocytic prophenoloxidases of Heliothis virescens. Arch. Insect Biochem. Physiol. 2008, 69, 127–142. [Google Scholar]
- Chamberlin, M.E. Changes in mitochondrial electron transport chain activity during insect metamorphosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R1016–R1022. [Google Scholar] [CrossRef]
- Micchelli, C.A.; Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 2006, 439, 475–479. [Google Scholar] [CrossRef]
- Braun, L.; Keddie, B.A. A new tissue technique for evaluating effects of Bacillus thuringiensis toxins on insect midgut epithelium. J. Invert. Pathol. 1997, 69, 92–104. [Google Scholar] [CrossRef]
- Rahman, M.M.; Gopinathan, K.P. Systemic and in vitro infection process of Bombyx mori nucleopolyhedrovirus. Virus Res. 2004, 101, 109–118. [Google Scholar] [CrossRef]
- Haas-Stapleton, E.J.; Washburn, J.O.; Volkman, L.E. Pathogenesis of Autographa californica M nucleopolyhedrovirus in fifth instar Spodoptera frugiperda. J. Gen. Virol. 2003, 84, 2033–2040. [Google Scholar] [CrossRef]
- Granados, R.R.; Lawler, K.A. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 1981, 108, 297–308. [Google Scholar] [CrossRef]
- Clem, R.J. Baculoviruses and apoptosis: A diversity of genes and responses. Curr. Drug Targets 2007, 8, 1069–1074. [Google Scholar] [CrossRef]
- Bushell, M.; McKendrick, L.; Janicke, R.U.; Clemens, M.J.; Morley, S.J. Caspase-3 is necessary and sufficient for cleavage of protein synthesis eukaryotic initiation factor 4G during apoptosis. FEBS Lett. 1999, 451, 332–336. [Google Scholar] [CrossRef]
- Clem, R.J.; Miller, L.K. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J. Virol. 1993, 67, 3730–3738. [Google Scholar]
- Miller, L.K. Baculovirus interaction with host apoptotic pathways. J. Cell. Physiol. 1997, 173, 178–182. [Google Scholar] [CrossRef]
- Chiou, S.K.; Jones, M.K.; Tarnawski, A.S. Survivin—An anti-apoptosis protein: Its biological roles and implications for cancer and beyond. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2003, 9, PI25–PI29. [Google Scholar]
- O'Driscoll, L.; Linehan, R.; Clynes, M. Survivin: Role in normal cells and in pathological conditions. Curr. Cancer Drug Targets 2003, 3, 131–152. [Google Scholar] [CrossRef]
- Dionne, K.R.; Zhuang, Y.; Leser, J.S.; Tyler, K.L.; Clarke, P. Daxx upregulation within the cytoplasm of reovirus-infected cells is mediated by interferon and contributes to apoptosis. J. Virol. 2013, 87, 3447–3460. [Google Scholar] [CrossRef]
- Yang, X.; Khosravi-Far, R.; Chang, H.Y.; Baltimore, D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 1997, 89, 1067–1076. [Google Scholar] [CrossRef]
- Zobalova, R.; Swettenham, E.; Chladova, J.; Dong, L.F.; Neuzil, J. Daxx inhibits stress-induced apoptosis in cardiac myocytes. Redox Rep. Commun. Free Radic. Res. 2008, 13, 263–270. [Google Scholar] [CrossRef]
- Miller, J.E.; Levine, R.B. Steroid hormone activation of wandering in the isolated nervous system of Manduca sexta. J. Comp. Physiol. A Neuro. Sens. Neur. Behav. Physiol. 2006, 192, 1049–1062. [Google Scholar] [CrossRef]
- O'Reilly, D.R.; Miller, L.K. A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyl transferase. Science 1989, 245, 1110–1112. [Google Scholar]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene 2006, 367, 17–37. [Google Scholar] [CrossRef]
- Cordin, O.; Beggs, J.D. RNA helicases in splicing. RNA Biol. 2013, 10, 83–95. [Google Scholar] [CrossRef]
- Berretta, M.F.; Deshpande, M.; Crouch, E.A.; Passarelli, A.L. Functional characterization of Bombyx mori nucleopolyhedrovirus late gene transcription and genome replication factors in the non-permissive insect cell line SF-21. Virology 2006, 348, 175–189. [Google Scholar] [CrossRef]
- Du, X.; Thiem, S.M. Responses of insect cells to baculovirus infection: Protein synthesis shutdown and apoptosis. J. Virol. 1997, 71, 7866–7872. [Google Scholar]
- Ludwig, A.; Valente, V.L.; Loreto, E.L. Multiple invasions of Errantivirus in the genus Drosophila. Insect Mol. Biol. 2008, 17, 113–124. [Google Scholar] [CrossRef]
- Terzian, C.; Pelisson, A.; Bucheton, A. Evolution and phylogeny of insect endogenous retroviruses. BMC Evolut. Biol. 2001. [Google Scholar] [CrossRef]
- Menzel, T.; Rohrmann, G.F. Diversity of errantivirus (retrovirus) sequences in two cell lines used for baculovirus expression, Spodoptera frugiperda and Trichoplusia ni. Virus Genes 2008, 36, 583–586. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Noland, J.E.; Breitenbach, J.E.; Popham, H.J.R.; Hum-Musser, S.M.; Vogel, H.; Musser, R.O. Gut Transcription in Helicoverpa zea is Dynamically Altered in Response to Baculovirus Infection. Insects 2013, 4, 506-520. https://doi.org/10.3390/insects4030506
Noland JE, Breitenbach JE, Popham HJR, Hum-Musser SM, Vogel H, Musser RO. Gut Transcription in Helicoverpa zea is Dynamically Altered in Response to Baculovirus Infection. Insects. 2013; 4(3):506-520. https://doi.org/10.3390/insects4030506
Chicago/Turabian StyleNoland, Jeffrey E., Jonathan E. Breitenbach, Holly J. R. Popham, Sue M. Hum-Musser, Heiko Vogel, and Richard O. Musser. 2013. "Gut Transcription in Helicoverpa zea is Dynamically Altered in Response to Baculovirus Infection" Insects 4, no. 3: 506-520. https://doi.org/10.3390/insects4030506




