Effects of Flight on Gene Expression and Aging in the Honey Bee Brain and Flight Muscle
Abstract
:1. Introduction
2. Experimental Section
2.1. Field Methods
2.2. Behavioral Groups
| Behavioral Group | Abbreviation | Age (days) | Days of flight |
|---|---|---|---|
| Young nurse | YN | 8 to 10 | < 1 |
| Precocious forager | PF | 8 to 10 | 2 to 3 |
| Older nurse | ON | 19 to 22 | < 1 |
| Forager-low flight | TL | 19 to 22 | 2 to 3 |
| Forager-high flight | TH | 19 to 22 | 7 to 9 |
| Forager-old | OF | 25 to 26 | 10 to 12 |
| Reverted nurse | RN | 25 to 26 | 7 to 9 |
2.3. RNA Extraction
2.4. Microarray Hybridization

2.5. Microarray Data Analysis
2.6. Quantitative Real-Time Polymerase Chain Reaction
3. Results and Discussion
3.1. Behavior Induces Dramatic Changes in Global Gene Expression of A. mellifera Brains and Thoraces

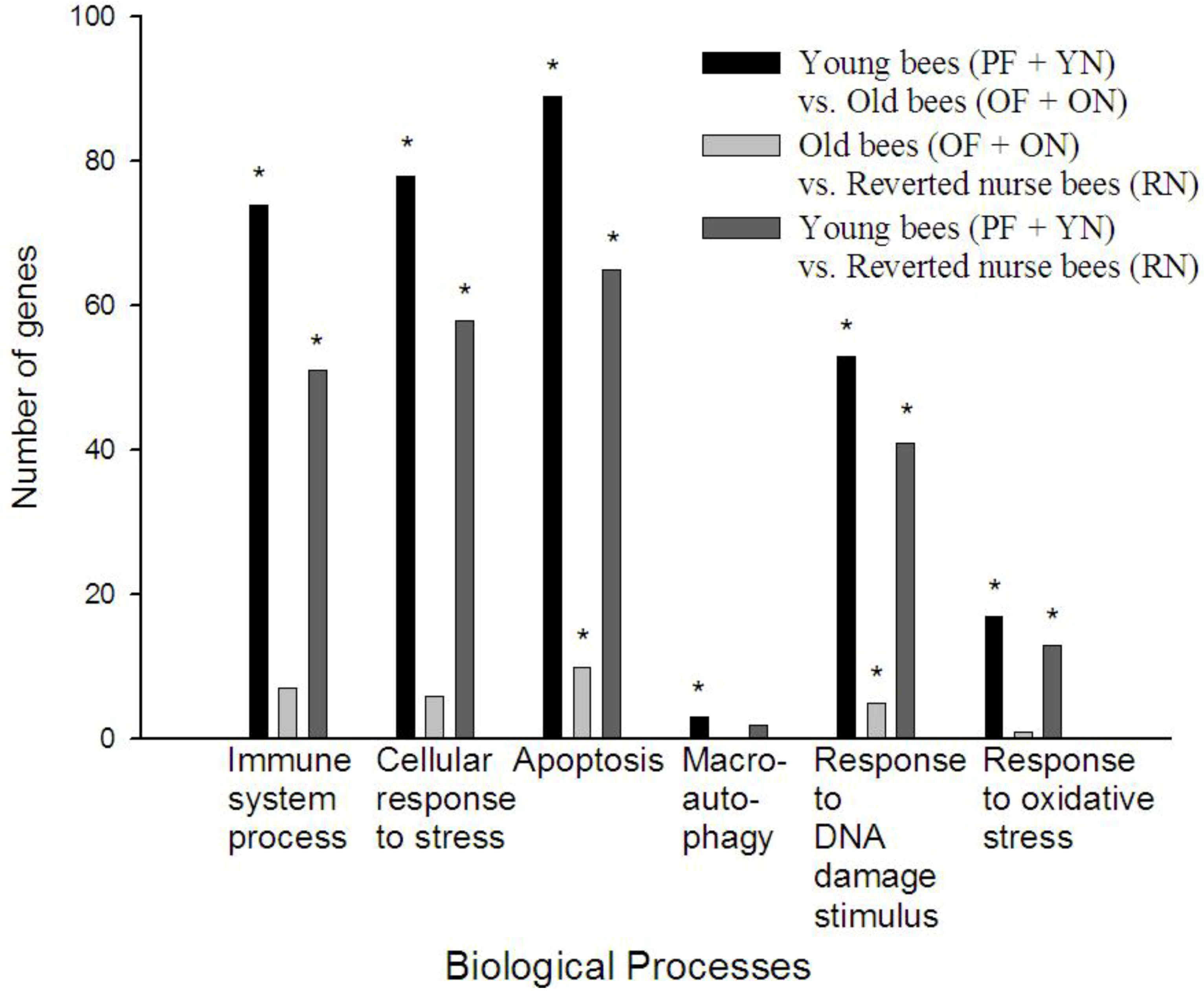
3.3. Gene Ontology Analysis Reveals Age Causes a Decrease in Certain Stress and Immune Processes in A. mellifera

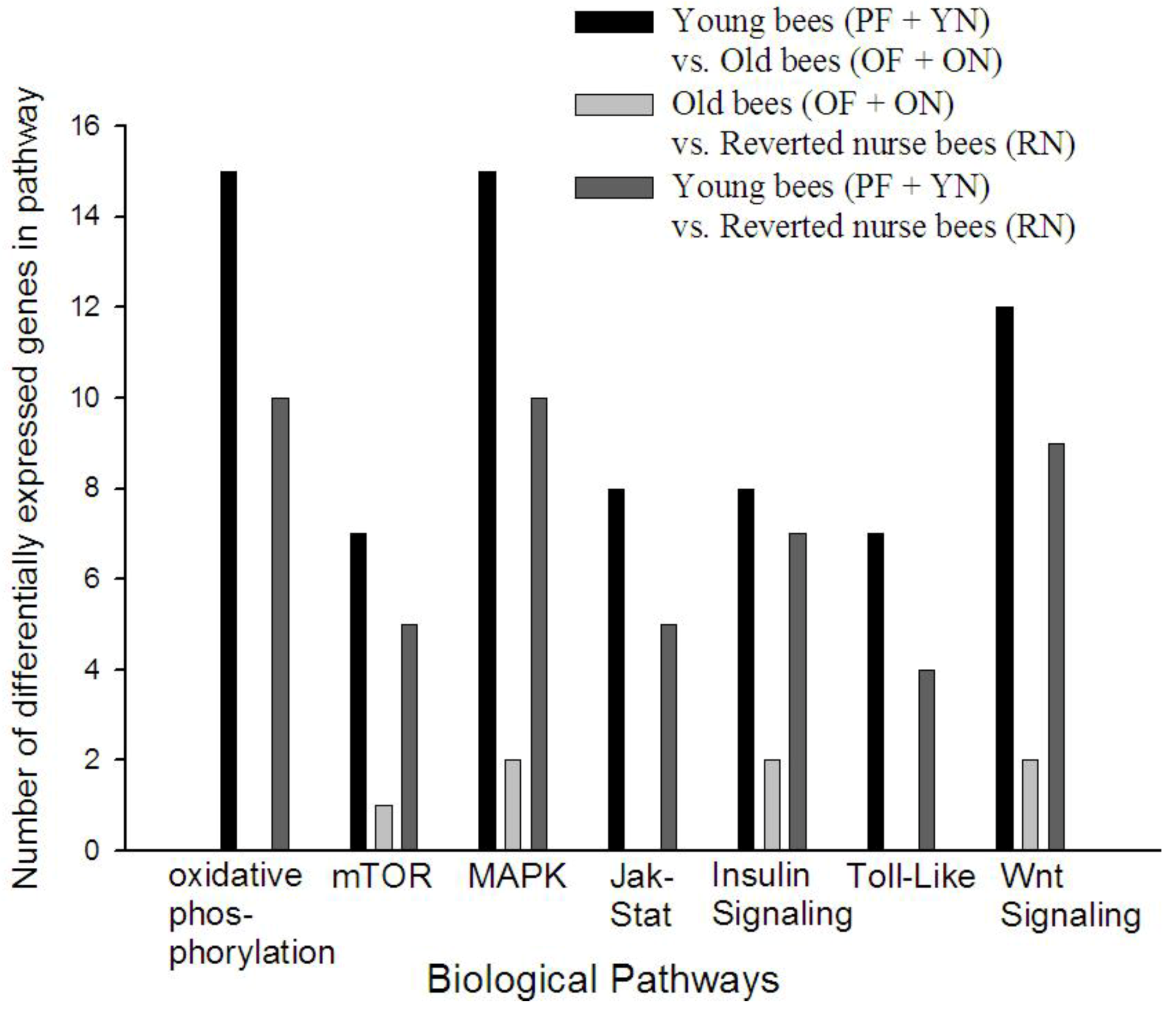
3.4. KEGG Pathway Analysis Reveals Age Alters Specific Signaling Pathways in A. mellifera
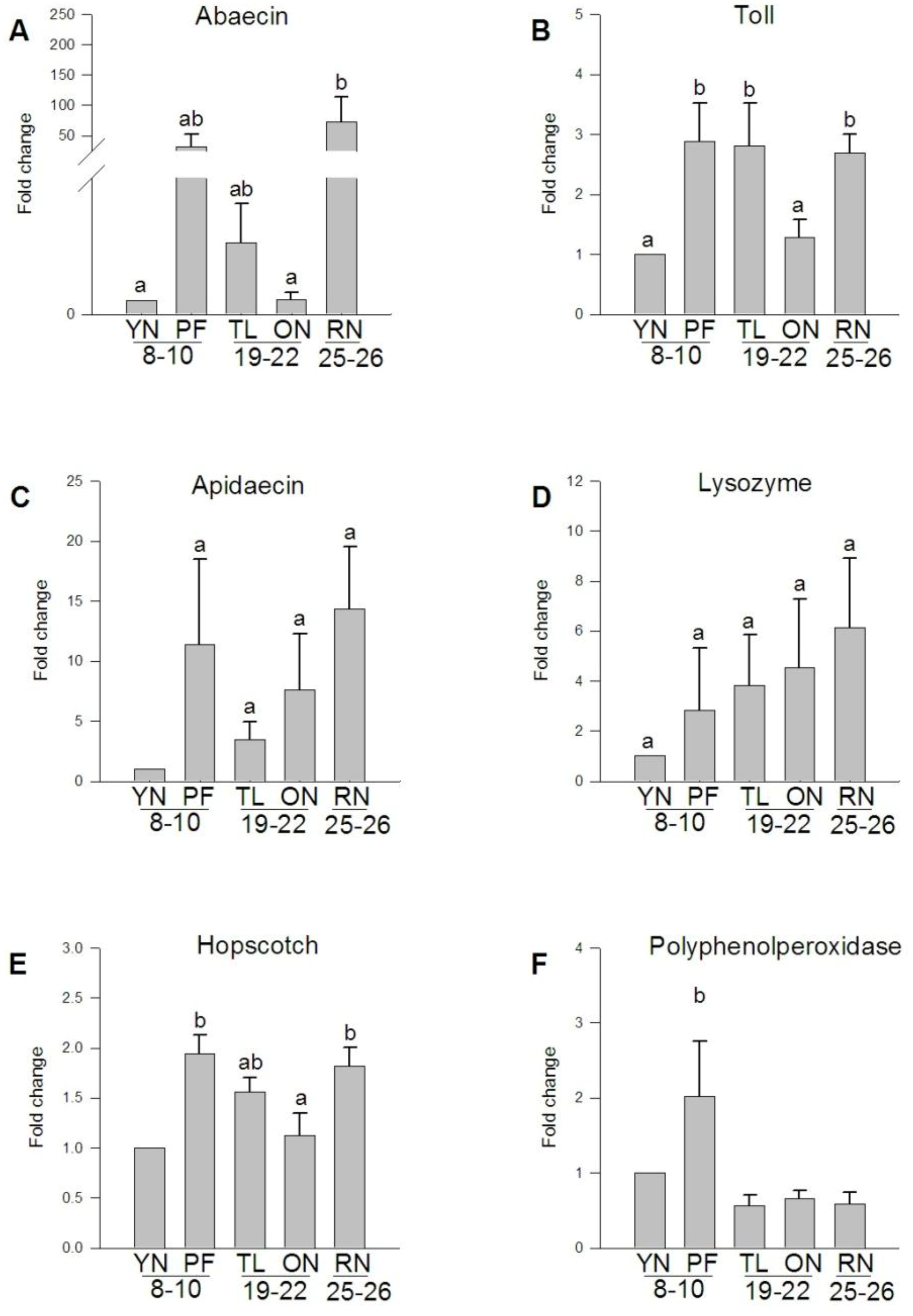
3.5. Expression of Transcripts Involved in the A. mellifera Immune Response
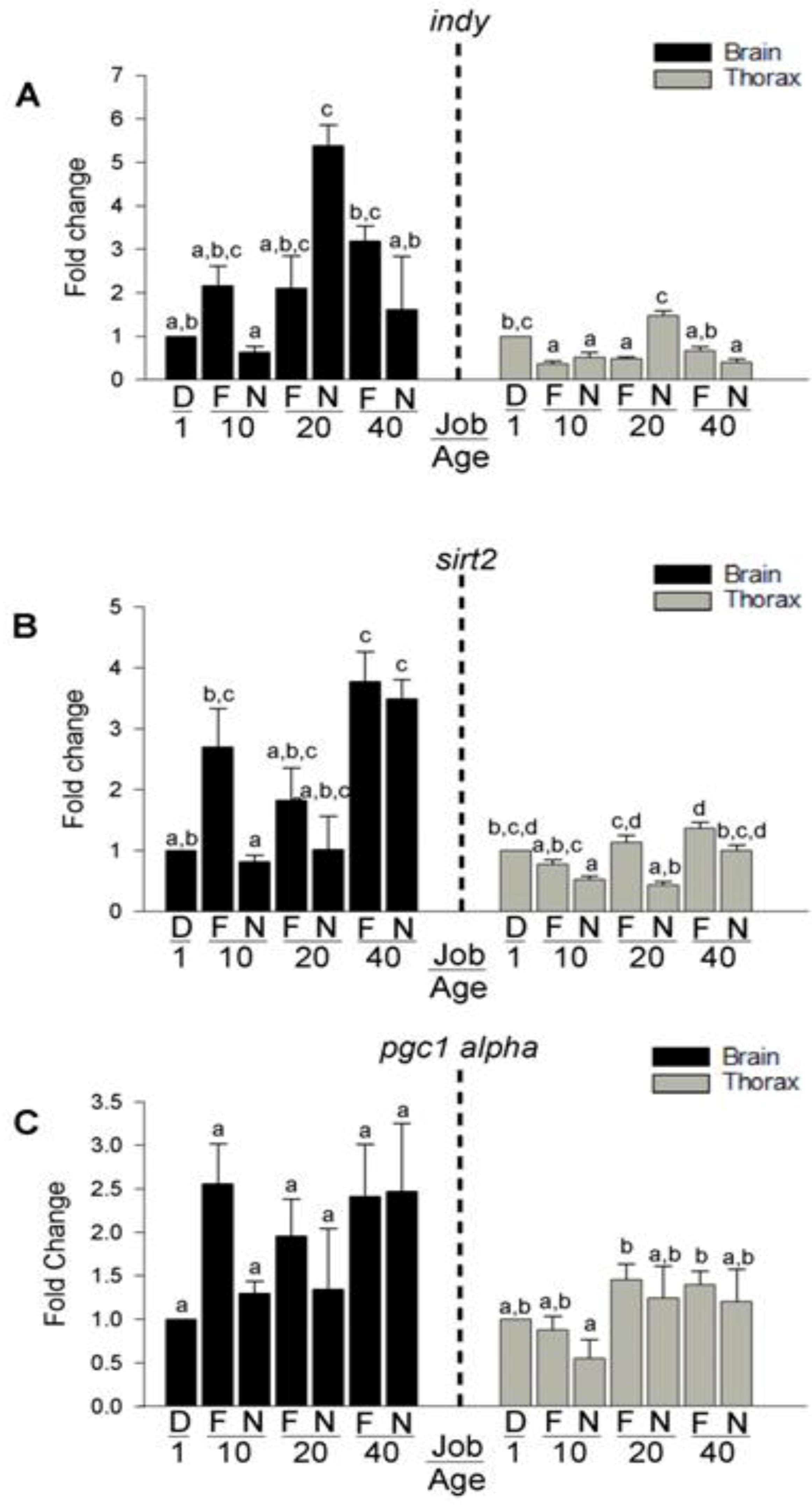
3.6. Expression of Longevity Mediating Transcripts
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
References and Notes
- Hulbert, A.J.; Pamplona, R.; Buffenstein, R.; Buttemer, W.A. Life and death: Metabolic rate, membrane composition, and life span of animals. Physiol. Rev. 2007, 87, 1175–1213. [Google Scholar] [CrossRef]
- Constantini, D. Oxidative stress in ecology and evolution: Lessons from avian studies. Ecol. Lett. 2008, 11, 1238–1251. [Google Scholar]
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 1956, 11, 298–300. [Google Scholar] [CrossRef]
- Lapointe, J.; Hekimi, S. When a theory of aging ages badly. Cell. Mol. Life Sci. 2010, 67, 1–8. [Google Scholar] [CrossRef]
- Salmon, A.B.; Richardson, A.; Pérez, V.L. Update on the oxidative stress theory of aging: Does oxidative stress play a role in aging or healthy aging? Free Radic. Biol. Med. 2010, 48, 642–655. [Google Scholar] [CrossRef]
- Sanz, A.; Stefanatos, R.K.A. The mitochondrial free radical theory of aging: A critical view. Curr. Aging Sci. 2008, 1, 10–21. [Google Scholar]
- Parker, J.D. What are social insects telling us about aging? Myrmecol. News 2010, 13, 103–110. [Google Scholar]
- Chen, J.H.; Hales, C.N.; Ozanne, S.E. DNA damage, cellular senescence and organismal ageing: Causal or correlative? Nucleic Acids Res. 2007, 35, 7417–7428. [Google Scholar] [CrossRef]
- Monaghan, P.; Charmantier, A.; Nussey, D.H.; Ricklefs, R.E. The evolutionary ecology of senescence. Funct. Ecol. 2008, 22, 371–378. [Google Scholar] [CrossRef]
- Metcalfe, N.; Alonso-Álvarez, C. Oxidative stress as a life-history constraint: The role of reactive oxygen species (ROS) in shaping phenotypes from conception to death. Funct. Ecol. 2010, 24, 984–996. [Google Scholar] [CrossRef]
- Von Schantz, T.; Bensch, S.; Grahn, M.; Hasselquist, D.; Wittzell, H. Good genes, oxidative stress and condition-dependent sexual signals. Proc. Roy. Soc. Lond. B Biol. Sci. 1999, 266, 1–12. [Google Scholar]
- Alonso-Álvarez, C.; Bertrand, S.; Faivre, B.; Chastel, O.; Sorci, G. Testosterone and oxidative stress: the oxidation handicap hypothesis. Proc. Roy. Soc. Lond. B Biol. Sci. 2007, 274, 819–825. [Google Scholar] [CrossRef]
- Robinson, G.E. Regulation of honey bee age polyethism by juvenile hormone. Behav. Ecol. Sociobiol. 1987, 20, 329–338. [Google Scholar] [CrossRef]
- Pankiw, T.; Huang, Z.Y.; Winston, M.L.; Robinson, G.E. Queen mandibular gland pheromone influences worker honey bee (Apis mellifera L.) foraging ontogeny and juvenile hormone titers. J. Insect Physiol. 1998, 44, 685–692. [Google Scholar] [CrossRef]
- Elekonich, M.M.; Roberts, S.P. Physiological underpinnings of behavioral development in honey bees. Comp. Biochem. Phys. A 2005, 141, 362–371. [Google Scholar] [CrossRef]
- Winston, M.L. The Biology of the Honey Bee; Harvard University Press: Cambridge, MA, USA, 1987. [Google Scholar]
- Suarez, R.K.; Lighton, J.R.B.; Joos, B.; Roberts, S.P.; Harrison, J.F. Energy metabolism, enzymatic flux capacities and metabolic flux rates in flying honeybees. Proc. Natl. Acad. Sci. USA 1996, 93, 12616–12620. [Google Scholar]
- Williams, J.B.; Roberts, S.P.; Elekonich, M.M. Age and natural metabolically-intensive behavior affect oxidative stress and antioxidant mechanisms. Exp. Gerontol. 2008, 43, 538–549. [Google Scholar] [CrossRef]
- Vance, J.T.; Williams, J.B.; Elekonich, M.M.; Roberts, S.P. The effects of age and behavioral development on honey bee (Apis mellifera) flight performance. J. Exp. Biol. 2009, 212, 2604–2611. [Google Scholar] [CrossRef]
- Visscher, P.K.; Dukas, R. Survivorship of foraging honey bees. Insectes Soc. 1997, 44, 1–5. [Google Scholar] [CrossRef]
- Rueppell, O.; Christine, S.; Mulcrone, C.; Groves, L. Aging without functional senescence in honey bee workers. Curr. Biol. 2007, 17, R274–R275. [Google Scholar] [CrossRef]
- Robinson, G.E.; Page, R.E., Jr.; Strambi, C.; Strambi, A. Colony integration in honey bees: Mechanisms of behavioral reversion. Ethology 1992, 90, 336–348. [Google Scholar]
- Huang, Z.Y.; Robinson, G.E. Regulation of honey bee division of labor by colony age demography. Behav. Ecol. Sociobiol. 1996, 39, 147–158. [Google Scholar] [CrossRef]
- Amdam, G.V.; Aase, A.L.; Seehuus, S.C.; Kim Fondrk, M.; Norberg, K.; Hartfelder, K. Social reversal of immunosenescence in honey bee workers. Exp. Gerontol. 2005, 40, 939–947. [Google Scholar] [CrossRef]
- Miojevic, B.D. A new interpretation of the social life of the honeybee. Bee World 1940, 21, 39–41. [Google Scholar]
- Amdam, G.V.; Simôes, Z.L.P.; Hagen, A.; Norberg, K.; Schrøder, K.; Mikkelsen, O.; Kirkwood, T.B.L.; Omholt, S.W. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp. Gerontol. 2004, 39, 767–773. [Google Scholar] [CrossRef]
- Baker, N.; Wolschin, F.; Amdam, G.V. Age-related learning deficits can be reversible in honeybees Apis mellifera. Exp. Gerontol. 2012, 47, 764–772. [Google Scholar] [CrossRef]
- Wolschin, F.; Amdam, G.V. Comparative proteomics reveal characteristics of life-history transitions in a social insect. Proteome Sci. 2007, 5, 10. [Google Scholar] [CrossRef] [Green Version]
- Bowling, A.C.; Beal, M.F. Bioenergetic and oxidative stress in neurodegenerative diseases. Life Sci. 1995, 56, 1151–1171. [Google Scholar] [CrossRef]
- Giray, T.; Robinson, G.E. Effects of intracolony variability in behavioral development on plasticity of division of labor in honey bee colonies. Behav. Ecol. Sociobiol. 1994, 35, 13–20. [Google Scholar] [CrossRef]
- Page, R.E.; Robinson, G.E.; Britton, D.S.; Fondrk, M.K. Genotypic variability for rates of behavioral development in worker honey bees (Apis mellifera). Behav. Ecol. 1992, 3, 173–180. [Google Scholar] [CrossRef]
- Imai, S.; Johnson, F.B.; Marciniak, R.A.; McVey, M.; Park, P.U.; Guarente, L. Sir2: An NAD dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 297–302. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Rogina, B.; Reenan, R.A.; Nielsen, S.P.; Helfand, S.L. Extended life-span conferred by cotransporter gene mutations inDrosophila. Science 2000, 290, 2137–2140. [Google Scholar] [CrossRef]
- Schulz, D.J.; Robinson, G.E. Biogenic amines and division of labor in honey bee colonies: Behaviorally related changes in the antennal lobes and age-related changes in the mushroom bodies. J. Comp. Physiol. A 1999, 184, 481–488. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Sharabash, N.M.; Whitfield, C.W.; Robinson, G.E. Pheromone-mediated gene expression in the honeybee brain. Proc. Natl. Acad. Sci. USA 2003, 100, 14519–14525. [Google Scholar]
- Kocher, S.D.; Richard, F.J.; Tarpy, D.R.; Grozinger, C.M. Genomic analysis of post-mating changes in the honey bee queen (Apis mellifera). BMC Genomics 2008, 9, 232. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing: Vienna, Austria, 2011.
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions using R and Bioconductor; Gentleman, V., Carey, S., Eds.; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Smyth, G.K.; Speed, T.P. Normalization of cDNA microarray data. Methods 2003, 31, 265–273. [Google Scholar] [CrossRef]
- Cui, X.; Churchill, G.A. Statistical tests for differential expression in cDNA Microarray experiments. Genome Biol. 2003, 4, 210. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Hwang, J.T.G.; Qiu, J.; Blades, N.; Churchill, G.A. Improved statistical tests for differential gene expression by shrinking variance components. Biostatistics 2005, 6, 59–75. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genome-wide experiments. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Tong, W.; Cao, X.; Harris, S.; Sun, H.; Fang, H.; Fuscoe, J.; Harris, A.; Hong, H.; Xie, Q.; Perkins, R. ArrayTrack-supporting toxicogenomic research at the U.S. Food and Drug Administration National Center for Toxicological Research. Enviro. Health Perspect. 2003, 111, 1819–1826. [Google Scholar] [CrossRef]
- Whitfield, C.W.; Cziko, A.M.; Robinson, G.E. Gene expression profiles in the brain predict behavior in individual honey bees. Science 2003, 302, 296–299. [Google Scholar] [CrossRef]
- Girardot, F.; Lasbleiz, C.; Monnier, V.; Tricoire, H. Specific age related signatures in Drosophila body parts transcriptome. BMC Genomics 2006, 7, 69. [Google Scholar] [CrossRef]
- Neukirch, A. Dependence of the life span of the honeybee (Apis mellifica) upon flight performance and energy consumption. J. Comp. Physiol. B 1982, 146, 35–40. [Google Scholar] [CrossRef]
- Behrends, A.; Scheiner, R.; Baker, N.; Amdam, G.V. Cognitive aging is linked to social role in honey bees (Apis mellifera). Exp. Gerontol. 2007, 42, 1146–1153. [Google Scholar] [CrossRef]
- Seehuus, S.C.; Krekling, T.; Amdam, G.V. Cellular senescence in honey bee brain is largely independent of chronological age. Exp. Gerontol. 2006, 41, 1117–1125. [Google Scholar] [CrossRef]
- Zheng, J.; Mutcherson, R., II; Helfand, S.L. Calorie restriction delays lipioxidative damage in Drosophila melanogaster. Aging Cell 2005, 4, 209–216. [Google Scholar] [CrossRef]
- Vicencio, J.M.; Galluzzi, L.; Tajeddine, N.; Ortiz, C.; Criollo, A.; Tasdemir, E.; Morselli, E.; Ben Younes, A.; Maiuri, M.C.; Lavandero, S. Senescence, apoptosis or autophagy? When a damaged cell must decide its path—A mini-review. Gerontology 2008, 54, 92–99. [Google Scholar] [CrossRef]
- Huang, Z.Y.; Robinson, G.E. Honey bee colony integration: Worker-worker interactions mediate hormonally regulated plasticity in division of labor. Proc. Natl. Acad. Sci. USA 1992, 89, 11726–11729. [Google Scholar] [CrossRef]
- Rutz, W.; Gerig, L.; Wille, H.; Luscher, M. A bioassay for juvenile hormone (JH) effects of insect growth regulators (IGR) on adult worker honeybees. Mitt. Schweiz. Entomol. Ges. 1974, 47, 307–313. [Google Scholar]
- Wille, H.; Rutz, W. Beziehungen zwischen Juvenil hormone titer und Hämozyten erwachsener Sommerbienen (Apis mellifera L.). Schweiz. Landwirtsch. Forsch. 1975, 14, 339–353. [Google Scholar]
- Bedick, J.C.; Tunaz, H.; Nor Aliza, A.R.; Putman, S.M.; Ellis, M.D.; Stanley, D.W. Eicosanoids act in nodulation reactions to bacterial infections in newly emerged adult honey bees, Apis mellifera, but not in older foragers. Comp. Biochem. Physiol. 2001, 130C, 107–117. [Google Scholar]
- Franssens, V.; Simonet, G.; Bronckaers, A.; Claeys, I.; de Loof, A.; Broeck, J.V. Eicosanoids mediate the laminarin-induced nodulation response in larvae of the flesh fly, Neobellieria bullata. Arch. Insect Biochem. Physiol. 2005, 59, 32–41. [Google Scholar] [CrossRef]
- Evans, J.; Aronstein, K.; Chen, Y.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect. Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef]
- Harrison, J.M. Caste specific changes in honeybee flight capacity. Physiol. Zool. 1986, 59, 175–187. [Google Scholar]
- Kim, S.J.; Hwang, S.G.; Shin, D.Y.; Kang, S.S.; Chun, J.S. p38 kinase regulates nitric oxide-induced apoptosis of articular chondrocytes by accumulating p53 via NFkappa B-dependent transcription and stabilization by serine 15 phosphorylation. J. Biol. Chem. 2002, 277, 33501–33508. [Google Scholar]
- Kohchi, C.; Inagawa, H.; Nishizawa, T.; Soma, G. ROS and innate immunity. Anticancer Res. 2009, 29, 817–821. [Google Scholar]
- Stokes, D.R. Insect muscle innervated by single motoneurons: Structural and biochemical features. Am. Zool. 1987, 27, 1001–1010. [Google Scholar]
- Harrison, J.F.; Fewell, J.H.; Roberts, S.P.; Hall, H.G. Achievement of thermal stability by varying metabolic heat production in flying honey bees. Science 1996, 274, 88–90. [Google Scholar] [CrossRef]
- Landis, G.N.; Abdueva, D.; Skvortsov, D.; Yang, J.; Rabin, B.E.; Carrick, J.; Tavaré, S.; Tower, J. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2004, 101, 7663–7668. [Google Scholar]
- Munch, D.; Amdam, G.V. The curious case of aging plasticity in honey bees. FEBS Lett. 2010, 584, 2496–2503. [Google Scholar] [CrossRef]
- Ferguson, M.; Mockett, R.J.; Shen, Y.; Orr, W.C.; Sohal, R.S. Age-associated decline in mitochondrial respiration and electron transport in Drosophila melanogaster. Biochem. J. 2005, 390, 501–511. [Google Scholar] [CrossRef]
- Lyons, C.N.; Mathieu-Costello, O.; Moyes, C.D. Regulation of skeletal muscle mitochondrial content during aging. J. Gerontol. A Biol. Sci. Med. Sci. 2006, 61, 3–13. [Google Scholar] [CrossRef]
- Schippers, M.P.; Dukas, R.; McClelland, G.B. Life-time- and caste-specific changes in flight metabolic rate and muscle biochemistry of honeybees, Apis mellifera. J. Comp. Phys. B 2010, 180, 45–55. [Google Scholar] [CrossRef]
- Correa-Fernandez, F.; Cruz-Landim, C. Differential flight muscle development in workers, queens and males of the eusocial bees, Apis mellifera and Scaptotrigona postica. J. Insect Sci. 2010, 10, 85. [Google Scholar]
- Fernandez-Winckler, F.; Cruz-Landim, C. A morphological view of the relationship between indirect flight muscle maturation and the flying needs of two species of advanced eusocial bees. Micron 2008, 39, 1235–1242. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Margotta, J.W.; Mancinelli, G.E.; Benito, A.A.; Ammons, A.; Roberts, S.P.; Elekonich, M.M. Effects of Flight on Gene Expression and Aging in the Honey Bee Brain and Flight Muscle. Insects 2013, 4, 9-30. https://doi.org/10.3390/insects4010009
Margotta JW, Mancinelli GE, Benito AA, Ammons A, Roberts SP, Elekonich MM. Effects of Flight on Gene Expression and Aging in the Honey Bee Brain and Flight Muscle. Insects. 2013; 4(1):9-30. https://doi.org/10.3390/insects4010009
Chicago/Turabian StyleMargotta, Joseph W., Georgina E. Mancinelli, Azucena A. Benito, Andrew Ammons, Stephen P. Roberts, and Michelle M. Elekonich. 2013. "Effects of Flight on Gene Expression and Aging in the Honey Bee Brain and Flight Muscle" Insects 4, no. 1: 9-30. https://doi.org/10.3390/insects4010009




