The Evolutionary Innovation of Nutritional Symbioses in Leaf-Cutter Ants
Abstract
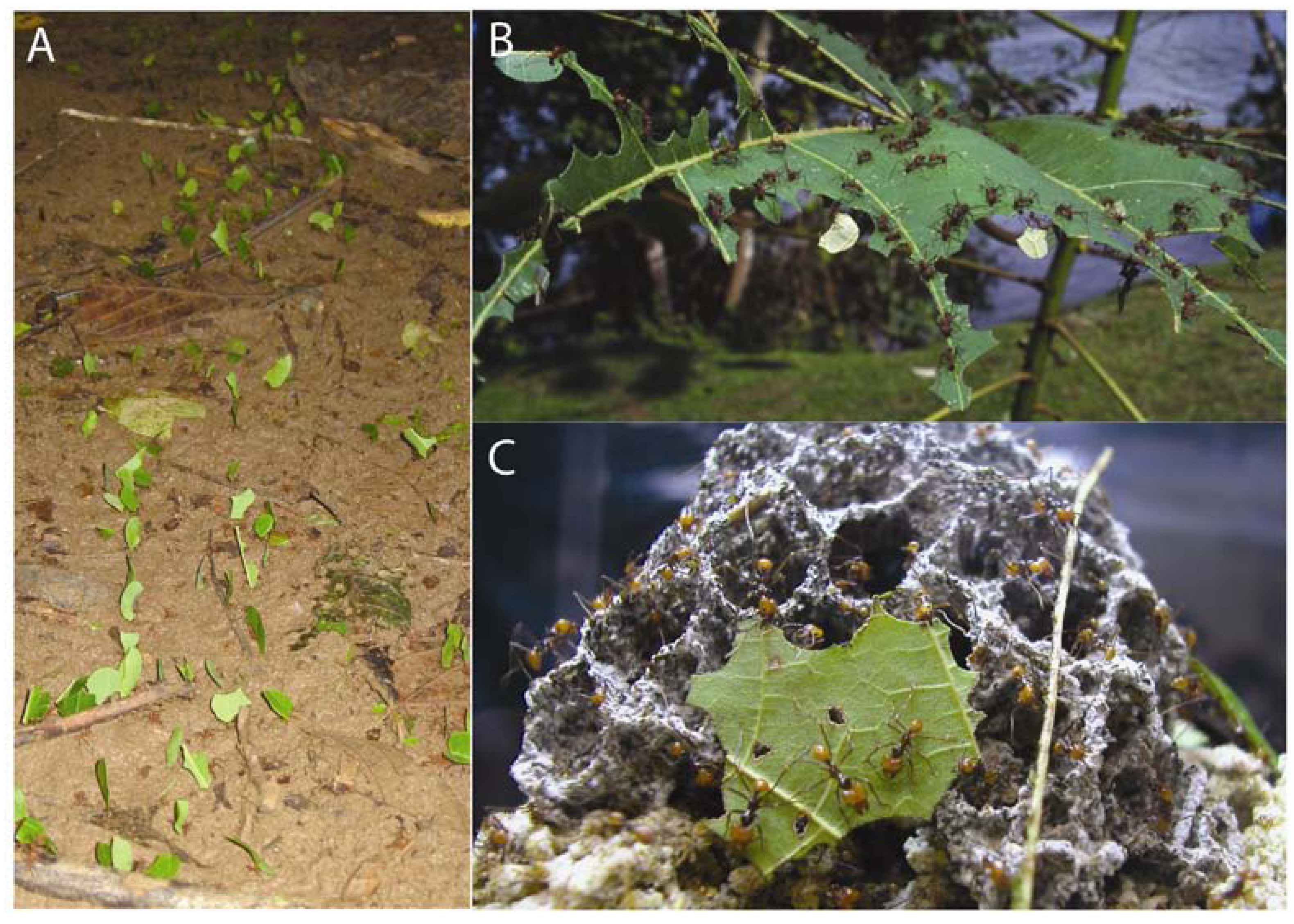
:1. Introduction
2. The Fungus Garden Ecosystem
3. The Ancillary Gut Hypothesis
| Ant Genera | Collection Location | Microbes Analyzed | Plant Polymers Analyzed | Methods | Principle Findings | Reference |
|---|---|---|---|---|---|---|
| Atta | Brazil | L. gongylophorus | Cellulose | Growth assays, enzymatic assays. | Evidence that L. gongylophorus can grow on cellulose in pure culture and hydrolyze this polymer efficiently. | [57] |
| Atta | Brazil | Bacteria | Gelatin, cellulose, cellobiose, casein | Directed culturing. | Isolation of plant polymer-degrading bacteria from fungus gardens. | [58] |
| Acromyrmex, Atta, Myrmicocrypta, Trachymyrmex, Cyphomyrmex | Brazil, Texas, Trinidad and Tobago | Microfungi, yeasts, | NA | Culturing, bioassays. | Isolation and characterization of microfungi and yeasts from fungus gardens, especially those of the genera Escovopsis, Fusarium, and Trichoderma; evidence that yeasts may antagonize potential garden pathogens; Identification of a novel Trichosporon species in fungus gardens. | [26,27,30,31,32,33,35,37,38,39] |
| Acromyrmex, Atta | Argentina | L. gongylophorus | Cellulose | Pure culture growth assays, estimation of lignin and cellulose content. | Indication that L. gongylophorus does not degrade cellulose in pure culture. | [59,60] |
| Acromyrmex, Atta | Brazil, Panama | Whole fungus garden, ants, larvae | Polysaccharides, heterosides, oligosaccharides | Enzymatic activity assays on workers, larvae, and fungus gardens. | Enzymatic activity profiles for the fungus garden and host ants were largely non-overlapping; xylanase, amylase, laminarinase, cellulase, and lichenanase activities identified in fungus garden samples; evidence for high variability in the enzymatic activities of fungus gardens between different nests and ant species. | [61,62] |
| Acromyrmex, Atta | Panama | L. gongylophorus | Pectin, CMC, ABTS (laccase substrate), protein | Isoelectric focusing, enzymatic assays. | Identification of fungal pectinases, CMCases, proteinases, and laccases concentrated by leaf-cutter ants in their fecal droplets. | [63] |
| Atta | Brazil | L. gongylophorus | Starch, pectin, xylan, cellulose, CMC | Growth and enzymatic assays. | Rapid growth of L. gongylophorus on xylan and starch, but poor growth on cellulose; Production of pectinases, xylanases, cellulases, and amylases by L. gongylophorus when grown in pure culture on different carbon sources; Identification of potential mechanisms for regulation of L. gongylophorus starch metabolism by the ant hosts. | [64,65,66] |
| Atta | Brazil | Whole fungus garden | All biomass | Estimation of cellulose and lignin content. | Evidence that the lignin:cellulose ratio is higher in fungus garden waste than leaf material. | [67] |
| Acromyrmex | Panama | L. gongylophorus | Xylan | Activity measurements of a xylanase, AZCL-based colorimetric assays. | Identification and characterization of an L. gongylophorus xylanase. | [68] |
| Acromyrmex | Brazil | Whole fungus garden, L. gongylophorus pure cultures | Numerous polysaccharides | Enzymatic activity assays on L. gongylophorus and whole fungus gardens, SEM. | Demonstration of broad lignocellulolytic capabilities of L. gongylophorus and whole fungus gardens, including activity against, laminarin, chitin, pectin, and CMC. | [69] |
| Cross phylogeny | Panama | Whole fungus gardens | Numerous polysaccharides | AZCL-based colorimetric assays. | Evidence for an evolutionary transition towards more efficient proteinase and amylase activity in leaf-cutter ant fungus gardens; evidence for broad lignocellulolytic capacity in lower attine fungus gardens. | [70] |
| Atta | Texas, Panama | L. gongylophorus | NA | Microsattelite profiling of L. gongylophorus from different ant nests and time points. | Confirms that a single strain of L. gongylophorus is cultured in leaf-cutter ant fungus gardens. | [23] |
| Atta, Acromyrmex | Panama, Costa Rica, Argentina | Pantoea, Klebsiella | NA | Directed culturing, stable isotope analysis, acetylene reduction analysis, phylogenetic comparisons. | Identification of nitrogen-fixing Klebsiella and Pantoea isolates in fungus gardens; Evidence that nitrogen fixed in fungus gardens is integrated into ant biomass. | [46] |
| Acromyrmex | Panama | L. gongylophorus | Pectin | Proteomics, RT-qPCR, enzymatic assays. | Identification of diverse fungal pectinases concentrated in the fecal droplets of the ants; evidence that L. gongylophorus produces enzymes specifically in the hyphal swellings fed to the ants. | [71] |
| Atta, Acromyrmex | Panama, Argentina | Primarily Gram-negative bacteria | NA | Lipid profiling using PLFA and FAME. | Evidence that ant fungus gardens and refuse dumps contain distinct microbial communities; evidence that the prokaryotic fraction of fungus gardens is dominated by Gram-negative bacteria. | [43] |
| Atta | Panama | γ-proteobacteria, primarily Klebsiella and Pantoea | Cellulose, hemicelluloses | Community metagenomics, 16S surveys, genome sequencing, enzymatic assays, sugar composition analysis. | Survey of bacterial diversity in fungus gardens; Identification of abundant Klebsiella and Pantoea populations; characterization of bacterial glycoside hydrolases; Evidence for significant amounts of cellulose degradation in fungus gardens. | [44] |
| Atta | Whole fungus garden | Numerous polysaccharides | AZCL-based colorimetric assays | Evidence that enzyme profiles in fungus gardens shift rapidly when integrated foliar biomass changes. | [72] | |
| Acromyrmex | Panama | Whole fungus garden | Pectin, xylose | Antibody and CBM-based polysaccharide microarray profiling, AZCL-based colorimetric assays. | Evidence for the degradation of xylan and pectin, but not cellulose, in fungus gardens; Indication that plant material is only partially degraded in these ecosystems. | [73] |
| Atta | Brazil | L. gongylophorus | All biomass | Dye and photomicrography of plant biomass. | Evidence for substantial degradation of all non-lignified plant tissues in fungus gardens; indication that L. gongylophorus may degrade a large quantity of cellulose in fungus gardens | [74] |
| Cross phylogeny | Panama | L. gongylophorus | Protein | Enzymatic assays, pH and buffering analysis. | Indication that the fungal cultivars of higher attines have evolved proteinases with activity optima at pH ~5, closer to the pH of fungus gardens; characterization of different proteinase classes and buffing capacities in different fungus gardens | [75] |
| Atta | Panama | Bacteria | Cellulose, hemicelluloses | Community metagenomics, 16S surveys, metaproteomics. | Identification of abundant Enterobacter population in fungus gardens; Further characterization of Klebsiella and Pantoea populations; Proteomic identification of bacterial glycoside hydrolases; Identification of bacteriophage in fungus gardens | [45] |
4. Evolution of Hygiene in the Attines
| Factors limiting diversity | Weeding and grooming of fungus gardens, application of glandular secretions, application of antimicrobials from Pseudonocardia, antibiotics produced by the fungal cultivar, fecal droplets of the ants |
| Factors promoting diversity | Complex, nutrient rich substrate |
| Potential sources of microbial groups | Maternal transmission from parent colony, phyllosphere microbes on foliar biomass, surrounding soil, the ants themselves |
5. Plant Biomass Degradation in Fungus Gardens
6. Reciprocal Adaptation of the Ant Genome
7. Conclusions and Future Outlook
Acknowledgements
References
- Moran, N.A. Symbiosis. Curr. Biol. 2006, 16, R866–R871. [Google Scholar] [CrossRef]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Douglas, A.E. The Symbiotic Habit; Princeton University Press: Princeton, NJ, USA, 2010. [Google Scholar]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef]
- Woyke, T.; Teeling, H.; Ivanova, N.N.; Huntemann, M.; Richter, M.; Gloeckner, F.O.; Boffelli, D.; Anderson, I.J.; Barry, K.W.; Shapiro, H.J.; et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 2006, 443, 950–955. [Google Scholar]
- Warnecke, F.; Luginbuhl, P.; Ivanova, N.; Ghassemian, M.; Richardson, T.H.; Stege, J.T.; Cayouette, M.; McHardy, A.C.; Djordjevic, G.; Aboushadi, N.; et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 2007, 450, 560–565. [Google Scholar]
- ope, P.B.; Denman, S.E.; Jones, M.; Tringe, S.G.; Barry, K.; Malfatti, S.A.; McHardy, A.C.; Cheng, J.F.; Hugenholtz, P.; McSweeney, C.S.; et al. Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc. Natl. Acad. Sci. USA 2010, 107, 14793–14798. [Google Scholar]
- Distel, D.L.; Roberts, S.J. Bacterial endosymbionts in the gills of the deep-sea wood-boring bivalves Xylophaga atlantica and Xylophaga washingtona. Biol. Bull. 1997, 192, 253–261. [Google Scholar] [CrossRef]
- Yang, J.C.; Madupu, R.; Durkin, A.S.; Ekborg, N.A.; Pedamallu, C.S.; Hostetler, J.B.; Radune, D.; Toms, B.S.; Henrissat, B.; Coutinho, P.M.; et al. The complete genome of Teredinibacter turnerae T7901: An intracellular endosymbiont of marine wood-boring bivalves (shipworms). PLoS One 2009, 4, e6085. [Google Scholar]
- Schultz, T.R.; Brady, S.G. Major evolutionary transitions in ant agriculture. Proc. Natl. Acad. Sci. USA 2008, 105, 5435–5440. [Google Scholar] [CrossRef]
- Mayhe-Nunes, A.J.; Jaffe, K. On the biogeography of Attini (Hymenoptera: Formicidae). Ecotropicos 1998, 11, 45–54. [Google Scholar]
- Hölldobler, B.; Wilson, E.O. The Ants; Harvard University Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Hölldobler, B.; Wilson, E.O. The Superorganism: The Beauty, Elegance, and Strangeness of Insect Societies; W.W. Norton & Company: New York, NY, USA, 2008. [Google Scholar]
- Costa, A.N.; Vasconcelos, H.L.; Vieira-Neto, E.H.M.; Bruna, E.M. Do herbivores exert top-down effects in Neotropical savannas? Estimates of biomass consumption by leaf-cutter ants. J. Veg. Sci. 2009, 19, 849–854. [Google Scholar]
- Hölldobler, B.; Wilson, E.O. The Leafcutter Ants: Civilization by Instinct; W.W. Norton and Company, Inc.: New York, NY, USA, 2010. [Google Scholar]
- Nygaard, S.; Zhang, G.; Schiott, M.; Li, C.; Wurm, Y.; Hu, H.; Zhou, J.; Ji, L.; Qiu, F.; Rasmussen, M.; et al. The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 2011, 21, 1339–1348. [Google Scholar] [CrossRef]
- Suen, G.; Teiling, C.; Li, L.; Holt, C.; Abouheif, E.; Bornberg-Bauer, E.; Bouffard, P.; Caldera, E.J.; Cash, E.; Cavanaugh, A.; et al. The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet. 2011, 7, e1002007. [Google Scholar]
- Chapela, I.H.; Rehner, S.A.; Schultz, T.R.; Mueller, U.G. Evolutionary history of the symbiosis between fungus-growing ants and their fungi. Science 1994, 266, 1691–1694. [Google Scholar]
- Villesen, P.; Mueller, U.G.; Schultz, T.R.; Adams, R.M.; Bouck, A.C. Evolution of ant-cultivar specialization and cultivar switching in Apterostigma fungus-growing ants. Evolution 2004, 58, 2252–2265. [Google Scholar]
- Weber, N.A. The fungus-culturing behavior of ants. Am. Zool. 1972, 12, 577–587. [Google Scholar]
- Weber, N.A. Fungus-growing ants. Science 1966, 153, 587–604. [Google Scholar]
- Mueller, U.G.; Scott, J.J.; Ishak, H.D.; Cooper, M.; Rodrigues, A. Monoculture of leafcutter ant gardens. PLoS One 2010, 5. [Google Scholar]
- Mikheyev, A.S.; Mueller, U.G.; Abbot, P. Comparative dating of attine ant and lepiotaceous cultivar phylogenies reveals coevolutionary synchrony and discord. Am. Nat. 2010, 175, E126–E133. [Google Scholar] [CrossRef]
- Currie, C.R.; Scott, J.A.; Summerbell, R.C.; Malloch, D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 1999, 398, 701–704. [Google Scholar] [CrossRef]
- Currie, C.R.; Mueller, U.G.; Malloch, D. The agricultural pathology of ant fungus gardens. Proc. Natl. Acad. Sci. USA 1999, 96, 7998–8002. [Google Scholar]
- Currie, C.R. Prevalence and impact of a virulent parasite on a tripartite mutualism. Oecologia 2001, 128, 99–106. [Google Scholar] [CrossRef]
- Caldera, E.J.; Poulsen, M.; Suen, G.; Currie, C.R. Insect symbioses: A case study of past, present, and future fungus-growing ant research. Environ. Entomol. 2009, 38, 78–92. [Google Scholar] [CrossRef]
- Currie, C.R. A community of ants, fungi, and bacteria: A multilateral approach to studying symbiosis. Annu. Rev. Microbiol. 2001, 55, 357–380. [Google Scholar] [CrossRef]
- Carreiro, S.C.; Pagnocca, F.C.; Bacci, M., Jr.; Bueno, O.C.; Hebling, M.J.; Middelhoven, W.J. Occurrence of killer yeasts in leaf-cutting ant nests. Folia Microbiol (Praha) 2002, 47, 259–262. [Google Scholar] [CrossRef]
- Rodrigues, A.; Pagnocca, F.C.; Bacci, M.J.; Hebling, M.J.; Bueno, O.C.; Pfenning, L.H. Variability of non-mutualistic filamentous fungi associated with Atta sexdens rubropilosa nests. Folia Microbiol (Praha) 2005, 50, 421–425. [Google Scholar] [CrossRef]
- Rodrigues, A.; Mueller, U.G.; Ishak, H.D.; Bacci, M., Jr.; Pagnocca, F.C. Ecology of microfungal communities in gardens of fungus-growing ants (Hymenoptera: Formicidae): A year-long survey of three species of attine ants in Central Texas. FEMS Microbiol. Ecol. 2011, 78, 244–255. [Google Scholar] [CrossRef]
- Rodrigues, A.; Cable, R.N.; Mueller, U.G.; Bacci, M., Jr.; Pagnocca, F.C. Antagonistic interactions between garden yeasts and microfungal garden pathogens of leaf-cutting ants. A. Van Leeuw. J. Microb. 2009, 96, 331–342. [Google Scholar] [CrossRef]
- Rodrigues, A.; Bacci, M., Jr.; Mueller, U.G.; Ortiz, A.; Pagnocca, F.C. Microfungal“weeds” in the leafcutter ant symbiosis. Microb. Ecol. 2008, 56, 604–614. [Google Scholar] [CrossRef]
- Pagnocca, F.C.; Legaspe, M.F.; Rodrigues, A.; Ruivo, C.C.; Nagamoto, N.S.; Bacci, M., Jr.; Forti, L.C. Yeasts isolated from a fungus-growing ant nest, including the description of Trichosporon chiarellii sp. nov., an anamorphic basidiomycetous yeast. Int. J. Syst. Evol. Microbiol. 2010, 60, 1454–1459. [Google Scholar] [CrossRef]
- Carreiro, S.C.; Pagnocca, F.C.; Bueno, O.C.; Bacci, M.J.; Hebling, M.J.; da Silva, O.A. Yeasts associated with nests of the leaf-cutting ant Atta sexdens rubropilosa Forel, 1908. A. Van Leeuw. J. Microb. 1997, 71, 243–248. [Google Scholar] [CrossRef]
- Carreiro, S.C.; Pagnocca, F.C.; Bacci, M., Jr.; Lachance, M.A.; Bueno, O.C.; Hebling, M.J.; Ruivo, C.C.; Rosa, C.A. Sympodiomyces attinorum sp. nov., a yeast species associated with nests of the leaf-cutting ant Atta sexdens. Int. J. Syst. Evol. Microbiol. 2004, 54, 1891–1894. [Google Scholar] [CrossRef]
- Fisher, P.J.; Stradling, D.J.; Sutton, B.C.; Petrini, L.E. Microfungi in the fungus gardens of the leaf-cutting ant Atta cephalotes: A preliminary study. Mycol. Res. 1996, 100, 541–546. [Google Scholar] [CrossRef]
- Silva, A.; Rodrigues, A.; Bacci, M., Jr.; Pagnocca, F.C.; Bueno, O.C. Susceptibility of the ant-cultivated fungus Leucoagaricus gongylophorus (Agaricales: Basidiomycota) towards microfungi. Mycopathologia 2006, 162, 115–119. [Google Scholar] [CrossRef]
- Mueller, U.; Gerardo, N.; Aanen, D.; Six, D.; Schultz, T. The evolution of agriculture in insects. Ann. Rev. Ecol. Evol. Syst. 2005, 36, 563–595. [Google Scholar] [CrossRef]
- Santos, A.V.; Dillon, R.J.; Dillon, V.M.; Reynolds, S.E.; Samuels, R.I. Ocurrence of the antibiotic producing bacterium Burkholderia sp. in colonies of the leaf-cutting ant Atta sexdens rubropilosa. FEMS Microbiol. Lett. 2004, 239, 319–323. [Google Scholar] [CrossRef]
- Haeder, S.; Wirth, R.; Herz, H.; Spiteller, D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 4742–4746. [Google Scholar]
- Scott, J.J.; Budsberg, K.J.; Suen, G.; Wixon, D.L.; Balser, T.C.; Currie, C.R. Microbial community structure of leaf-cutter ant fungus gardens and refuse dumps. PLoS One 2010, 5, e9922. [Google Scholar]
- Suen, G.; Scott, J.J.; Aylward, F.O.; Adams, S.M.; Tringe, S.G.; Pinto-Tomas, A.A.; Foster, C.E.; Pauly, M.; Weimer, P.J.; Barry, K.W.; et al. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet 2010, 6. [Google Scholar]
- Aylward, F.O.; Burnum, K.E.; Scott, J.J.; Suen, G.; Tringe, S.G.; Adams, S.M.; Barry, K.W.; Nicora, C.D.; Piehowski, P.D.; Purvine, S.O.; et al. Metagenomic and metaproteomic insights into leaf-cutter ant fungus gardens. ISME J. 2011. submitted.. [Google Scholar]
- Pinto-Tomas, A.A.; Anderson, M.A.; Suen, G.; Stevenson, D.M.; Chu, F.S.; Cleland, W.W.; Weimer, P.J.; Currie, C.R. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 2009, 326, 1120–1123. [Google Scholar]
- Backhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar]
- Brookman, J.L.; Mennim, G.; Trinci, A.P.; Theodorou, M.K.; Tuckwell, D.S. Identification and characterization of anaerobic gut fungi using molecular methodologies based on ribosomal ITS1 and 185 rRNA. Microbiology 2000, 146, 393–403. [Google Scholar]
- Liggenstoffer, A.S.; Youssef, N.H.; Couger, M.B.; Elshahed, M.S. Phylogenetic diversity and community structure of anaerobic gut fungi (phylum Neocallimastigomycota) in ruminant and non-ruminant herbivores. ISME J. 2010, 4, 1225–1235. [Google Scholar] [CrossRef]
- Teunissen, M.J.; Op den Camp, H.J. Anaerobic fungi and their cellulolytic and xylanolytic enzymes. Antonie Van Leeuwenhoek 1993, 63, 63–76. [Google Scholar]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar]
- Hess, M.; Sczyrba, A.; Egan, R.; Kim, T.W.; Chokhawala, H.; Schroth, G.; Luo, S.; Clark, D.S.; Chen, F.; Zhang, T.; et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 2011, 331, 463–467. [Google Scholar]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar]
- Brulc, J.M.; Antonopoulos, D.A.; Miller, M.E.; Wilson, M.K.; Yannarell, A.C.; Dinsdale, E.A.; Edwards, R.E.; Frank, E.D.; Emerson, J.B.; Wacklin, P. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. USA 2009, 106, 1948–1953. [Google Scholar]
- Bacci, M.; Anversa, M.M.; Pagnocca, F.C. Cellulose degradation by Leucocoprinus gongylophorus, the fungus cultured by the leaf-cutting ant Atta sexdens rubropilosa. A. Van Leeuw. J. Microb. 1995, 67, 385–386. [Google Scholar] [CrossRef]
- Bacci, M.; Ribeiro, S.B.; Casarotto, M.E.F.; Pagnocca, F.C. Biopolymer-degrading bacteria from nests of the leaf-cutting ant Atta sexdens Rubropilosa. Braz. J. Med. Biol. Res. 1995, 28, 79–82. [Google Scholar]
- Abril, A.B.; Bucher, E.H. Evidence that the fungus cultured by leaf-cutting ants does not metabolize cellulose. Ecol. Lett. 2002, 5, 325–328. [Google Scholar] [CrossRef]
- Abril, A.B.; Bucher, E.H. Nutritional sources of the fungus cultured by leaf-cutting ants. App. Soil Ecol. 2004, 26, 243–247. [Google Scholar] [CrossRef]
- D’Ettorre, P.; Mora, P.; Dibangou, V.; Rouland, C.; Errard, C. The role of the symbiotic fungus in the digestive metabolism of two species of fungus-growing ants. J. Comp. Physiol. B 2002, 172, 169–176. [Google Scholar] [CrossRef]
- Richard, F.J.; Mora, P.; Errard, C.; Rouland, C. Digestive capacities of leaf-cutting ants and the contribution of their fungal cultivar to the degradation of plant material. J. Comp. Physiol. B 2005, 175, 297–303. [Google Scholar] [CrossRef]
- Ronhede, S.; Boomsma, J.J.; Rosendahl, S. Fungal enzymes transferred by leaf-cutting ants in their fungus gardens. Mycol. Res. 2004, 108, 101–106. [Google Scholar] [CrossRef]
- Silva, A.; Bacci, M., Jr.; Pagnocca, F.C.; Bueno, O.C.; Hebling, M.J. Production of polysaccharidases in different carbon sources by Leucoagaricus gongylophorus Moller (Singer), the symbiotic fungus of the leaf-cutting ant Atta sexdens Linnaeus. Curr. Microbiol. 2006, 53, 68–71. [Google Scholar]
- Silva, A.; Bacci, M., Jr.; Pagnocca, F.C.; Bueno, O.C.; Hebling, M.J. Starch metabolism in Leucoagaricus gongylophorus, the symbiotic fungus of leaf-cutting ants. Microbiol. Res. 2006, 161, 299–303. [Google Scholar] [CrossRef]
- Gomes De Siqueira, C.; Bacci, M., Jr.; Pagnocca, F.C.; Bueno, O.C.; Hebling, M.J.A. Metabolism of plant polysaccharides by Leucoagaricus gongylophorus, the symbiotic fungus of the leaf-cutting ant Atta sexdens. Appl. Environ. Microbiol. 1998, 64, 4820–4822. [Google Scholar]
- Sousa-Souto, L.; Guerra, M.B.B.; Schoereder, J.H.; Ernesto, C.; Schaefer, G.R.; d. Silva, W.L. Determination of the conversion factor in colonies of Atta sexdens rubropilosa (Hymenoptera: Formicidae) and its relationship with the quality of harvested leaf substrate. Revista Arvore 2007, 31, 163–166. [Google Scholar] [CrossRef]
- Schiott, M.; de Fine Licht, H.H.; Lange, L.; Boomsma, J.J. Towards a molecular understanding of symbiont function: Identification of a fungal gene for the degradation of xylan in the fungus gardens of leaf-cutting ants. BMC Microbiol. 2008, 8. [Google Scholar]
- Erthal, M., Jr.; Silva, C.P.; Cooper, R.M.; Samuels, R.I. Hydrolytic enzymes of leaf-cutting ant fungi. Comp. Biochem. Physiol. B 2009, 152, 54–59. [Google Scholar] [CrossRef]
- De Fine Licht, H.H.; Schiott, M.; Mueller, U.G.; Boomsma, J.J. Evolutionary transitions in enzyme activity of ant fungus gardens. Evolution 2010, 64, 2055–2069. [Google Scholar]
- Schiott, M.; Rogowska-Wrzesinska, A.; Roepstorff, P.; Boomsma, J.J. Leaf-cutting ant fungi produce cell wall degrading pectinase complexes reminiscent of phytopathogenic fungi. BMC Biol. 2010, 8, 156. [Google Scholar] [Green Version]
- Kooij, P.W.; Schiott, M.; Boomsma, J.J.; de Fine Licht, H.H. Rapid shifts in Atta cephalotes fungus-garden enzyme activity after a change in fungal substrate (Attini, Formicidae). InsectesSoc. 2011, 58, 145–151. [Google Scholar]
- Moller, I.E.; de Fine Licht, H.H.; Harholt, J.; Willats, W.G.; Boomsma, J.J. The dynamics of plant cell-wall polysaccharide decomposition in leaf-cutting ant fungus gardens. PLoS One 2011, 6. [Google Scholar]
- Nagamoto, N.S.; Garcia, M.G.; Forti, L.C.; Verza, S.S.; Noronha, N.C.; Rodella, R.A. Microscopic evidence supports the hypothesis of high cellulose degradation capacity by the symbiotic fungus of leaf-cutting ants. Journal of Biological Research-Thessaloniki 2011, 16, 308–312. [Google Scholar]
- Semenova, T.A.; Hughes, D.P.; Boomsma, J.J.; Schiott, M. Evolutionary patterns of proteinase activity in attine ant fungus gardens. BMC Microbiol 2011, 11, 15. [Google Scholar] [CrossRef]
- Currie, C.R.; Stuart, A.E. Weeding and grooming of pathogens in agriculture by ants. Proc. R. Soc. London Ser. B Biol. Sci. 2001, 268, 1033–1039. [Google Scholar] [CrossRef]
- Martin, M.M.; Gieselmann, M.J.; Martin, J.S. Rectal enzymes of attine ants. α-Amylase and chitinase. J. Insect Physiol. 1970, 19, 1409–1416. [Google Scholar]
- Rodrigues, A.; Carletti, C.D.; BuenoI, O.C.; PagnoccaI, F.C. Leaf-cutting ant faecal fluid and mandibular gland secretion: Effects on microfungi spore germination. Braz. J. Microbiol. 2008, 39, 64–67. [Google Scholar] [CrossRef]
- Beattie, A.J.; Turnbull, C.L.; Hough, T.; Knox, R.B. Antibiotic production: A possible function for the metapleural glands of ants (Hymenoptera: Formicidae). Ann. Entomol. Soc. Am. 1986, 79, 448–450. [Google Scholar]
- Veal, D.A.; Trimble, J.E.; Beattie, A.J. Antimicrobial properties of secretions from the metapleural glands of Myrmeciagulosa (the Australian bull ant). J. Appl. Bacteriol. 1992, 72, 188–194. [Google Scholar] [CrossRef]
- Hölldobler, B.; Engel-Siegel, H. On the metapleural gland of ants. Psyche 1984, 91, 201–224. [Google Scholar]
- Fernandez-Marin, H.; Zimmerman, J.K.; Rehner, S.A.; Wcislo, W.T. Active use of the metapleural glands by ants in controlling fungal infection. Proc. Biol. Sci. 2006, 273, 1689–1695. [Google Scholar] [CrossRef]
- Fernandez-Marin, H.; Zimmerman, J.K.; Nash, D.R.; Boomsma, J.J.; Wcislo, W.T. Reduced biological control and enhanced chemical pest management in the evolution of fungus farming in ants. Proc. R. Soc. B 2009, 276, 2263–2269. [Google Scholar] [CrossRef]
- Ortius-Lechner, D.; Maile, R.; Morgan, E.D.; Boomsma, J.J. Metapleural gland secretion of the leaf-cutter ant Acromyrmex octospinosus: New compounds and their functional significance. J. Chem. Ecol. 2000, 26, 1667–1683. [Google Scholar] [CrossRef]
- Do Nascimento, R.R.; Schoeters, E.; Morgan, E.D.; Billen, J.; Stradling, D.J. Chemistry of metapleural gland secretions of three attine ants, Atta sexdens rubropilosa, Atta cephalotes, and Acromyrmex octospinosus (Hymenoptera: Formicidae). J. Chem. Ecol. 1996, 22, 987–1000. [Google Scholar]
- Mendonca Ade, L.; da Silva, C.E.; de Mesquita, F.L.; Campos Rda, S.; Do Nascimento, R.R.; Ximenes, E.C.; Sant'Ana, A.E. Antimicrobial activities of components of the glandular secretions of leaf cutting ants of the genus Atta. A. Van Leeuw. J. Microb. 2009, 95, 295–303. [Google Scholar] [CrossRef]
- Bot, A.; Ortius-Lechner, D.; Finster, K.; Maile, R.; Boomsma, J. Variable sensitivity of fungi and bacteria to compounds produced by the metapleural glands of leaf-cutting ants. Insectes Soc. 2002, 49, 363–370. [Google Scholar] [CrossRef]
- Lorenzen, K.; Anke, T. Basidiomycetes as a source for new bioactive natural products. Curr. Org. Chem. 1998, 2, 329–364. [Google Scholar]
- Huff, T.; Kuball, H.G.; Anke, T. 7-Chloro-4,6-dimethoxy-1(3H)-isobenzofuranone and basidalin: antibiotic secondary metabolites from Leucoagaricus carneifolia Gillet (basidiomycetes) [corrected]. Z Naturforsch C 1994, 49, 407–410. [Google Scholar]
- Mead, M.I.; Khan, A.H.; Nickless, G.; Greally, B.R.; Tainton, D.; Shallcross, D.E. Leaf cutter ants: A possible missing source of biogenic halocarbons. Environ. chem. 2008, 5, 5–10. [Google Scholar] [CrossRef]
- De Jong, E.; Field, J.A. Sulfur tuft and turkey tail: Biosynthesis and biodegradation of organohalogens by Basidiomycetes. Annu. Rev. Microbiol. 1997, 51, 375–414. [Google Scholar] [CrossRef]
- Gerardo, N.M.; Jacobs, S.R.; Currie, C.R.; Mueller, U.G. Ancient host-pathogen associations maintained by specificity of chemotaxis and antibiosis. PLoSBiol. 2006, 4, e235. [Google Scholar]
- Currie, C.R.; Bot, A.N.M.; Boomsma, J.J. Experimental evidence of a tripartite mutualism: Bacteria protect ant fungus gardens from specialized parasites. Oikos 2003, 101, 91–102. [Google Scholar] [CrossRef]
- Poulsen, M.; Cafaro, M.; Erhardt, D.; Little, A.; NM, G.; Tebbets, B.; Klein, B.; Currie, C. Variation in Pseudonocardia antibiotic defence helps govern parasite-induced morbidity in Acromyrmex leaf-cutting ants. Environ. Microbiol. Rep. 2010, 2, 534–540. [Google Scholar]
- Cafaro, M.J.; Poulsen, M.; Little, A.E.; Price, S.L.; Gerardo, N.M.; Wong, B.; Stuart, A.E.; Larget, B.; Abbot, P.; Currie, C.R. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc. Biol. Sci. 2011, 278, 1814–1822. [Google Scholar] [CrossRef]
- Sen, R.; Ishak, H.D.; Estrada, D.; Dowd, S.E.; Hong, E.; Mueller, U.G. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc. Natl. Acad. Sci. USA 2009, 106, 17805–17810. [Google Scholar]
- Mueller, U.G.; Dash, D.; Rabeling, C.; Rodrigues, A. Coevolution between attine ants and actinomycete bacteria: A reevaluation. Evolution 2008, 62, 2894–2912. [Google Scholar] [CrossRef]
- Oh, D.C.; Poulsen, M.; Currie, C.R.; Clardy, J. Dentigerumycin: A bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 2009, 5, 391–393. [Google Scholar] [CrossRef]
- Barke, J.; Seipke, R.F.; Gruschow, S.; Heavens, D.; Drou, N.; Bibb, M.J.; Goss, R.J.; Yu, D.W.; Hutchings, M.I. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 2010, 8, 109. [Google Scholar] [Green Version]
- Seipke, R.F.; Barke, J.; Brearley, C.; Hill, L.; Yu, D.W.; Goss, R.J.; Hutchings, M.I. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS One 2011, 6. [Google Scholar]
- Martin, M.M.; Carman, R.M.; Macconnell, J.G. Nutrients derived from the fungus cultured by the fungus-growing ant Atta colombica tonsipes. Ann. Entom. Soc. Am. 1969, 62, 11–13. [Google Scholar]
- Mueller, U.G.; Schultz, T.R.; Currie, C.R.; Adams, R.M.; Malloch, D. The origin of the attine ant-fungus mutualism. Q. Rev. Biol. 2001, 76, 169–197. [Google Scholar]
- Martin, M.M.; Weber, N.A. The cellulose-utilizing capability of the fungus cultured by the attine ant Atta colombica tonsipes. Ann. Entomol. Soc. Am. 1969, 62, 1386–1387. [Google Scholar]
- Wirth, R.; Herz, H.; Ryel, R.J.; Beyschlag, W.; Hoelldobler, B. Herbivoryof Leaf-Cutting Ants: A Case Study on Atta Colombica in the Tropical Rainforest of Panama; Springer: New York, NY, USA, 2003. [Google Scholar]
- De Fine Licht, H.H.; Boomsma, J. Forage collection, substrate preparation, and diet composition in fungus-growing ants. Ecol. Ent. 2010, 35, 259–269. [Google Scholar] [CrossRef]
- Rockwood, L.L. The effects of seasonality on foraging in two species of leaf-cutting ants (Atta) in Guanacaste province, Costa Rica. Biotropica 1975, 7, 176–193. [Google Scholar] [CrossRef]
- Martin, M.M.; Martin, J.S. The presence of protease activity in the rectal fluid of primitive attine ants. J. Insect Physiol. 1971, 17, 1897–1906. [Google Scholar] [CrossRef]
- Martin, J.S.; Martin, M.M. The presence of protease activity in the rectal fluid of attine ants. J. Insect Physiol. 1970, 16, 227–232. [Google Scholar] [CrossRef]
- Martin, M.M.; Gieselmann, M.J.; Martin, J.S. Rectal enzymes of attine ants. α-Amylase and chitinase. J. Insect Physiol. 1970, 19, 1409–1416. [Google Scholar]
- Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef]
- International Aphid Genome Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010, 8. [Google Scholar]
- Shigenobu, S.; Watanabe, H.; Hattori, M.; Sakaki, Y.; Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 2000, 407, 81–86. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Aylward, F.O.; Currie, C.R.; Suen, G. The Evolutionary Innovation of Nutritional Symbioses in Leaf-Cutter Ants. Insects 2012, 3, 41-61. https://doi.org/10.3390/insects3010041
Aylward FO, Currie CR, Suen G. The Evolutionary Innovation of Nutritional Symbioses in Leaf-Cutter Ants. Insects. 2012; 3(1):41-61. https://doi.org/10.3390/insects3010041
Chicago/Turabian StyleAylward, Frank O., Cameron R. Currie, and Garret Suen. 2012. "The Evolutionary Innovation of Nutritional Symbioses in Leaf-Cutter Ants" Insects 3, no. 1: 41-61. https://doi.org/10.3390/insects3010041




