Non-Conserved Residues in Clostridium acetobutylicum tRNAAla Contribute to tRNA Tuning for Efficient Antitermination of the alaS T Box Riboswitch
Abstract
:1. Introduction
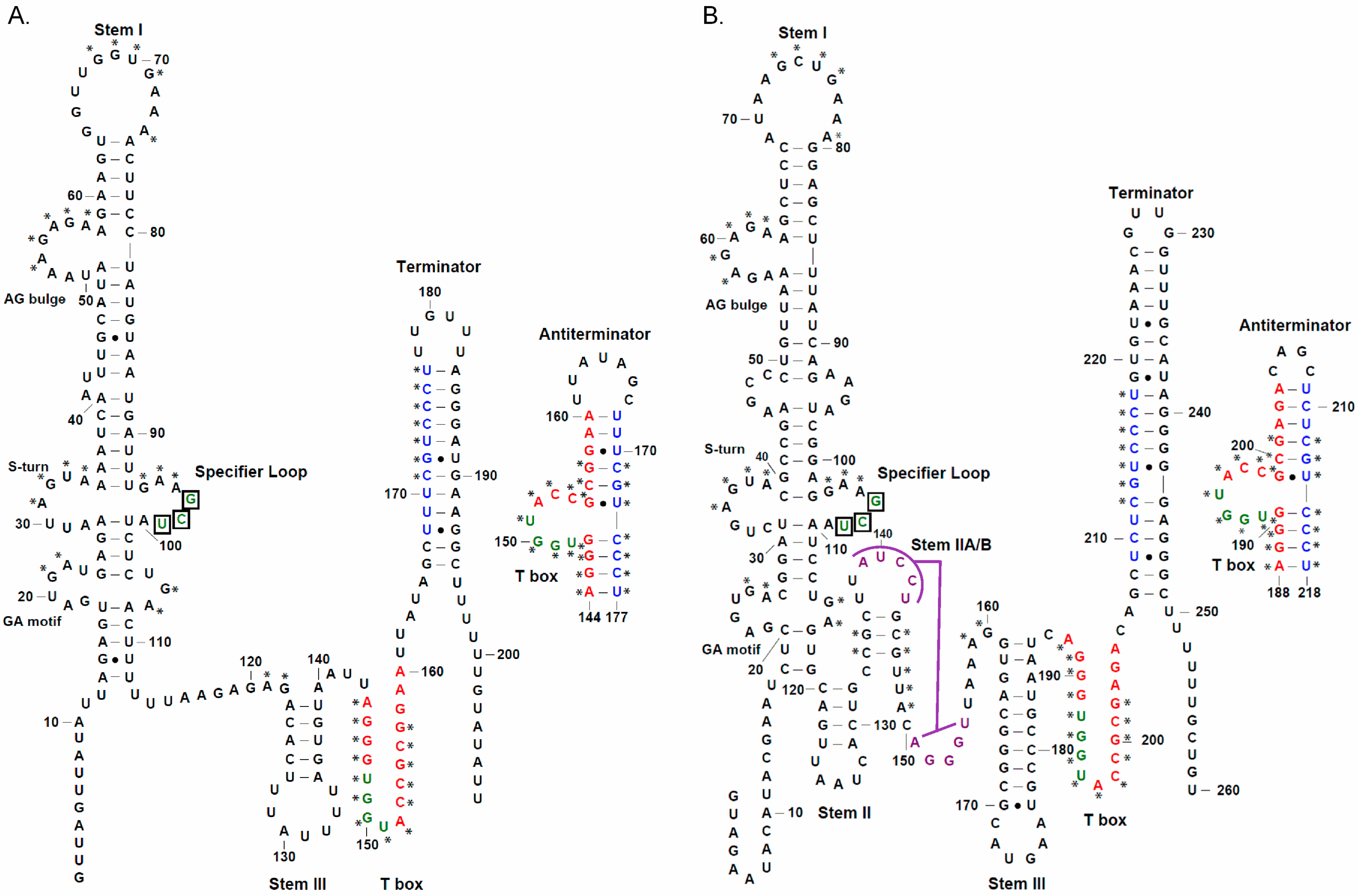
2. Experimental Section
2.1. Generation of DNA Templates
2.2. T7 RNAP Transcription
2.3. In Vitro Transcription Termination Assays
2.4. Genetic Techniques
2.5. Bacterial Growth Conditions and β-galactosidase Assays
3. Results
3.1. tRNAAla-Directed alaS Antitermination in Vitro

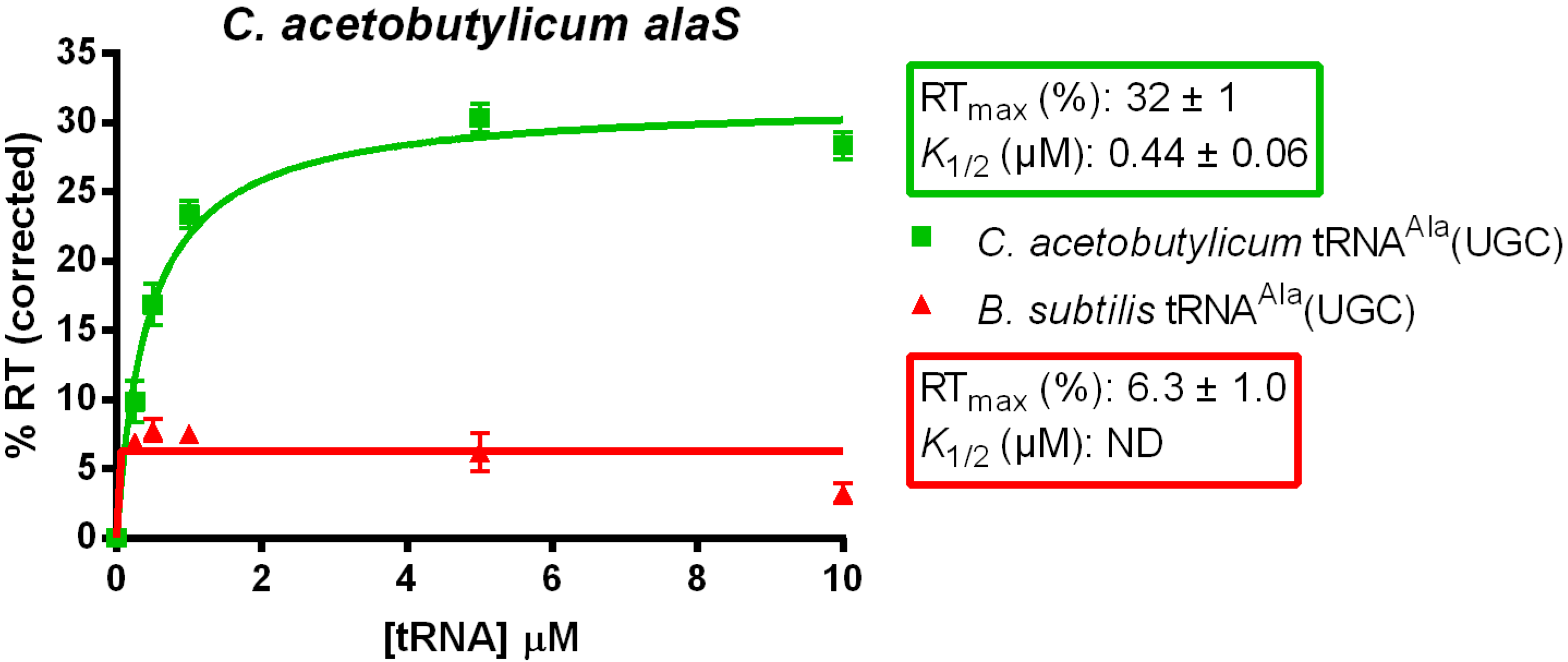
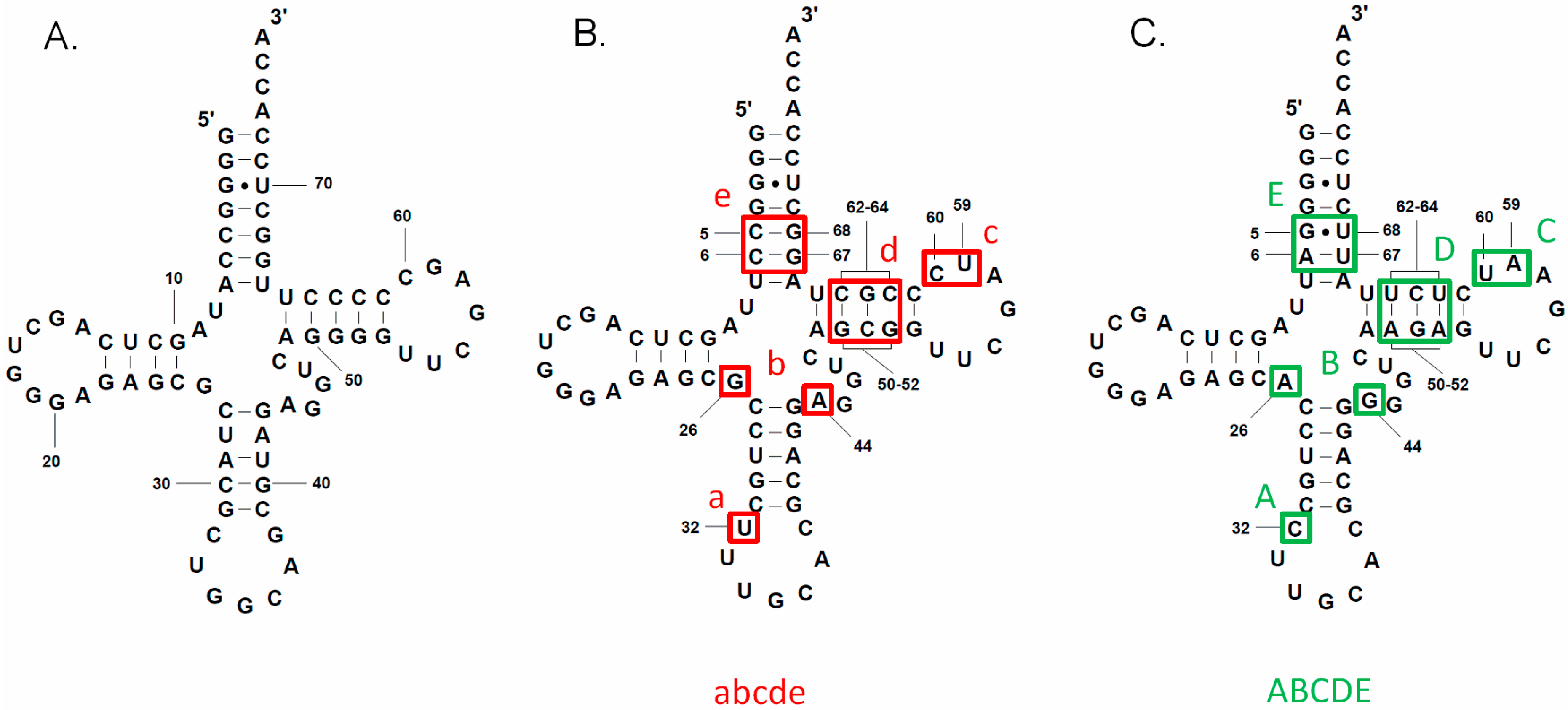
3.2. tRNAAla Requirements for alaS Antitermination
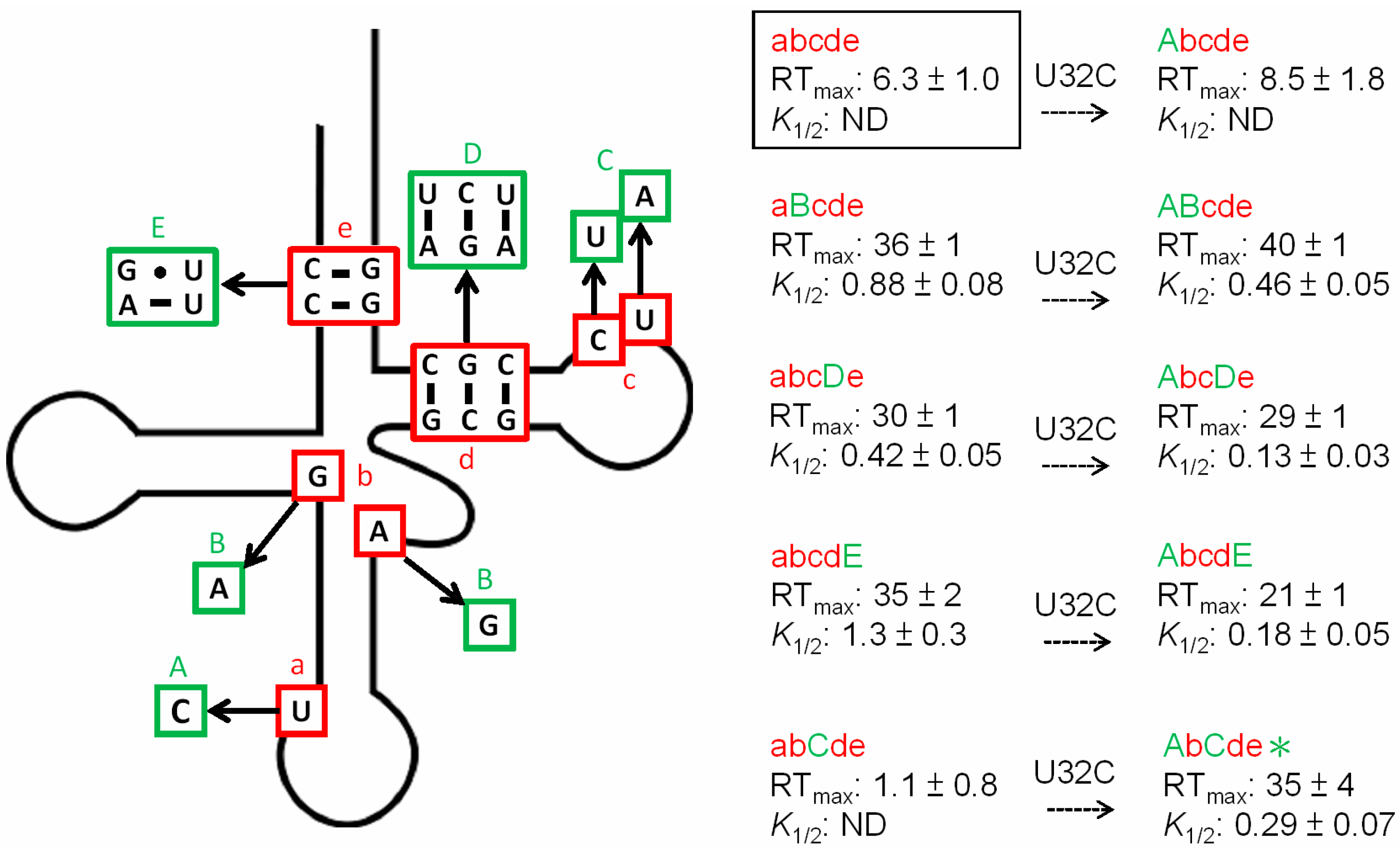
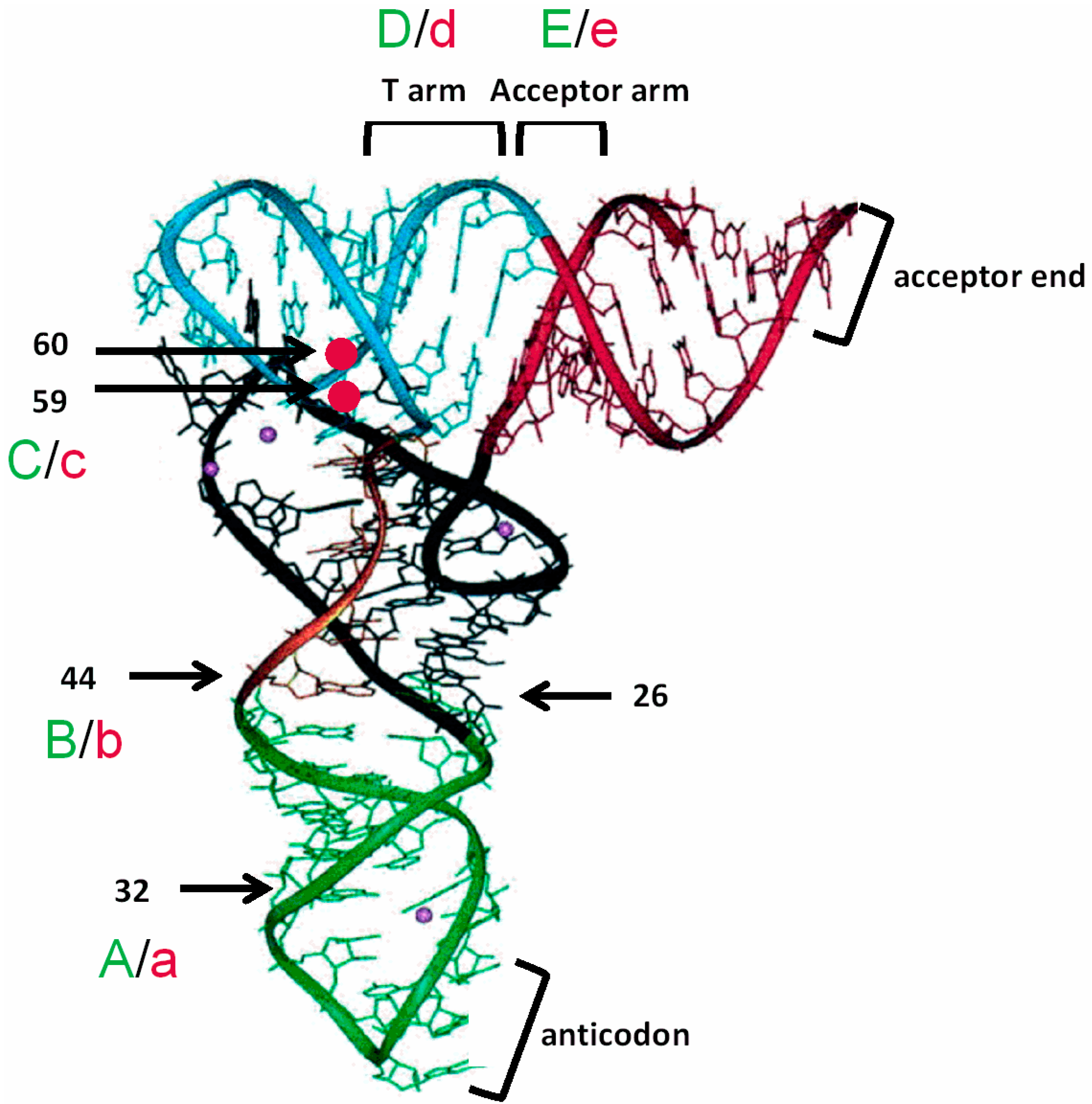
3.3. Non-Conserved Residues Contribute to tRNAAla Tuning for alaS Antitermination
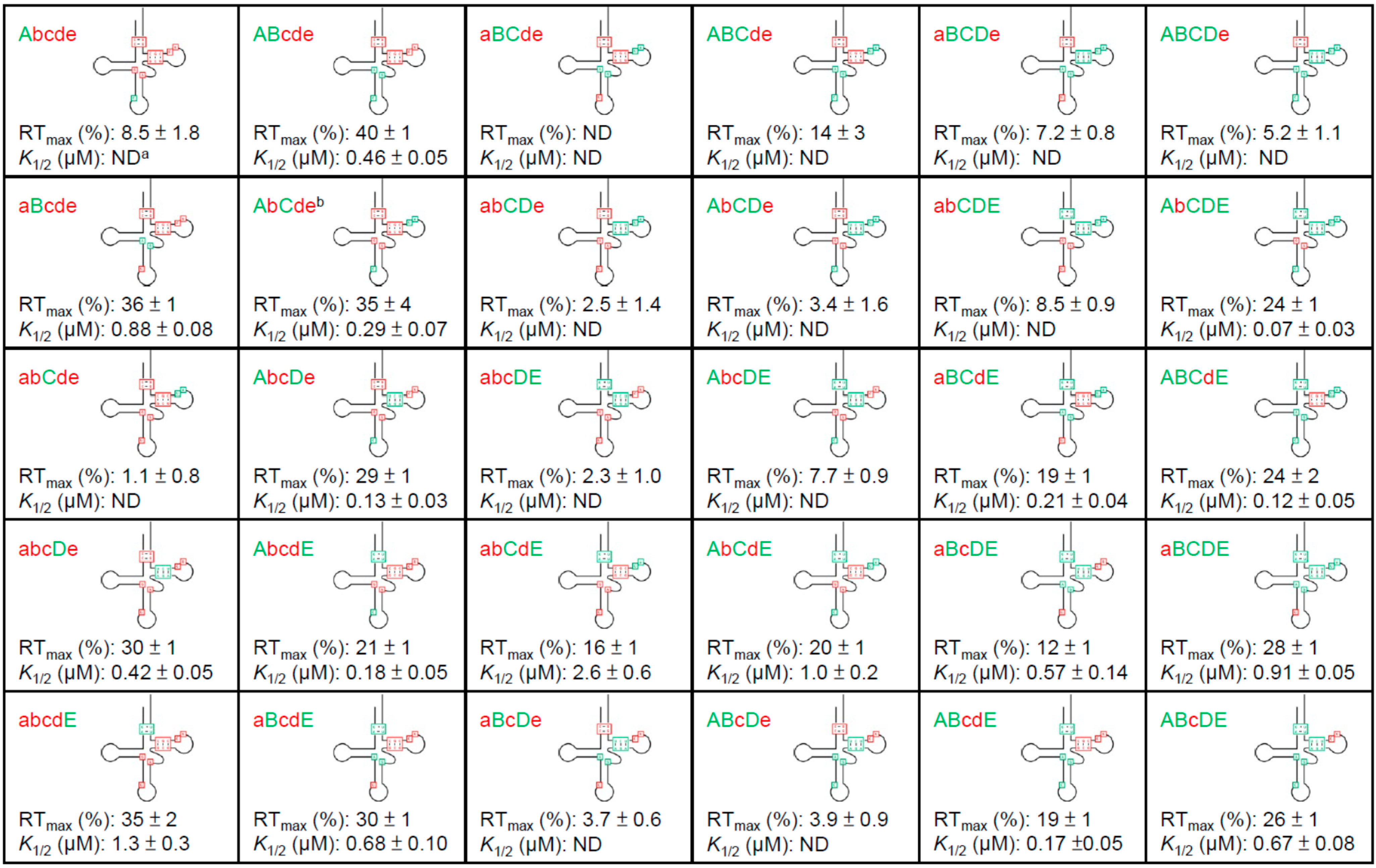
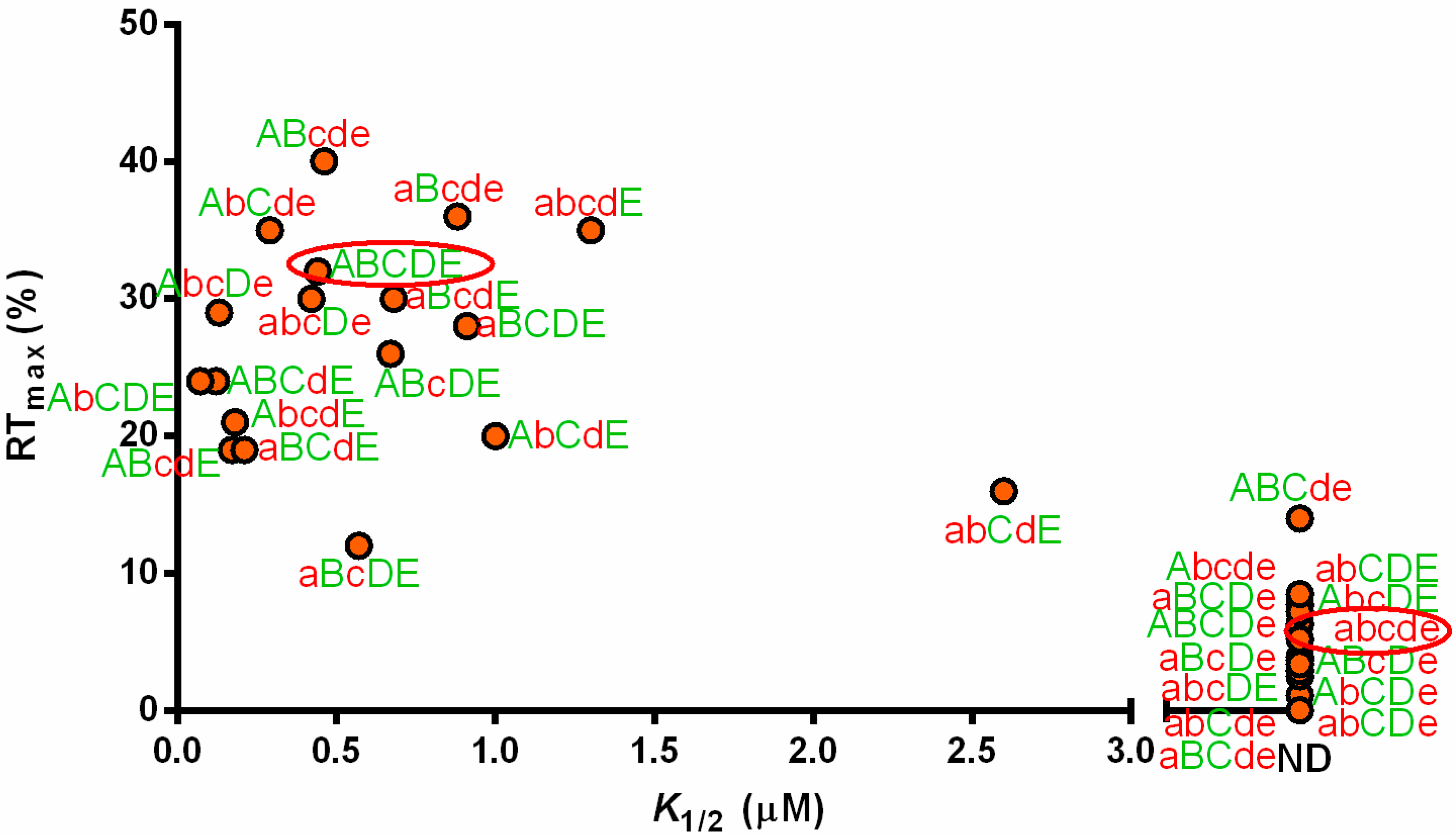
3.4. Expression of alaS-lacZ Fusions in B. subtilis
| β-galactosidase activity (Miller units) a | induction ratio | ||
|---|---|---|---|
| uninduced | induced | ||
| B. subtilis alaS-lacZ | 11 ± 1b | 22 ± 1 | 2.0 |
| C. acetobutylicum alaS-lacZ | 5.9 ± 1.7 | 6.4 ± 1.3 | 1.1 |
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Gutierrez-Preciado, A.; Henkin, T.M.; Grundy, F.J.; Yanofsky, C.; Merino, E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol. Mol. Biol. Rev. 2009, 73, 36–61. [Google Scholar] [CrossRef] [PubMed]
- Green, N.J.; Grundy, F.J.; Henkin, T.M. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010, 584, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Grundy, F.J.; Henkin, T.M. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 1993, 74, 475–482. [Google Scholar] [CrossRef]
- Grundy, F.J.; Rollins, S.M.; Henkin, T.M. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: A new role for the discriminator base. J. Bacteriol. 1994, 176, 4518–4526. [Google Scholar] [PubMed]
- Sherwood, A.V.; Grundy, F.J.; Henkin, T.M. T box riboswitches in Actinobacteria: Translational regulation via novel tRNA interactions. Proc. Natl. Acad. Sci. USA 2015, 112, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Rollins, S.M.; Grundy, F.J.; Henkin, T.M. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 1997, 25, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Grundy, F.J.; Winkler, W.C.; Henkin, T.M. tRNA-mediated transcription antitermination in vitro: Codon-anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. USA 2002, 99, 11121–11126. [Google Scholar] [CrossRef] [PubMed]
- Yousef, M.R.; Grundy, F.J.; Henkin, T.M. tRNA requirements for glyQS antitermination: A new twist on tRNA. RNA 2003, 9, 1148–1156. [Google Scholar] [CrossRef] [PubMed]
- Grundy, F.J.; Henkin, T.M. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J. Bacteriol. 2004, 186, 5392–5399. [Google Scholar] [CrossRef] [PubMed]
- Grundy, F.J.; Yousef, M.R.; Henkin, T.M. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J. Mol. Biol. 2005, 346, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Yousef, M.R.; Grundy, F.J.; Henkin, T.M. Structural transitions induced by the interaction between tRNAGlyand the Bacillus subtilis glyQS T box leader RNA. J. Mol. Biol. 2005, 349, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.R.; Henkin, T.M.; Agris, P.F. tRNA regulation of gene expression: Interactions of an mRNA 5'-UTR with a regulatory tRNA. RNA 2006, 12, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Grundy, F.J.; Murphy, B.A.; Henkin, T.M. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 2001, 7, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Lescoute, A.; Leontis, N.B.; Massire, C.; Westhof, E. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005, 33, 2395–2409. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, K.T.; McPhee, S.A.; Ouellet, J.; Lilley, D.M. A structural database for k-turn motifs in RNA. RNA 2010, 16, 1463–1468. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Matsugi, J.; Ishikura, H.; Murao, K. Bacillus subtilis tRNAPr° with the anticodon mo5UGG can recognize the codon CCC. Biochim. Biophys. Acta. 2005, 1728, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Kanaya, S.; Yamada, Y.; Kudo, Y.; Ikemura, T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: Gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 1999, 238, 143–155. [Google Scholar] [CrossRef]
- Saks, M.E.; Conery, J.S. Anticodon-dependent conservation of bacterial tRNA gene sequences. RNA 2007, 13, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.C.; Grundy, F.J.; Henkin, T.M. Conserved elements in tRNAGly contribute to efficient antitermination of the Bacillus subtilis glyQS T box gene. To be submitted for publication.
- Grundy, F.J.; Collins, J.A.; Rollins, S.M.; Henkin, T.M. tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene. RNA 2000, 6, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ferre-D’Amare, A.R. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 2013, 500, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Grigg, J.C.; Chen, Y.; Grundy, F.J.; Henkin, T.M.; Pollack, L.; Ke, A. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc. Natl. Acad. Sci. USA 2013, 110, 7240–7245. [Google Scholar] [CrossRef] [PubMed]
- Sprinzl, M.; Horn, C.; Brown, M.; Ioudovitch, A.; Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998, 26, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Grundy, F.J.; Waters, D.A.; Allen, S.H.; Henkin, T.M. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J. Bacteriol. 1993, 175, 7348–7355. [Google Scholar] [PubMed]
- Zuber, P.; Losick, R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 1987, 169, 2223–2230. [Google Scholar] [PubMed]
- Nakano, M.M.; Zuber, P. Cloning and characterization of srfB, a regulatory gene involved in surfactin production and competence in Bacillus subtilis. J. Bacteriol. 1989, 171, 5347–5353. [Google Scholar] [PubMed]
- Anagnostopoulos, C.; Spizizen, J. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 1961, 81, 741–746. [Google Scholar] [PubMed]
- Putzer, H.; Condon, C.; Brechemier-Baey, D.; Brito, R.; Grunberg-Manago, M. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 2002, 30, 3026–3033. [Google Scholar] [CrossRef] [PubMed]
- Grundy, F.J.; Hodil, S.E.; Rollins, S.M.; Henkin, T.M. Specificity of tRNA-mRNA interactions in Bacillus subtilis tyrS antitermination. J. Bacteriol. 1997, 179, 2587–2594. [Google Scholar] [PubMed]
- Grundy, F.J.; Moir, T.R.; Haldeman, M.T.; Henkin, T.M. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: Disparity between conservation and functional requirements. Nucleic Acids Res. 2002, 30, 1646–1655. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res 2004, 32, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M.; Schimmel, P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 1988, 333, 140–145. [Google Scholar] [CrossRef] [PubMed]
- McClain, W.H.; Foss, K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science 1988, 240, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.; Varani, G. Structure of the acceptor stem of Escherichia coli tRNAAla: Role of the G3.U70 base pair in synthetase recognition. Nucleic Acids Res. 1997, 25, 2083–2090. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Voorhees, R.M.; Kelley, A.C.; Ramakrishnan, V. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat. Struct. Mol. Biol. 2011, 18, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Valle, M.; Sengupta, J.; Swami, N.K.; Grassucci, R.A.; Burkhardt, N.; Nierhaus, K.H.; Agrawal, R.K.; Frank, J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002, 21, 3557–3567. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.; Yarus, M. Transfer RNA structure and coding specificity. II. A D-arm tertiary interaction that restricts coding range. J. Mol. Biol. 1989, 206, 503–511. [Google Scholar] [CrossRef]
- Smith, D.; Yarus, M. Transfer RNA structure and coding specificity. I. Evidence that a D-arm mutation reduces tRNA dissociation from the ribosome. J. Mol. Biol. 1989, 206, 489–501. [Google Scholar] [CrossRef]
- Schultz, D.W.; Yarus, M. tRNA structure and ribosomal function: I. tRNA nucleotide 27-43 mutations enhance first position wobble. J. Mol. Biol. 1994, 235, 1381–1394. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.W.; Yarus, M. tRNA structure and ribosomal function. II. Interaction between anticodon helix and other tRNA mutations. J. Mol. Biol. 1994, 235, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Guigou, L.; Mirande, M. Determinants in tRNA for activation of arginyl-tRNA synthetase: Evidence that tRNA flexibility is required for the induced-fit mechanism. Biochemistry 2005, 44, 16540–16548. [Google Scholar] [CrossRef] [PubMed]
- Cochella, L.; Green, R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science 2005, 308, 1178–1180. [Google Scholar] [CrossRef] [PubMed]
- Yarus, M.; Valle, M.; Frank, J. A twisted tRNA intermediate sets the threshold for decoding. RNA 2003, 9, 384–385. [Google Scholar] [CrossRef] [PubMed]
- Zagryadskaya, E.I.; Kotlova, N.; Steinberg, S.V. Key elements in maintenance of the tRNA L-shape. J. Mol. Biol. 2004, 340, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Cavallo, L.; Tramontano, A. Accurate energies of hydrogen bonded nucleic acid base pairs and triplets in tRNA tertiary interactions. Nucleic Acids Res. 2006, 34, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Rich, A.; RajBhandary, U.L. Transfer RNA: Molecular structure, sequence, and properties. Annu. Rev. Biochem. 1976, 45, 805–860. [Google Scholar] [CrossRef] [PubMed]
- Olejniczak, M.; Dale, T.; Fahlman, R.P.; Uhlenbeck, O.C. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 2005, 12, 788–793. [Google Scholar] [CrossRef] [PubMed]
- Ledoux, S.; Olejniczak, M.; Uhlenbeck, O.C. A sequence element that tunes Escherichia coli tRNAAlaGGC to ensure accurate decoding. Nat. Struct. Mol. Biol. 2009, 16, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Fender, A.; Sissler, M.; Florentz, C.; Giegé, R. Functional idiosyncrasies of tRNA isoacceptors in cognate and noncognate aminoacylation systems. Biochimie 2004, 86, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, S.; Leclerc, F.; Cedergren, R. Structural rules and conformational compensations in the tRNA L-form. J. Mol. Biol. 1997, 266, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.M.; Uhlenbeck, O.C. Is the sequence-specific binding of aminoacyl-tRNAs by EF-Tu universal among bacteria? Nucl Acids Res 2011, 39, 9746–9758. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.M.; Chapman, S.J.; Uhlenbeck, O.C. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc. Natl. Acad. Sci. USA 2011, 108, 5215–5220. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Campbell, F.E.; Zahler, N.H.; Harris, M.E. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 2006, 25, 3998–4007. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.-C.; Grundy, F.J.; Henkin, T.M. Non-Conserved Residues in Clostridium acetobutylicum tRNAAla Contribute to tRNA Tuning for Efficient Antitermination of the alaS T Box Riboswitch. Life 2015, 5, 1567-1582. https://doi.org/10.3390/life5041567
Liu L-C, Grundy FJ, Henkin TM. Non-Conserved Residues in Clostridium acetobutylicum tRNAAla Contribute to tRNA Tuning for Efficient Antitermination of the alaS T Box Riboswitch. Life. 2015; 5(4):1567-1582. https://doi.org/10.3390/life5041567
Chicago/Turabian StyleLiu, Liang-Chun, Frank J. Grundy, and Tina M. Henkin. 2015. "Non-Conserved Residues in Clostridium acetobutylicum tRNAAla Contribute to tRNA Tuning for Efficient Antitermination of the alaS T Box Riboswitch" Life 5, no. 4: 1567-1582. https://doi.org/10.3390/life5041567





