Acetate Metabolism in Anaerobes from the Domain Archaea
Abstract
:1. Introduction
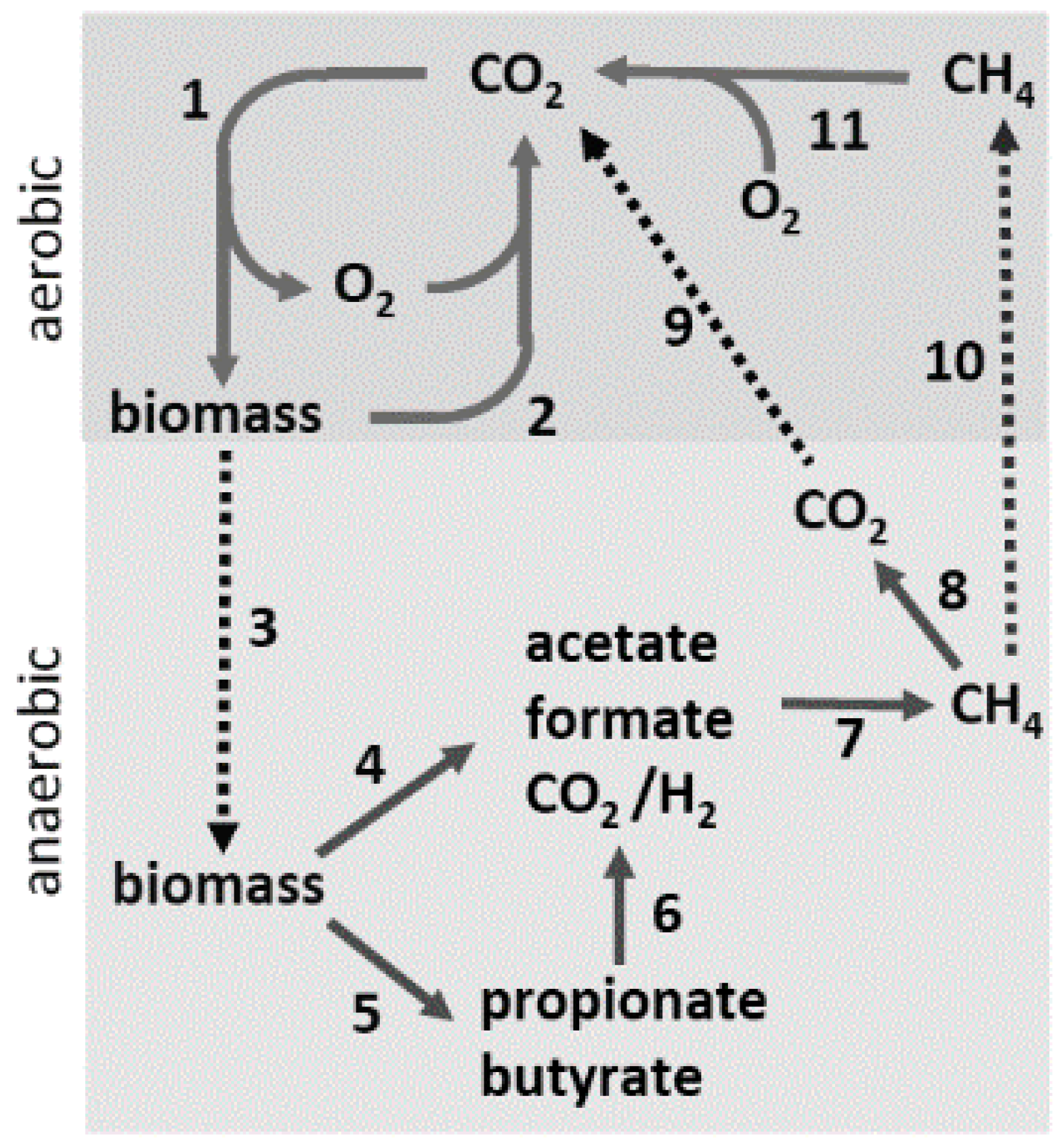
2. Acetate Production
2.1. Heterotrophic Energy-Converting Pathways Producing Acetate

2.2. Chemolithotrophic Energy-Converting Pathways Producing Acetate
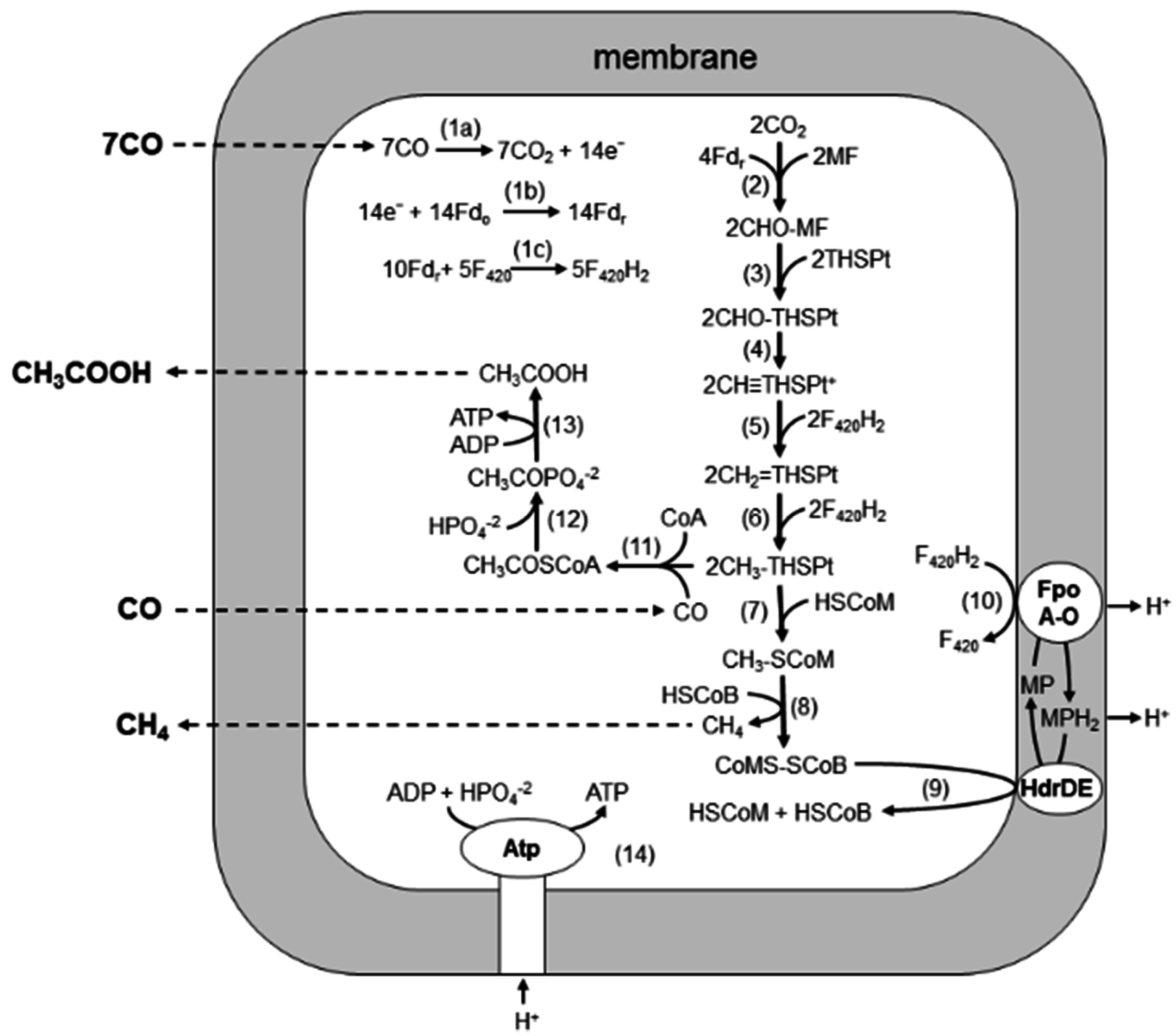
3. Acetate Utilization
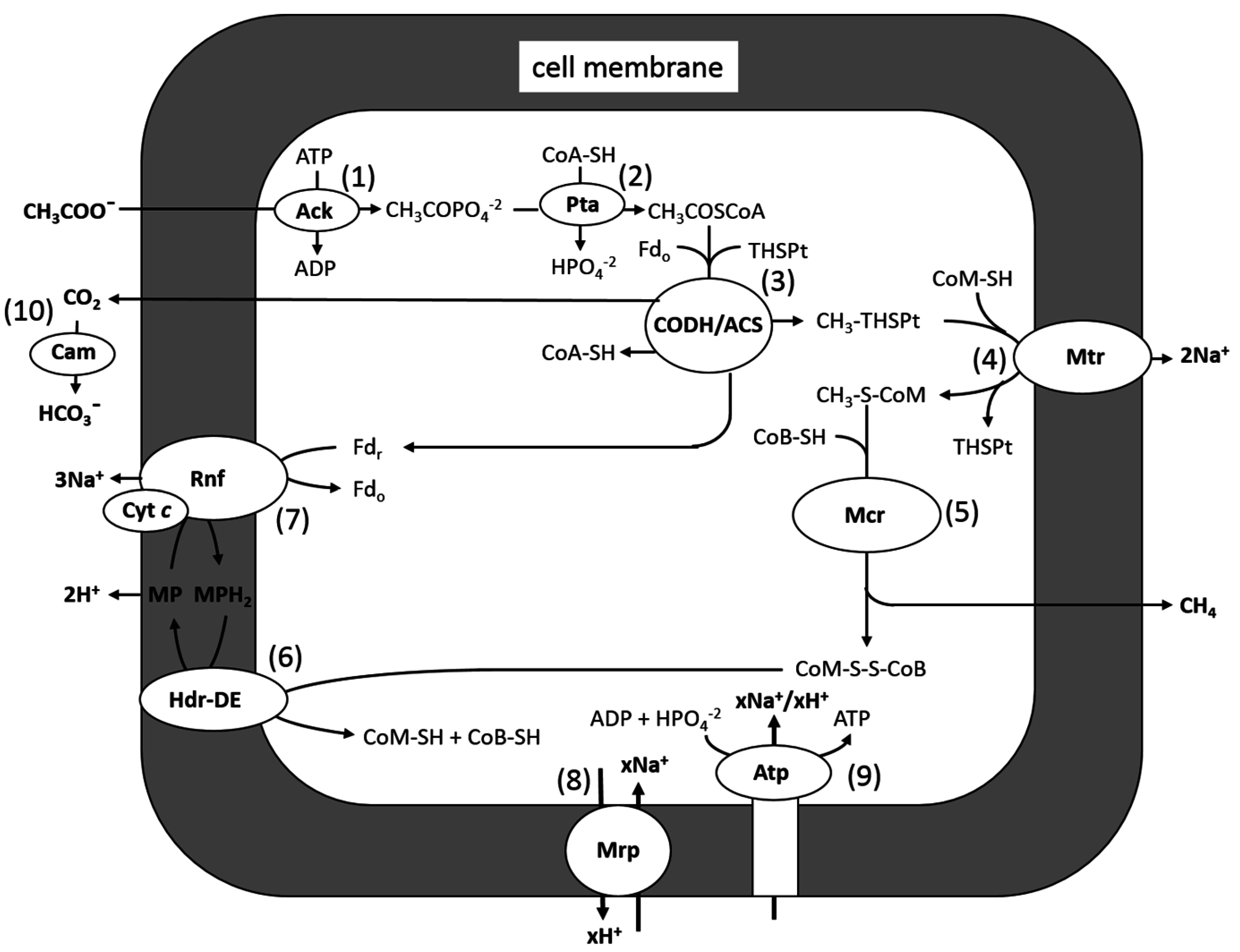
3.1. Acetotrophic Energy-Converting Pathways Producing Methane
3.2. Acetotrophic Energy-Converting Pathways Reducing Exogenous Electron Acceptors
4. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The Unseen Majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef] [PubMed]
- Berg, I.A.; Kockelkorn, D.; Ramos-Vera, W.H.; Say, R.F.; Zarzycki, J.; Hugler, M.; Alber, B.E.; Fuchs, G. Autotrophic Carbon Fixation in Archaea. Nat. Rev. Microbiol. 2010, 8, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Thauer, R.K. Functionalization of Methane in Anaerobic Microorganisms. Angew. Chem. Int. Ed. Engl. 2010, 49, 6712–6713. [Google Scholar] [CrossRef] [PubMed]
- Sapra, R.; Bagramyan, K.; Adams, M.W. A Simple Energy-Conserving System: Proton Reduction Coupled to Proton Translocation. Proc. Natl. Acad. Sci. USA 2003, 100, 7545–7550. [Google Scholar] [CrossRef] [PubMed]
- Selig, M.; Xavier, K.B.; Santos, H.; Schonheit, P. Comparative Analysis of Embden-Meyerhof and Entner-Doudoroff Glycolytic Pathways in Hyperthermophilic Archaea and the Bacterium Thermotoga. Arch. Microbiol. 1997, 167, 217–232. [Google Scholar] [PubMed]
- Perevalova, A.A.; Svetlichny, V.A.; Kublanov, I.V.; Chernyh, N.A.; Kostrikina, N.A.; Tourova, T.P.; Kuznetsov, B.B.; Bonch-Osmolovskaya, E.A. Desulfurococcus Fermentans Sp. Nov., A Novel Hyperthermophilic Archaeon from a Kamchatka Hot Spring, and Emended Description of the Genus Desulfurococcus. Int. J. Syst. Evol. Microbiol. 2005, 55, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Anderson, I.J.; Dharmarajan, L.; Rodriguez, J.; Hooper, S.; Porat, I.; Ulrich, L.E.; Elkins, J.G.; Mavromatis, K.; Sun, H.; Land, M.; et al. The Complete Genome Sequence of Staphylothermus Marinus Reveals Differences in Sulfur Metabolism among Heterotrophic Crenarchaeota. BMC Genomics 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Labes, A.; Schonheit, P. Sugar Utilization in the Hyperthermophilic, Sulfate-Reducing Archaeon Archaeoglobus Fulgidus Strain 7324: Starch Degradation to Acetate and CO2 via a Modified Embden-Meyerhof Pathway and Acetyl-Coa Synthetase (Adp-Forming). Arch. Microbiol. 2001, 176, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Mukund, S.; Adams, M.W. Glyceraldehyde-3-Phosphate Ferredoxin Oxidoreductase, a Novel Tungsten-Containing Enzyme with a Potential Glycolytic Role in the Hyperthermophilic Archaeon Pyrococcus furiosus. J. Biol. Chem. 1995, 270, 8389–8392. [Google Scholar] [CrossRef] [PubMed]
- Blamey, J.M.; Adams, M.W.W. Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1993, 1161, 19–27. [Google Scholar] [CrossRef]
- Ma, K.; Hutchins, A.; Sung, S.J.; Adams, M.W. Pyruvate Ferredoxin Oxidoreductase from the Hyperthermophilic Archaeon, Pyrococcus furiosus, Functions as a Coa-Dependent Pyruvate Decarboxylase. Proc. Natl. Acad. Sci. USA 1997, 94, 9608–9613. [Google Scholar] [CrossRef] [PubMed]
- Musfeldt, M.; Selig, M.; Schonheit, P. Acetyl Coenzyme a Synthetase (ADP Forming) from the Hyperthermophilic Archaeon Pyrococcus furiosus: Identification, Cloning, Separate Expression of the Encoding Genes, Acdai and Acdbi, in Escherichia Coli, and in vitro Reconstitution of the Active Heterotetrameric Enzyme from Its Recombinant Subunits. J. Bacteriol. 1999, 181, 5885–5888. [Google Scholar] [PubMed]
- Glasemacher, J.; Bock, A.K.; Schmid, R.; Schonheit, P. Purification and Properties of Acetyl-Coa Synthetase (Adp-Forming), an Archaeal Enzyme of Acetate Formation and Atp Synthesis, from the Hyperthermophile Pyrococcus furiosus. Eur. J. Biochem. 1997, 244, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Schafer, T.; Selig, M.; Schonheit, P. Acetyl-Coa Synthetase (Adp Forming) in Archaea, a Novel Enzyme Involved in Acetate Formation and Atp Synthesis. Arch. Microbiol. 1993, 159, 72–83. [Google Scholar] [CrossRef]
- Brasen, C.; Schmidt, M.; Grotzinger, J.; Schonheit, P. Reaction Mechanism and Structural Model of ADP-Forming Acetyl-Coa Synthetase from the Hyperthermophilic Archaeon Pyrococcus furiosus: Evidence for a Second Active Site Histidine Residue. J. Biol. Chem. 2008, 283, 15409–15418. [Google Scholar] [CrossRef] [PubMed]
- Lessner, D.J.; Li, L.; Li, Q.; Rejtar, T.; Andreev, V.P.; Reichlen, M.; Hill, K.; Moran, J.J.; Karger, B.L.; Ferry, J.G. An Unconventional Pathway for Reduction of CO2 to Methane in Co-Grown Methanosarcina acetivorans Revealed by Proteomics. Proc. Natl. Acad. Sci. USA 2006, 103, 17921–17926. [Google Scholar] [CrossRef] [PubMed]
- Rother, M.; Oelgeschlager, E.; Metcalf, W.M. Genetic and Proteomic Analyses of Co Utilization by Methanosarcina acetivorans. Arch. Microbiol. 2007, 188, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Ferry, J.G. Fundamentals of Methanogenic Pathways That Are Key to the Biomethanation of Complex Biomass. Curr. Opin. Biotechnol. 2011, 22, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Ferry, J.G. How to Make a Living Exhaling Methane. Annu. Rev. Microbiol. 2010, 64, 453–473. [Google Scholar] [CrossRef] [PubMed]
- Thauer, R.K.; Kaster, A.K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic Archaea: Ecologically Relevant Differences in Energy Conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Whitman, W.B. Metabolic, Phylogenetic, and Ecological Diversity of the Methanogenic Archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Welte, C.; Deppenmeier, U. Re-Evaluation of the Function of the F420 Dehydrogenase in Electron Transport of Methanosarcina Mazei. FEBS J. 2011, 278, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Vepachedu, V.R.; Ferry, J.G. Role of the Fused Corrinoid/Methyl Transfer Protein Cmta during Co-Dependent Growth of Methanosarcina acetivorans. J. Bacteriol. 2012, 194, 4161–4168. [Google Scholar] [CrossRef] [PubMed]
- Moller-Zinkhan, D.; Thauer, R.K. Anaerobic Lactate Oxidation to 3 CO2 by Archaeoglobus Fulgidus via The Carbon Monoxide Dehydrogenase Pathway. Demonstration of the Acetyl-Coa Carbon-Carbon Cleavage Reaction in Cell Extracts. Arch. Microbiol. 1990, 153, 215–218. [Google Scholar] [CrossRef]
- Gorris, L.; Voet, A.; van der Drift, C. Structural Characteristics of Methanogenic Cofactors in the Non-Methanogenic Archaebacterium Archaeoglobus fulgidus. Biofactors 1991, 3, 29–35. [Google Scholar] [PubMed]
- Schworer, B.; Breitung, J.; Klein, A.R.; Stetter, K.O.; Thauer, R.K. Formylmethanofuran: Tetrahydromethanopterin Formyltransferase and N5,N10-Methylenetetrahydromethanopterin Dehydrogenase from the Sulfate-Reducing Archaeoglobus Fulgidus: Similarities with the Enzymes from Methanogenic Archaea. Arch. Microbiol. 1993, 159, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Kunow, J.; Linder, D.; Stetter, K.O.; Thauer, R.K. F420h2:Quinone Oxidoreductase from Archaeoglobus Fulgidus. Characterization of a Membrane-Bound Multisubunit Complex Containing Fad and Iron-Sulfur Clusters. Eur. J. Biochem. 1994, 223, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.R.; Reed, D.W.; Millstein, J.H.; Hartzell, P.L.; Grahame, D.A.; Demoll, E. Acetyl-Coa Decarbonylase/Synthase Complex from Archaeoglobus fulgidus. Arch. Microbiol. 1998, 169, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Ingram-Smith, C.; Smith, K.S. Amp-Forming Acetyl-Coa Synthetases in Archaea Show Unexpected Diversity in Substrate Utilization. Archaea 2007, 2, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Henstra, A.M.; Dijkema, C.; Stams, A.J. Archaeoglobus Fulgidus Couples CO Oxidation to Sulfate Reduction and Acetogenesis with Transient Formate Accumulation. Environ. Microbiol. 2007, 9, 1836–1841. [Google Scholar] [CrossRef] [PubMed]
- Musfeldt, M.; Schonheit, P. Novel Type of Adp-Forming Acetyl Coenzyme a Synthetase in Hyperthermophilic Archaea: Heterologous Expression and Characterization of Isoenzymes from the Sulfate Reducer Archaeoglobus fulgidus and the Methanogen Methanococcus jannaschii. J. Bacteriol. 2002, 184, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Battistuzzi, F.U.; Feijao, A.; Hedges, S.B. A Genomic Timescale of Prokaryote Evolution: Insights into the Origin of Methanogenesis, Phototrophy, and the Colonization of Land. BMC Evol. Biol. 2004, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, Y.; Yamada, K.; Yoshida, N.; Maruyama, S.; Isozaki, Y. Evidence from Fluid Inclusions for Microbial Methanogenesis in the Early Archaean Era. Nature 2006, 440, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Rothman, D.H.; Fournier, G.P.; French, K.L.; Alm, E.J.; Boyle, E.A.; Cao, C.; Summons, R.E. Methanogenic Burst in the End-Permian Carbon Cycle. Proc. Natl. Acad. Sci. USA. 2014, 111, 5462–5467. [Google Scholar] [CrossRef] [PubMed]
- Buss, K.A.; Cooper, D.R.; Ingram-Smith, C.; Ferry, J.G.; Sanders, D.A.; Hasson, M.S. Urkinase: Structure of Acetate Kinase, a Member of the Askha Superfamily of Phosphotransferases. J. Bacteriol. 2001, 183, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, P.A.; Chang, B. The Evolution of Acetyl-Coa Synthase. Orig. Life Evol. Biosph. 2001, 31, 403–434. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, G.; Thauer, R.K. The Na+ Translocating Methyltransferase Complex from Methanogenic Archaea. Biochim. Biophys. Acta 2001, 1505, 28–36. [Google Scholar] [CrossRef]
- Shima, S.; Warkentin, E.; Thauer, R.K.; Ermler, U. Structure and Function of Enzymes Involved in the Methanogenic Pathway Utilizing Carbon Dioxide and Molecular Hydrogen. J. Biosci. Bioeng. 2002, 93, 519–530. [Google Scholar] [CrossRef]
- Suharti, S.; Wang, M.; de Vries, S.; Ferry, J.G. Characterization of the RNFB and RNFG Subunits of the RNF Complex from the Archaeon Methanosarcina acetivorans. PLoS ONE 2014, 9, E97966. [Google Scholar] [CrossRef] [PubMed]
- Gorrell, A.; Lawrence, S.H.; Ferry, J.G. Structural and Kinetic Analyses of Arginine Residues in the Active-Site of the Acetate Kinase from Methanosarcina thermophila. J. Biol. Chem. 2005, 280, 10731–10742. [Google Scholar] [CrossRef] [PubMed]
- Miles, R.D.; Gorrell, A.; Ferry, J.G. Evidence for a Transition State Analog, Mgadp-Aluminum Fluoride-Acetate, in Acetate Kinase from Methanosarcina thermophila. J. Biol. Chem. 2002, 277, 22547–22552. [Google Scholar] [CrossRef] [PubMed]
- Ingram-Smith, C.; Gorrell, A.; Lawrence, S.H.; Iyer, P.; Smith, K.; Ferry, J.G. Identification of the Acetate Binding Site in the Methanosarcina thermophila Acetate Kinase. J. Bacteriol. 2005, 187, 2386–2394. [Google Scholar] [CrossRef] [PubMed]
- Miles, R.D.; Iyer, P.P.; Ferry, J.G. Site-Directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina thermophila. J. Biol. Chem. 2001, 276, 45059–45064. [Google Scholar] [CrossRef] [PubMed]
- Ingram-Smith, C.; Barber, R.D.; Ferry, J.G. The Role of Histidines in the Acetate Kinase from Methanosarcina thermophila. J. Biol. Chem. 2000, 275, 33765–33770. [Google Scholar] [CrossRef] [PubMed]
- Singh-Wissmann, K.; Miles, R.D.; Ingram-Smith, C.; Ferry, J.G. Identification of Essential Arginines in the Acetate Kinase from Methanosarcina thermophila. Biochemistry 2000, 39, 3671–3677. [Google Scholar] [CrossRef] [PubMed]
- Singh-Wissmann, K.; Ingram-Smith, C.; Miles, R.D.; Ferry, J.G. Identification of Essential Glutamates in the Acetate Kinase From Methanosarcina thermophila. J. Bacteriol. 1998, 180, 1129–1134. [Google Scholar] [PubMed]
- Gorrell, A.; Ferry, J.G. Investigation of the Methanosarcina thermophila Acetate Kinase Mechanism by Fluorescence Quenching. Biochemistry 2007, 46, 14170–14176. [Google Scholar] [CrossRef] [PubMed]
- Ingram-Smith, C.; Wharton, J.; Reinholz, C.; Doucet, T.; Hesler, R.; Smith, K. The Role of Active Site Residues in Atp Binding and Catalysis in the Methanosarcina thermophila Acetate Kinase. Life 2015, 5, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Iyer, P.P.; Lawrence, S.H.; Luther, K.B.; Rajashankar, K.R.; Yennawar, H.P.; Ferry, J.G.; Schindelin, H. Crystal Structure of Phosphotransacetylase from the Methanogenic Archaeon Methanosarcina thermophila. Structure 2004, 12, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Abbanat, D.R.; Ferry, J.G. Synthesis of Acetyl-Coa by the Carbon Monoxide Dehydrogenase Complex from Acetate-Grown Methanosarcina thermophila. J. Bacteriol. 1990, 172, 7145–7150. [Google Scholar] [PubMed]
- Raybuck, S.A.; Ramer, S.E.; Abbanat, D.R.; Peters, J.W.; Orme-Johnson, W.H.; Ferry, J.G.; Walsh, C.T. Demonstration of Carbon-Carbon Bond Cleavage of Acetyl Coenzyme a by Using Isotopic Exchange Catalyzed by the Co Dehydrogenase Complex from Acetate-Grown Methanosarcina thermophila. J. Bacteriol. 1991, 173, 929–932. [Google Scholar] [PubMed]
- Terlesky, K.C.; Nelson, M.J.K.; Ferry, J.G. Isolation of an Enzyme Complex with Carbon Monoxide Dehydrogenase Activity Containing a Corrinoid and Nickel from Acetate-Grown Methanosarcina thermophila. J. Bacteriol. 1986, 168, 1053–1058. [Google Scholar] [PubMed]
- Eggen, R.I.L.; Vankranenburg, R.; Vriesema, A.J.M.; Geerling, A.C.M.; Verhagen, M.F.J.M.; Hagen, W.R.; Devos, W.M. Carbon Monoxide Dehydrogenase from Methanosarcina Frisia Gö1. Characterization of the Enzyme and the Regulated Expression of Two Operon-Like Cdh Gene Clusters. J. Biol. Chem. 1996, 271, 14256–14263. [Google Scholar] [PubMed]
- Jetten, M.S.M.; Hagen, W.R.; Pierik, A.J.; Stams, A.J.M.; Zehnder, A.J.B. Paramagnetic Centers and Acetyl-Coenzyme a CO Exchange Activity of Carbon Monoxide Dehydrogenase from Methanothrix soehngenii. Eur. J. Biochem. 1991, 195, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Grahame, D.A. Substrate and Cofactor Reactivity of A Carbon Monoxide Dehydrogenase Corrinoid Enzyme Complex. Stepwise Reduction of Iron Sulfur and Corrinoid Centers, the Corrinoid Co2+/1+ Redox Midpoint Potential, and Overall Synthesis of Acetyl-Coa. Biochemistry 1993, 32, 10786–10793. [Google Scholar] [CrossRef] [PubMed]
- Grahame, D.A.; Demoll, E. Substrate and Accessory Protein Requirements and Thermodynamics of Acetyl-Coa Synthesis and Cleavage in Methanosarcina barkeri. Biochemistry 1995, 34, 4617–4624. [Google Scholar] [CrossRef] [PubMed]
- Jetten, M.S.M.; Stams, A.J.M.; Zehnder, A.J.B. Purification and Characterization of an Oxygen-Stable Carbon Monoxide Dehydrogenase of Methanothrix soehngenii. FEBS Lett. 1989, 181, 437–441. [Google Scholar] [CrossRef]
- Eggen, R.I.L.; Geerling, A.C.M.; Jetten, M.S.M.; Devos, W.M. Cloning, Expression, and Sequence Analysis of the Genes for Carbon Monoxide Dehydrogenase of Methanothrix soehngenii. J. Biol. Chem. 1991, 266, 6883–6887. [Google Scholar] [PubMed]
- Jetten, M.S.M.; Pierik, A.J.; Hagen, W.R. Epr Characterization of a High-Spin System in Carbon Monoxide Dehydrogenase from Methanothrix soehngenii. Eur. J. Biochem. 1991, 202, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Terlesky, K.C.; Ferry, J.G. Ferredoxin Requirement for Electron Transport from the Carbon Monoxide Dehydrogenase Complex to a Membrane-Bound Hydrogenase in Acetate-Grown Methanosarcina thermophila. J. Biol. Chem. 1988, 263, 4075–4079. [Google Scholar] [PubMed]
- Terlesky, K.C.; Ferry, J.G. Purification and Characterization of a Ferredoxin from Acetate-Grown Methanosarcina thermophila. J. Biol. Chem. 1988, 263, 4080–4082. [Google Scholar] [PubMed]
- Fischer, R.; Thauer, R.K. Ferredoxin-Dependent Methane Formation from Acetate in Cell Extracts of Methanosarcina barkeri (Strain Ms). FEBS Lett. 1990, 269, 368–372. [Google Scholar] [CrossRef]
- Ferry, J.G.; House, C.H. The Stepwise Evolution of Early Life Driven by Energy Conservation. Mol. Biol. Evol. 2006, 23, 1286–1292. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.; Russell, M.J. On the Origin of Biochemistry at an Alkaline Hydrothermal Vent. Philos. Trans. R. Soc. B 2006, 362, 1887–1926. [Google Scholar] [CrossRef] [PubMed]
- Abbanat, D.R.; Ferry, J.G. Resolution of Component Proteins in an Enzyme Complex from Methanosarcina thermophila Catalyzing the Synthesis or Cleavage of Acetyl-Coa. Proc. Natl. Acad. Sci. USA 1991, 88, 3272–3276. [Google Scholar] [CrossRef] [PubMed]
- Clements, A.P.; Ferry, J.G. Cloning, Nucleotide Sequence, and Transcriptional Analyses of the Gene Encoding a Ferredoxin from Methanosarcina thermophila. J. Bacteriol. 1992, 174, 5244–5250. [Google Scholar] [PubMed]
- Gong, W.; Hao, B.; Wei, Z.; Ferguson, D.J., Jr.; Tallant, T.; Krzycki, J.A.; Chan, M.K. Structure of the A2e2 Ni-Dependent Co Dehydrogenase Component of the Methanosarcina barkeri Acetyl-Coa Decarbonylase/Synthase Complex. Proc. Natl. Acad. Sci. USA 2008, 105, 9558–9563. [Google Scholar] [CrossRef] [PubMed]
- Murakami, E.; Ragsdale, S.W. Evidence for Intersubunit Communication during Acetyl-Coa Cleavage by the Multienzyme Co Dehydrogenase/Acetyl-Coa Synthase Complex from Methanosarcina thermophila Evidence That the Beta Subunit Catalyzes C-C and C-S Bond Cleavage. J. Biol. Chem. 2000, 275, 4699–4707. [Google Scholar] [CrossRef] [PubMed]
- Grahame, D.A.; Demoll, E. Partial Reactions Catalyzed by Protein Components of the Acetyl-Coa Decarbonylase Synthase Enzyme Complex from Methanosarcina barkeri. J. Biol. Chem. 1996, 271, 8352–8358. [Google Scholar] [PubMed]
- Funk, T.; Gu, W.W.; Friedrich, S.; Wang, H.X.; Gencic, S.; Grahame, D.A.; Cramer, S.P. Chemically Distinct Ni Sites in the A-Cluster in Subunit Beta of the Acetyl-Coa Decarbonylase/Synthase Complex from Methanosarcina thermophila: Ni L-Edge Absorption And X-Ray Magnetic Circular Dichroism Analyses. J. Am. Chem. Soc. 2004, 126, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Gencic, S.; Grahame, D.A. Nickel in Subunit B of the Acetyl-Coa Decarbonylase/Synthase Multienzyme Complex in Methanogens. J. Biol. Chem. 2003, 278, 6101–6110. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.W.; Gencic, S.; Cramer, S.P.; Grahame, D.A. The A-Cluster in Subunit Beta of the Acetyl-Coa Decarbonylase/Synthase Complex from Methanosarcina thermophila: Ni and Fe K-Edge Xanes and Exafs Analyses. J. Am. Chem. Soc. 2003, 125, 15343–15351. [Google Scholar] [CrossRef] [PubMed]
- Ragsdale, S.W. Nickel and the Carbon Cycle. J. Inorg. Biochem. 2007, 101, 1657–1666. [Google Scholar] [CrossRef] [PubMed]
- Gencic, S.; Grahame, D.A. Two Separate One-Electron Steps in the Reductive Activation of the a Cluster in Subunit Beta of the Acds Complex in Methanosarcina thermophila. Biochemistry 2008, 47, 5544–5455. [Google Scholar] [CrossRef] [PubMed]
- Gencic, S.; Duin, E.C.; Grahame, D.A. Tight Coupling of Partial Reactions in the Acetyl-Coa Decarbonylase/Synthase (Acds) Multienzyme Complex from Methanosarcina thermophila: Acetyl C-C Bond Fragmentation at the a Cluster Promoted by Protein Conformational Changes. J. Biol. Chem. 2010, 285, 15450–15463. [Google Scholar] [CrossRef] [PubMed]
- Gencic, S.; Kelly, K.; Ghebreamlak, S.; Duin, E.C.; Grahame, D.A. Different Modes of Carbon Monoxide Binding to Acetyl-Coa Synthase and the Role of a Conserved Phenylalanine in the Coordination Environment of Nickel. Biochemistry 2013, 52, 1705–1716. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, P.E.; Lu, W.P.; Ragsdale, S.W.; Ferry, J.G. Characterization of the Metal Centers of the Corrinoid/Iron- Sulfur Component of the Co Dehydrogenase Enzyme Complex from Methanosarcina thermophila by Epr Spectroscopy and Spectroelectrochemistry. J. Biol. Chem. 1993, 268, 325–329. [Google Scholar] [PubMed]
- Grahame, D.A. Catalysis of Acetyl-Coa Cleavage and Tetrahydrosarcinapterin Methylation by a Carbon Monoxide Dehydrogenase-Corrinoid Enzyme Complex. J. Biol. Chem. 1991, 266, 22227–22233. [Google Scholar] [PubMed]
- Maupin-Furlow, J.; Ferry, J.G. Characterization of the Cdhd and Cdhe Genes Encoding Subunits of the Corrinoid Iron-Sulfur Enzyme of the Co Dehydrogenase Complex from Methanosarcina thermophila. J. Bacteriol. 1996, 178, 340–346. [Google Scholar] [PubMed]
- Liu, J.S.; Marison, I.W.; von Stockar, U. Microbial Growth by a Net Heat Up-Take: A Calorimetric and Thermodynamic Study on Acetotrophic Methanogenesis by Methanosarcina barkeri. Biotechnol. Bioeng. 2001, 75, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Alber, B.E.; Ferry, J.G. A Carbonic Anhydrase from the Archaeon Methanosarcina thermophila. Proc. Natl. Acad. Sci. USA 1994, 91, 6909–6913. [Google Scholar] [CrossRef] [PubMed]
- Macauley, S.R.; Zimmerman, S.A.; Apolinario, E.E.; Evilia, C.; Hou, Y.; Ferry, J.G.; Sowers, K.R. The Archetype G-Class Carbonic Anhydrase (Cam) Contains Iron when Synthesized in vivo. Biochemistry 2009, 48, 817–819. [Google Scholar] [CrossRef] [PubMed]
- Tripp, B.C.; Bell, C.B.; Cruz, F.; Krebs, C.; Ferry, J.G. A Role for Iron in an Ancient Carbonic Anhydrase. J. Biol. Chem. 2004, 279, 6683–6687. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.A.; Ferry, J.G. Proposal for a Hydrogen Bond Network in the Active Site of the Prototypic G-Class Carbonic Anhydrase. Biochemistry 2006, 45, 5149–5157. [Google Scholar] [CrossRef] [PubMed]
- Hovey, R.; Lentes, S.; Ehrenreich, A.; Salmon, K.; Saba, K.; Gottschalk, G.; Gunsalus, R.P.; Deppenmeier, U. DNA Microarray Analysis of Methanosarcina Mazei Go1 Reveals Adaptation to Different Methanogenic Substrates. Mol. Genet. Genomics 2005, 273, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Q.; Rohlin, L.; Kim, U.; Salmon, K.; Rejtar, T.; Gunsalus, R.P.; Karger, B.L.; Ferry, J.G. Quantitative Proteomic and Microarray Analysis of the Archaeon Methanosarcina acetivorans Grown With Acetate versus Methanol. J. Proteome Res. 2007, 6, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, L.; Rejtar, T.; Lessner, D.J.; Karger, B.L.; Ferry, J.G. Electron Transport in the Pathway of Acetate Conversion to Methane in the Marine Archaeon Methanosarcina acetivorans. J. Bacteriol. 2006, 188, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Pisa, K.Y.; Weidner, C.; Maischak, H.; Kavermann, H.; Muller, V. The Coupling Ion in the Methanoarchaeal ATP Synthases: H+ vs. Na+ in the A1A0 ATP Synthase from the Archaeon Methanosarcina mazei Gö1. FEMS Microbiol. Lett. 2007, 277, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Hedderich, R. Energy-Converting [Nife] Hydrogenases from Archaea and Extremophiles: Ancestors of Complex I. J. Bioenerg. Biomembr. 2004, 36, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Meuer, J.; Kuettner, H.C.; Zhang, J.K.; Hedderich, R.; Metcalf, W.W. Genetic Analysis of the Archaeon Methanosarcina barkeri Fusaro Reveals a Central Role for ECH Hydrogenase and Ferredoxin in Methanogenesis and Carbon Fixation. Proc. Natl. Acad. Sci. USA 2002, 99, 5632–5637. [Google Scholar] [CrossRef] [PubMed]
- Heiden, S.; Hedderich, R.; Setzke, E.; Thauer, R.K. Purification of a Cytochrome-B Containing H2-Heterodisulfide Oxidoreductase Complex from Membranes of Methanosarcina barkeri. Eur. J. Biochem. 1993, 213, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Deppenmeier, U. The Membrane-Bound Electron Transport System of Methanosarcina Species. J. Bioenerg. Biomembr. 2004, 36, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Ueno, A.; Naganuma, T.; Kaneko, K. Methanosarcina subterranea sp. nov., a methanogenic archaeon isolated from a deep subsurface diatomaceous shale formation of northernmost japan. Int. J. Syst. Evol. Microbiol. 2015, 65, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; Schirmack, J.; Ganzert, L.; Morozova, D.; Mangelsdorf, K. Methanosarcina soligelidi sp. nov., a desiccation and freeze-thaw resistant methanogenic archaeon isolated from a siberian permafrost-affected soil. Int. J. Syst. Evol. Microbiol. 2013, 64, 3478–3484. [Google Scholar]
- Von Klein, D.; Arab, H.; Volker, H.; Thomm, M. Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles 2002, 6, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Blotevogel, K.H.; Fischer, U. Transfer of Methanococcus Frisius to the Genus Methanosarcina as Methanosarcina Frisia Comb. Nov. Int. J. Syst. Bacteriol. 1989, 39, 91–92. [Google Scholar] [CrossRef]
- Zinder, S.H.; Sowers, K.R.; Ferry, J.G. Methanosarcina thermophila sp. nov., a Thermophilic, Acetotrophic, Methane-Producing Bacterium. Int. J. Syst. Bacteriol. 1985, 35, 522–523. [Google Scholar] [CrossRef]
- Shimizu, S.; Upadhye, R.; Ishijima, Y.; Naganuma, T. Methanosarcina horonobensis sp. nov., a methanogenic archaeon isolated from a deep subsurface miocene formation. Int. J. Syst. Evol. Microbiol. 2011, 61, 2503–2507. [Google Scholar] [CrossRef] [PubMed]
- Lyimo, T.J.; Pol, A.; den Camp, H.J.O.; Harhangi, H.R.; Vogels, G.D. Methanosarcina semesiae sp. nov., a dimethylsulfide-utilizing methanogen from mangrove sediment. Int. J. Syst. Evol. Microbiol. 2000, 50, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Cabrol, N.; Villemur, R.; Perrier, J.; Jacob, F.; Foullet, B.; Chambon, P. Isolation of a methanogenic bacterium, Methanosarcina sp. strain FR, for its abiilty to degrade high concentrations of perchloroethylene. Can. J. Microbiol. 1998, 44, 1142–1147. [Google Scholar] [CrossRef]
- Zhilina, T.N.; Zavarzin, G.A. Comparative cytology of Methanosarcinae and description of Methanosarcina vacuolata sp. nova. Microbiology 1979, 48, 223–228. [Google Scholar]
- Sowers, K.R.; Baron, S.F.; Ferry, J.G. Methanosarcina acetivorans sp. nov., An Acetotrophic Methane-Producing Bacterium Isolated from Marine Sediments. Appl. Environ. Microbiol. 1984, 47, 971–978. [Google Scholar] [PubMed]
- Ganzert, L.; Schirmack, J.; Alawi, M.; Mangelsdorf, K.; Sand, W.; Hillebrand-Voiculescu, A.; Wagner, D. Methanosarcina spelaei sp. nov., a Methanogenic Archaeon Isolated from a Floating Biofilm of a Subsurface Sulphurous Lake. Int. J. Syst. Evol. Microbiol. 2014, 64, 3478–3484. [Google Scholar] [CrossRef] [PubMed]
- Simankova, M.V.; Parshina, S.N.; Tourova, T.P.; Kolganova, T.V.; Zehnder, A.J.; Nozhevnikova, A.N. Methanosarcina lacustris sp. nov., a New Psychrotolerant Methanogenic Archaeon from Anoxic Lake Sediments. Syst. Appl. Microbiol. 2001, 24, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, L.; Rejtar, T.; Lessner, D.J.; Karger, B.L.; Ferry, J.G. Electron Transport in the Pathway of Acetate Conversion to Methane in the Marine Archaeon Methanosarcina acetivorans. J. Bacteriol. 2006, 188, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tomb, J.F.; Ferry, J.G. Electron Transport in Acetate-Grown Methanosarcina acetivorans. BMC Microbiol. 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Hreha, T.N.; Mezic, K.G.; Herce, H.D.; Duffy, E.D.; Bourges, A.; Pryshchep, S.; Juarez, O.; Barquera, B. The Complete Topology of the RNF Complex from Vibrio Cholerae. Biochemistry 2015, 54, 2443–2455. [Google Scholar] [CrossRef] [PubMed]
- Saeki, K.; Kumagai, H. The Rnf Gene Products in Rhodobacter Capsulatus Play an Essential Role in Nitrogen Fixation during Anaerobic Dmso-Dependent Growth in the Dark. Arch. Microbiol. 1998, 169, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Muller, V.; Imkamp, F.; Biegel, E.; Schmidt, S.; Dilling, S. Discovery of a Ferredoxin: Nad+-Oxidoreductase (RNF) in Acetobacterium Woodii: A Novel Potential Coupling Site in Acetogens. Ann. N. Y. Acad. Sci. 2008, 1125, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Seedorf, H.; Fricke, W.F.; Veith, B.; Bruggemann, H.; Liesegang, H.; Strittmatter, A.; Miethke, M.; Buckel, W.; Hinderberger, J.; Li, F.; et al. The Genome of Clostridium kluyveri, a Strict Anaerobe with Unique Metabolic Features. Proc. Natl. Acad. Sci. USA 2008, 105, 2128–2133. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Kohler, J.; Hurek, T.; Reinhold-Hurek, B. A Novel Regulatory Role of the RNF Complex of Azoarcus SP. Strain Bh72. Mol. Microbiol. 2011, 83, 408–422. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, P.L.; Zhang, T.; Dar, S.A.; Leang, C.; Lovley, D.R. The RNF Complex of Clostridium Ljungdahlii Is a Proton-Translocating Ferredoxin: Nad+ Oxidoreductase Essential for Autotrophic Growth. mBio 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Kuehl, J.V.; Price, M.N.; Ray, J.; Deutschbauer, A.M.; Arkin, A.P.; Stahl, D.A. The Energy-Conserving Electron Transfer System Used by Desulfovibrio alaskensis Strain G20 During Pyruvate Fermentation Involves Reduction of Endogenously Formed Fumarate and cytoplasmic and membrane-bound complexes, Hdr-Flox and RNF. Environ. Microbiol. 2014, 16, 3463–3486. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, K.; Welte, C.; Deppenmeier, U.; Muller, V. Electron Transport During Aceticlastic Methanogenesis by Methanosarcina acetivorans Involves a Sodium-Translocating RNF Complex. FEBS J. 2012, 279, 4444–4452. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, K.; Leone, V.; Faraldo-Gomez, J.D.; Muller, V. Promiscuous Archaeal ATP Synthase Concurrently Coupled to Na+ And H+ Translocation. Proc. Natl. Acad. Sci. USA 2012, 109, 947–952. [Google Scholar] [CrossRef] [PubMed]
- Jasso-Chavez, R.; Apolinario, E.E.; Sowers, K.R.; Ferry, J.G. MRPA Functions in Energy Conversion during Acetate-Dependent Growth of Methanosarcina acetivorans. J. Bacteriol. 2013, 195, 3987–3994. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.S.; Ingram-Smith, C. Methanosaeta, the Forgotten Methanogen? Trends Microbiol. 2007, 7, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.; Welte, C.; Deppenmeier, U. Acetate Activation in Methanosaeta Thermophila: Characterization of the Key Enzymes Pyrophosphatase and Acetyl-Coa Synthetase. Archaea 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jetten, M.S.M.; Stams, A.J.M.; Zehnder, A.J.B. Isolation and Characterization of Acetyl-Coenzyme a Synthetase from Methanothrix soehngenii. J. Bacteriol. 1989, 171, 5430–5435. [Google Scholar] [PubMed]
- Eggen, R.I.L.; Geerling, A.C.M.; Boshoven, A.B.P.; Devos, W.M. Cloning, Sequence Analysis, and Functional Expression of the Acetyl Coenzyme a Synthetase Gene from Methanothrix soehngenii in Escherichia Coli. J. Bacteriol. 1991, 173, 6383–6389. [Google Scholar] [PubMed]
- Aceti, D.J.; Ferry, J.G. Purification and Characterization of Acetate Kinase from Acetate-Grown Methanosarcina thermophila. J. Biol. Chem. 1988, 263, 15444–15448. [Google Scholar] [PubMed]
- Tor, J.M.; Kashefi, K.; Lovley, D.R. Acetate Oxidation Coupled to Fe(III) Reduction in Hyperthermophilic Microorganisms. Appl. Environ. Microbiol. 2001, 67, 1363–1365. [Google Scholar] [CrossRef] [PubMed]
- Slobodkina, G.B.; Kolganova, T.V.; Querellou, J.; Bonch-Osmolovskaya, E.A.; Slobodkin, A.I. Geoglobus Acetivorans sp. nov., an Iron(III)-Reducing Archaeon from a Deep-Sea Hydrothermal Vent. Int. J. Syst. Evol. Microbiol. 2009, 59, 2880–2883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slobodkin, A.; Campbell, B.; Cary, S.C.; Bonch-Osmolovskaya, E.; Jeanthon, C. Evidence for the Presence of Thermophilic Fe(III)-Reducing Microorganisms in Deep-Sea Hydrothermal Vents at 13 Degrees N (East Pacific Rise). FEMS Microbiol. Ecol. 2001, 36, 235–243. [Google Scholar] [PubMed]
- Seyler, L.M.; Mcguinness, L.M.; Kerkhof, L.J. Crenarchaeal Heterotrophy in Salt Marsh Sediments. ISME J. 2014, 8, 1534–1543. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferry, J.G. Acetate Metabolism in Anaerobes from the Domain Archaea. Life 2015, 5, 1454-1471. https://doi.org/10.3390/life5021454
Ferry JG. Acetate Metabolism in Anaerobes from the Domain Archaea. Life. 2015; 5(2):1454-1471. https://doi.org/10.3390/life5021454
Chicago/Turabian StyleFerry, James G. 2015. "Acetate Metabolism in Anaerobes from the Domain Archaea" Life 5, no. 2: 1454-1471. https://doi.org/10.3390/life5021454




