The Routes of Emergence of Life from LUCA during the RNA and Viral World: A Conspectus
Abstract
:1. Introduction
2. Horizontal Gene Transfer
3. RNA-Dependent RNA Polymerases
4. On the Development of RNA
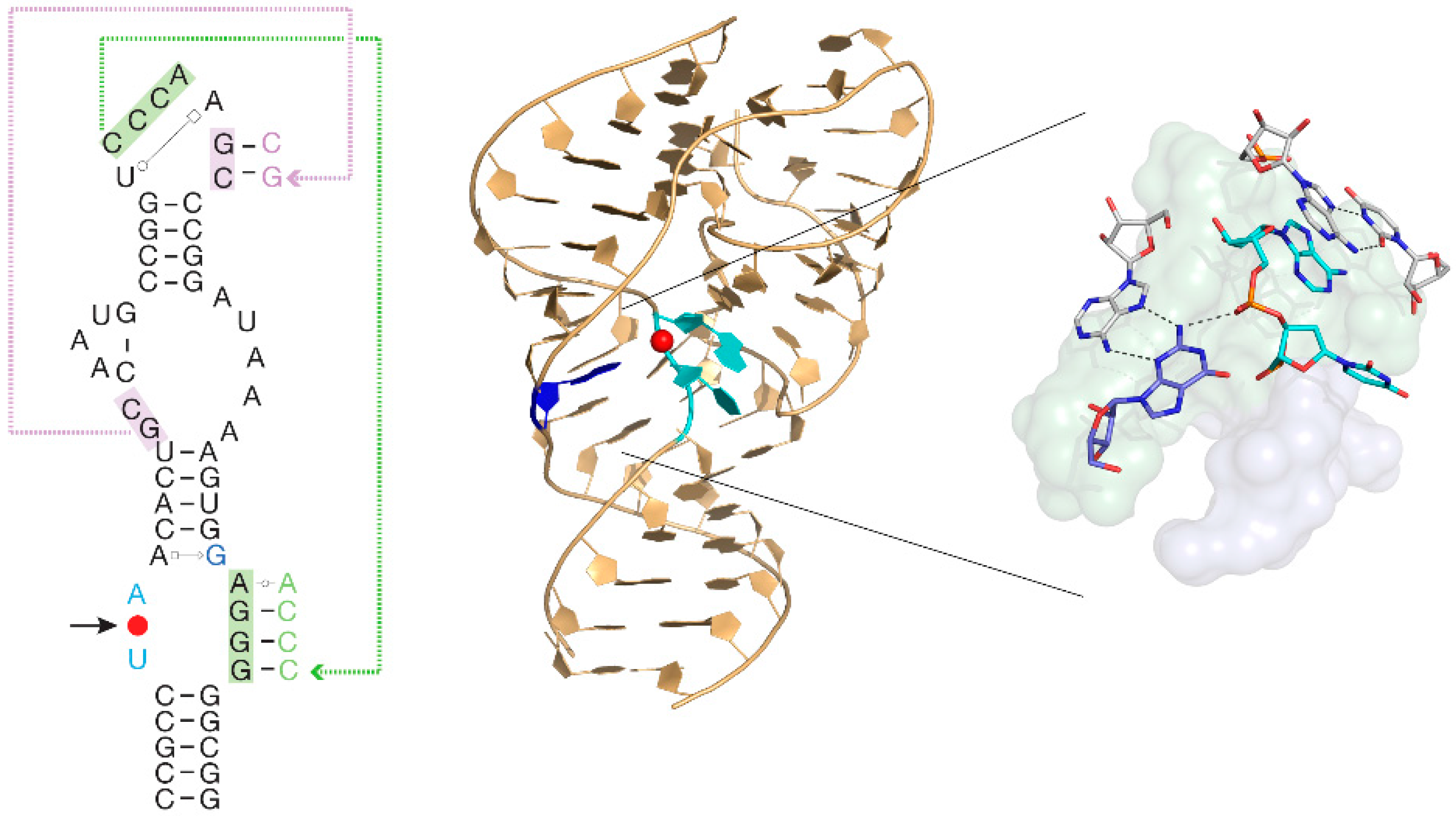
5. Alternative Hypotheses
6. Photosynthesis and Eukaryotes
7. What Next?
Acknowledgments
Conflicts of Interest
References
- Miller, S.L. A Production of amino acids under possible primitive earth conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef] [PubMed]
- Gribaldo, S.; Poole, A.M.; Daubin, V.; Forterre, P.; Brochier-Armanet, C. The origin of eukaryotes and their relationship with the Archaea: Are we at a phylogenomic impasse? Nat. Rev. Microbiol. 2010, 8, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Forterre, P. The two ages of the RNA world, and the transition to the DNA world: A story of viruses and cells. Biochimie 2005, 87, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Jheeta, S. Horizontal gene transfer and its part in the reorganisation of genetics during the LUCA epoch. Life 2013, 3, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.M.; Jeffares, D.C.; Penny, D. Relics from the RNA world. J. Mol. Evol. 1998, 46, 18–36. [Google Scholar] [CrossRef]
- Kotnik, T. Lightning-triggered electroporation and electrofusion as possible contributors to natural horizontal gene transfer. Phys. Life Rev. 2013, 13, 351–370. [Google Scholar] [CrossRef] [PubMed]
- Kotnik, T. Prokaryotic diversity, electrified DNA, lightning waveforms, abiotic gene transfer, and the Drake equation: Assessing the hypothesis of lightning-driven evolution. Phys. Life Rev. 2013, 13, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Marjanovic, I.; Kotnik, T. An experimental system for controlled exposure of biological samples to electrostatic discharges. Bioelectrochemistry 2013, 94, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Tuller, T.; Girshovich, Y.; Sella, Y.; Kreimer, A.; Freilich, S.; Kupiec, M.; Gophna, U.; Ruppin, E. Association between translation efficiency and horizontal gene transfer within microbial communities. Nucleic Acids Res. 2011, 39, 4743–4755. [Google Scholar] [CrossRef] [PubMed]
- Tuller, T. The Effect of Codon Usage on the Success of Horizontal Gene Transfer. In Lateral Gene Transfer in Evolution; Springer: New York, NY, USA, 2013; pp. 147–158. [Google Scholar]
- Tuller, T.; Zur, H. Multiple roles of the coding sequence 5' end in gene expression regulation. Nucleic Acids Res. 2015, 43, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Lazcano, A.; Forterre, P. The molecular search for the last common ancestor. J. Mol. Evol. 1999, 49, 411–412. [Google Scholar] [PubMed]
- Forterre, P. The Common Ancestor of Archaea and Eukarya Was Not an Archaeon. Archaea. 2013, 2013, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, M.; Gauliard, E.; Schouten, S.; Houel-Renault, L.; Lenormand, P.; Marguet, E.; Forterre, P. Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ. Microbiol. Rep. 2013, 5, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Hynes, A.P.; Mercer, R.G.; Watton, D.E.; Buckley, C.B.; Lang, A.S. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. J. Mol. Microbiol. 2012, 85, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Aswad, A.; Katzourakis, A. The first endogenous herpesvirus, identified in the tarsier genome, and novel sequences from primate rhadinoviruses and lymphocryptoviruses. PLoS Genet. 2014, 10, e1004332. [Google Scholar] [CrossRef] [PubMed]
- Overballe-Petersen, S.; Willerslev, E. Horizontal transfer of short and degraded DNA has evolutionary implications for microbes and eukaryotic sexual reproduction. Bioessays 2014, 36, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Cnossen, J.P.; Dulin, D.; Dekker, N.H. An optimized software framework for real-time, high-throughput tracking of spherical beads. Rev. Sci. Instrum. 2014, 85, 103712. [Google Scholar] [CrossRef] [PubMed]
- Dulin, D.; Vilfan, I.D.; Berghuis, B.A.; Hage, S.; Bamford, D.H.; Poranen, M.M.; Depken, M.; Dekker, N.H. Elongation-Competent Pauses Govern the Fidelity of a Viral RNA-Dependent RNA Polymerase. Cell. Rep. 2015, 10, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Sarkies, P.; Miska, E.A. Small RNAs break out: The molecular cell biology of mobile small RNAs. Mol. Cell. Biol. 2014, 15, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, R. RNA as an emergent entity: An understanding gained through studying its nonfunctional alternatives. Synlett 2014, 25, 1511–1518. [Google Scholar] [CrossRef]
- Powner, M.W.; Sutherland, J.D.; Szostak, J.W. Chemoselective multicomponent one-pot assembly of purine precursors in water. J. Am. Chem. Soc. 2010, 132, 16677–16688. [Google Scholar] [CrossRef] [PubMed]
- Powner, M.W.; Sutherland, J.D. Phosphate-mediated interconversion of Ribo- and Arabino-configured prebiotic nucleotide intermediates. Angew. Chem. 2010, 122, 4745–4747. [Google Scholar] [CrossRef]
- Bowler, F.R.; Chan, C.K.W.; Duffy, C.D.; Gerland, B.; Islam, S.; Powner, M.W.; Sutherland, J.D.; Xu, J. Prebiotically plausible oligoribonucleotide ligation facilitated by chemoselective acetylation. Nat. Chem. 2013, 5, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Attwater, J.; Wochner, A.; Pinheiro, V.B.; Coulson, A.; Holliger, P. Ice as a protocellular medium for RNA replication. Nat. Commun. 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- Wochner, A.; Attwater, J.; Coulson, A.; Holliger, P. Ribozyme-catalyzed transcription of an active ribozyme. Science 2011, 332, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wilson, T.J.; McPhee, S.A.; Lilley, D.M.J. Crystal structure and mechanistic investigation of the twister ribozyme. Nat. Chem. Biol. 2014, 10, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Farias, S.T.; Rêgo, T.G.; Jose, M.V. Origin and evolution of the Peptidyl Transferase Center from proto-tRNAs. FEBS Open Bio. 2014, 4, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Segre, D.B.; Ben-Eli, D.; Lancet, D. Compositional genomes: Prebiotic information transfer in mutually catalytic noncovalent assemblies. Proc. Natl. Acad. Sci. USA 2000, 97, 4112–4117. [Google Scholar] [CrossRef] [PubMed]
- Markovitch, O.; Lancet, D. Multispecies population dynamics of prebiotic compositional assemblies. J. Theor. Biol. 2014, 357, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Markovitch, O.; Lancet, D. Excess mutual catalysis is required for effective evolvability. Artif. Life 2012, 18, 243–266. [Google Scholar] [CrossRef] [PubMed]
- Gross, R.; Fouxon, I.; Lancet, D.; Markovitch, O. Quasispecies in population of compositional assemblies. BMC Evol. Biol. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Ichihashi, N.; Usui, K.; Kazuta, Y.; Sunami, T.; Matsuura, T.; Yomo, T. Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Saladino, R.; Crestini, C.; Costanzo, G.; Negri, R.; di Mauro, E. A possible prebiotic synthesis of purine, adenine, cytosine, and 4(3H)-pyrimidinone from formamide: Implications for the origin of life. Bioorganic Med. Chem. Lett. 2001, 9, 1249–1253. [Google Scholar] [CrossRef]
- Mulkidjanian, A.Y.; Bychkov, A.Y.; Dibrova, D.V.; Galperin, M.Y.; Koonin, E.V. Open questions on the origin of life at anoxic geothermal fields. Orig. Life Evol. Biosph. 2012, 42, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Saladino, R.; Botta, G.; Pino, S.; Costanzo, G.; di Mauro, E. Genetics first or metabolism first? The formamide clue. Chem. Soc. Rev. 2012, 41, 5526–5565. [Google Scholar] [CrossRef] [PubMed]
- Pino, S.; Costanzo, G.; Giorgi, A.; Šponer, J.; Šponer, J.E.; di Mauro, E. Ribozyme activity of RNA nonenzymatically polymerized from 3',5'-Cyclic GMP. Entropy 2013, 15, 5362–5383. [Google Scholar] [CrossRef]
- Sorci, L.; Martynowski, D.; Rodionova, D.A.; Eyobo, Y.; Zogaj, X.; Klose, K.E.; Nikolaev, E.V.; Magni, G.; Zhang, H.; Osterman, A.L. Nicotinamide mononucleotide synthetase is the key enzyme for an alternative route of NAD biosynthesis in Francisella tularensis. Proc. Natl. Acad. Sci. USA 2009, 106, 3083–3088. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.F. Redox homeostasis in the emergence of life. On the constant internal environment of nascent living cells. J. Cosmol. 2010, 10, 3362–3373. [Google Scholar]
- Nowack, E.C.M.; Melkonian, M.; Glöckner, G. Chromatophore genome sequence of paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 2008, 18, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Nowack, E.C.M.; Vogel, H.; Groth, M.; Grossman, A.R.; Melkonian, M.; Glöckner, G. Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora. Mol. Biol. Evol. 2011, 28, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Nowack, E.C.M.; Grossman, A.R. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc. Natl. Acad. Sci. USA 2012, 109, 5340–5345. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jheeta, S. The Routes of Emergence of Life from LUCA during the RNA and Viral World: A Conspectus. Life 2015, 5, 1445-1453. https://doi.org/10.3390/life5021445
Jheeta S. The Routes of Emergence of Life from LUCA during the RNA and Viral World: A Conspectus. Life. 2015; 5(2):1445-1453. https://doi.org/10.3390/life5021445
Chicago/Turabian StyleJheeta, Sohan. 2015. "The Routes of Emergence of Life from LUCA during the RNA and Viral World: A Conspectus" Life 5, no. 2: 1445-1453. https://doi.org/10.3390/life5021445