Effects of Elevated Carbon Dioxide and Salinity on the Microbial Diversity in Lithifying Microbial Mats
Abstract
:1. Introduction
2. Experimental Section
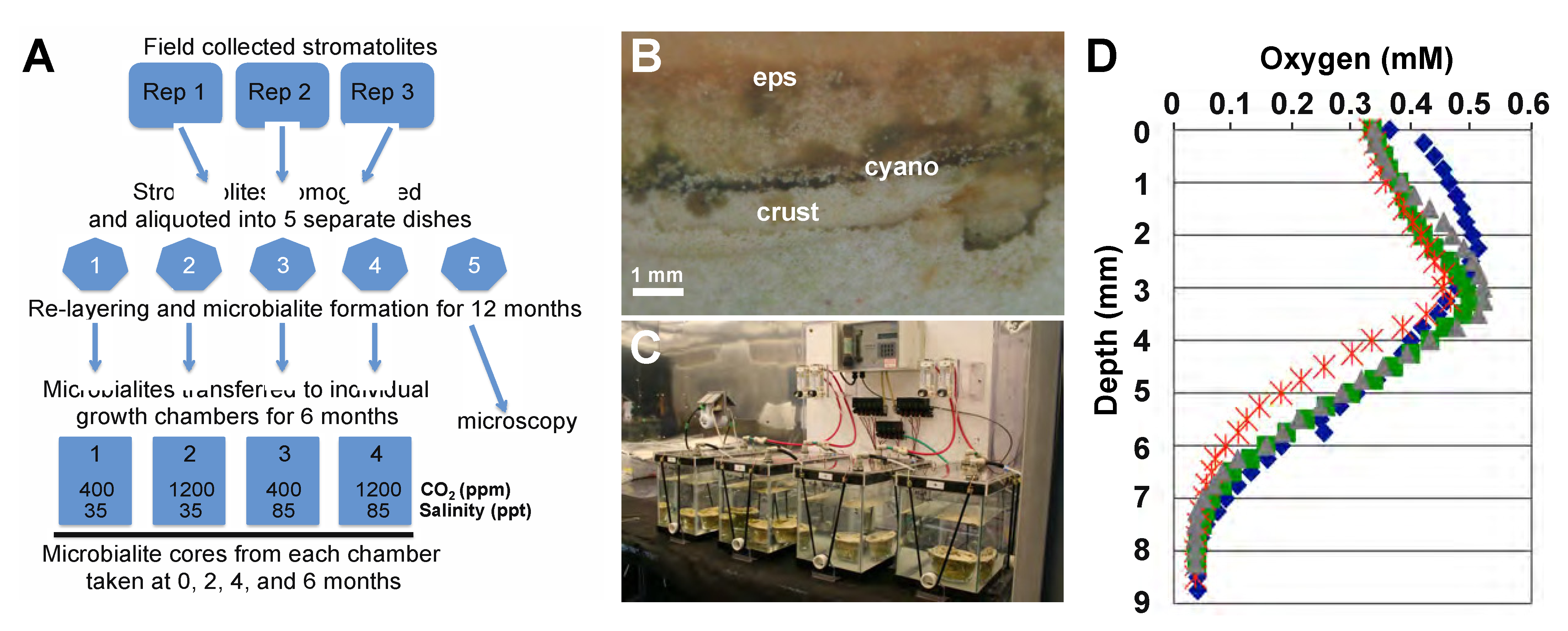
2.1. Lithifying Microbial Mat Cultivation
2.2. Construction of Environmental Manipulation Chambers
2.3. Microelectrode Depth Profiling of Oxygen
2.4. Sampling and DNA Extraction from Microbialites
2.5. Amplicon Library Synthesis, Barcode Labeling and Sequencing
2.6. Analysis of Pyrosequencing Sequencing Data
2.7. Community Comparisons between Treatments
3. Results
3.1. Microbialite Incubations under Variable Environmental Conditions
| Chamber | [CO2] (ppm) | Salinity (‰) | pH 1 (± std) | Temperature 2 (°C) |
|---|---|---|---|---|
| 1 | 400 | 35 | 8.47 (± 0.18) | 30 |
| 2 | 1200 | 35 | 8.13 (± 0.05) | 30 |
| 3 | 400 | 85 | 8.01 (± 0.12) | 30 |
| 4 | 1200 | 85 | 7.83 (± 0.19) | 30 |
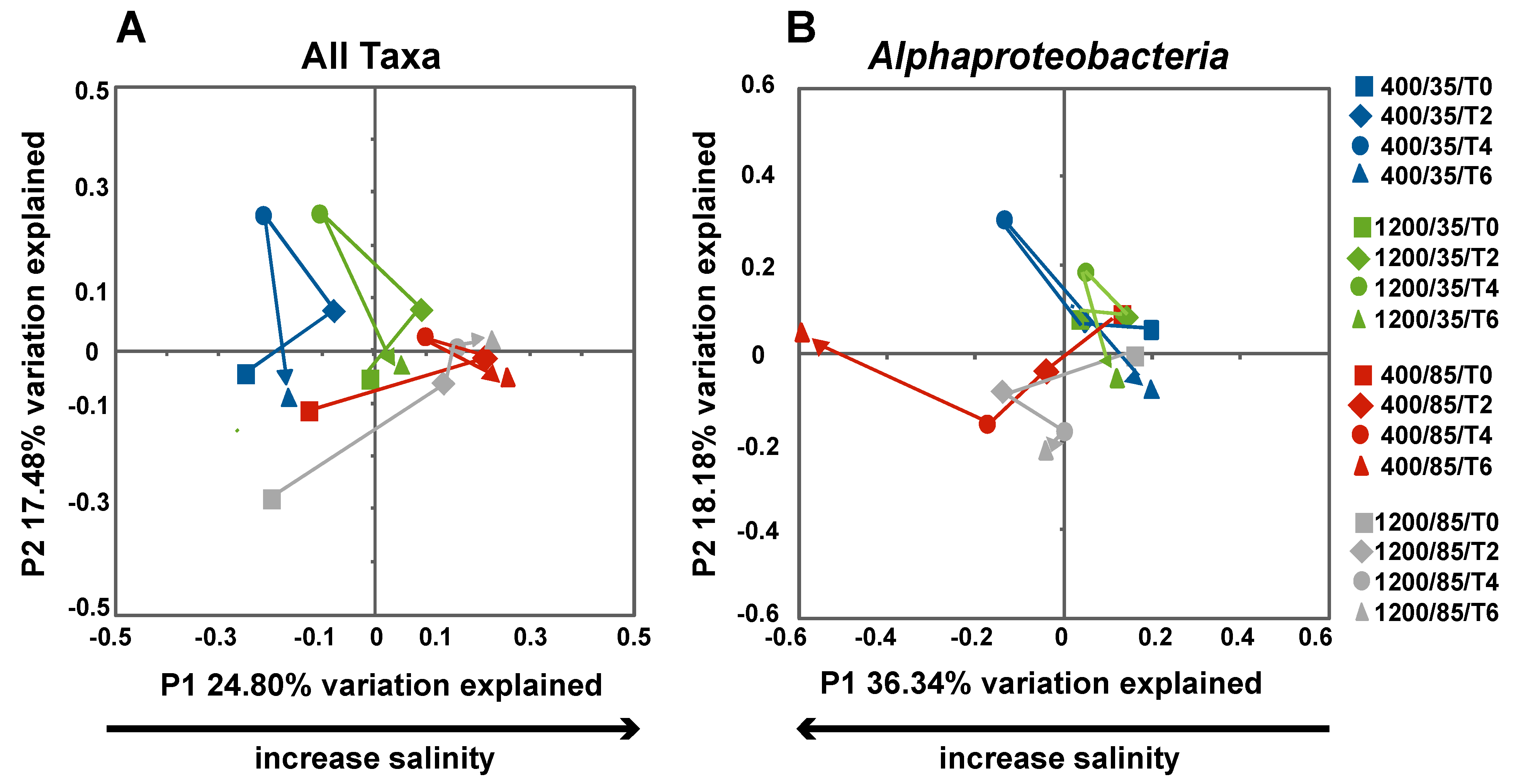
3.2. Comparison of Small Subunit rRNA Gene Diversity in the Microbialites Exposed to Environmental Changes
| Chamber | Chamber 1 (400/35) 1 | Chamber 2 (1200/35) | ||||||
|---|---|---|---|---|---|---|---|---|
| Time (Months) | T0 2 | T2 | T4 | T6 | T0 | T2 | T4 | T6 |
| Sequences | 2106 | 3503 | 4266 | 3910 | 1526 | 4126 | 2670 | 6894 |
| Base Pairs 3 | 239 | 309 | 314 | 296 | 240 | 316 | 309 | 315 |
| OTUs 4 | 369 | 865 | 631 | 611 | 453 | 683 | 517 | 1200 |
| Singlets 4 | 202 | 472 | 329 | 318 | 260 | 304 | 271 | 614 |
| Doublets 4 | 62 | 132 | 92 | 85 | 71 | 103 | 82 | 201 |
| OTUs 4,5 > 5% (Sum) | 2 (22%) | 0 (0%) | 3 (22%) | 2 (18%) | 0 (0%) | 0 (0%) | 1 (7%) | 1 (7%) |
| OTUs 4,5 > 1% (Sum) | 17 (58%) | 15 (20%) | 17 (47%) | 18 (45%) | 22 (36%) | 15 (27%) | 21 (44%) | 11 (30%) |
| Equalized sequences 6 | 1502 | 1502 | 1502 | 1502 | 1502 | 1502 | 1502 | 1502 |
| Shannon (confidence) | 4.30 (0.09) | 5.69 (0.06) | 4.78 (0.08) | 4.87 (0.08) | 5.35 (0.07) | 5.36 (0.06) | 4.97 (0.08) | 5.32 (0.08) |
| Evenness | 0.76 | 0.91 | 0.81 | 0.82 | 0.88 | 0.89 | 0.84 | 0.86 |
| Chao1 (confidence) 7 | 559 (468/698) | 1092 (938/1304) | 730 (610/907) | 766 (640/950) | 912 (775/1106) | 741 (641/887) | 772 (644/961) | 1066 (910/1281) |
| % Coverage 8 | 89% | 80% | 86% | 86% | 83% | 86% | 86% | 80% |
| Chamber | Chamber 3 (400/85) 1 | Chamber 4 (1200/85) | ||||||
| Time (Months) | T0 2 | T2 | T4 | T6 | T0 | T2 | T4 | T6 |
| Sequences | 2380 | 3789 | 5705 | 8371 | 2086 | 3958 | 5169 | 5242 |
| Base Pairs 3 | 235 | 318 | 298 | 313 | 241 | 315 | 308 | 314 |
| OTUs 4 | 440 | 584 | 482 | 452 | 257 | 426 | 721 | 618 |
| Singlets 4 | 226 | 300 | 242 | 214 | 143 | 230 | 388 | 299 |
| Doublets 4 | 75 | 104 | 63 | 48 | 35 | 60 | 113 | 87 |
| OTUs 4,5 > 5% (Sum) | 2 (22%) | 0 (0%) | 1 (8%) | 3 (26%) | 2 (45%) | 2 (24%) | 2 (16%) | 2 (13%) |
| OTUs 4,5 > 1% (Sum) | 21 (55%) | 26 (52%) | 28 (68%) | 21 (67%) | 14 (68%) | 20 (62%) | 16 (47%) | 19 (47%) |
| Equalized sequences 6 | 1502 | 1502 | 1502 | 1502 | 1502 | 1502 | 1502 | 1502 |
| Shannon (confidence) | 4.57 (0.09) | 4.90 (0.07) | 4.35 (0.07) | 4.01 (0.08) | 3.41 (0.11) | 4.16 (0.09) | 4.74 (0.08) | 4.75 (0.08) |
| Evenness | 0.78 | 0.84 | 0.80 | 0.76 | 0.64 | 0.75 | 0.81 | 0.83 |
| Chao1 (confidence) 7 | 676 (564/844) | 749 (616/948) | 490 (391/653) | 345 (283/453) | 449 (349/620) | 514 (416/673) | 850 (689/1087) | 626 (518/792) |
| % Coverage 8 | 88% | 87% | 91% | 94% | 92% | 91% | 85% | 89% |
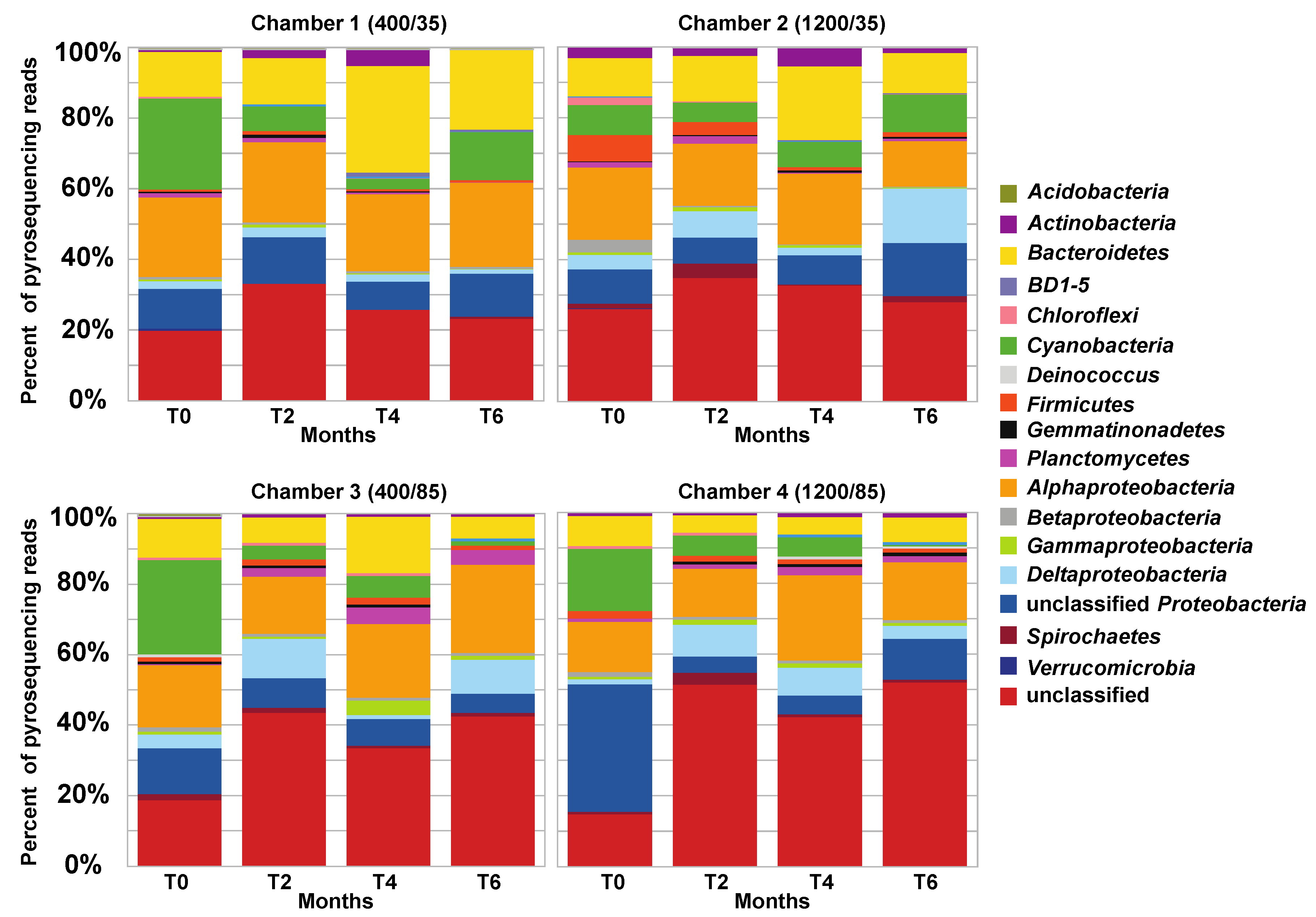
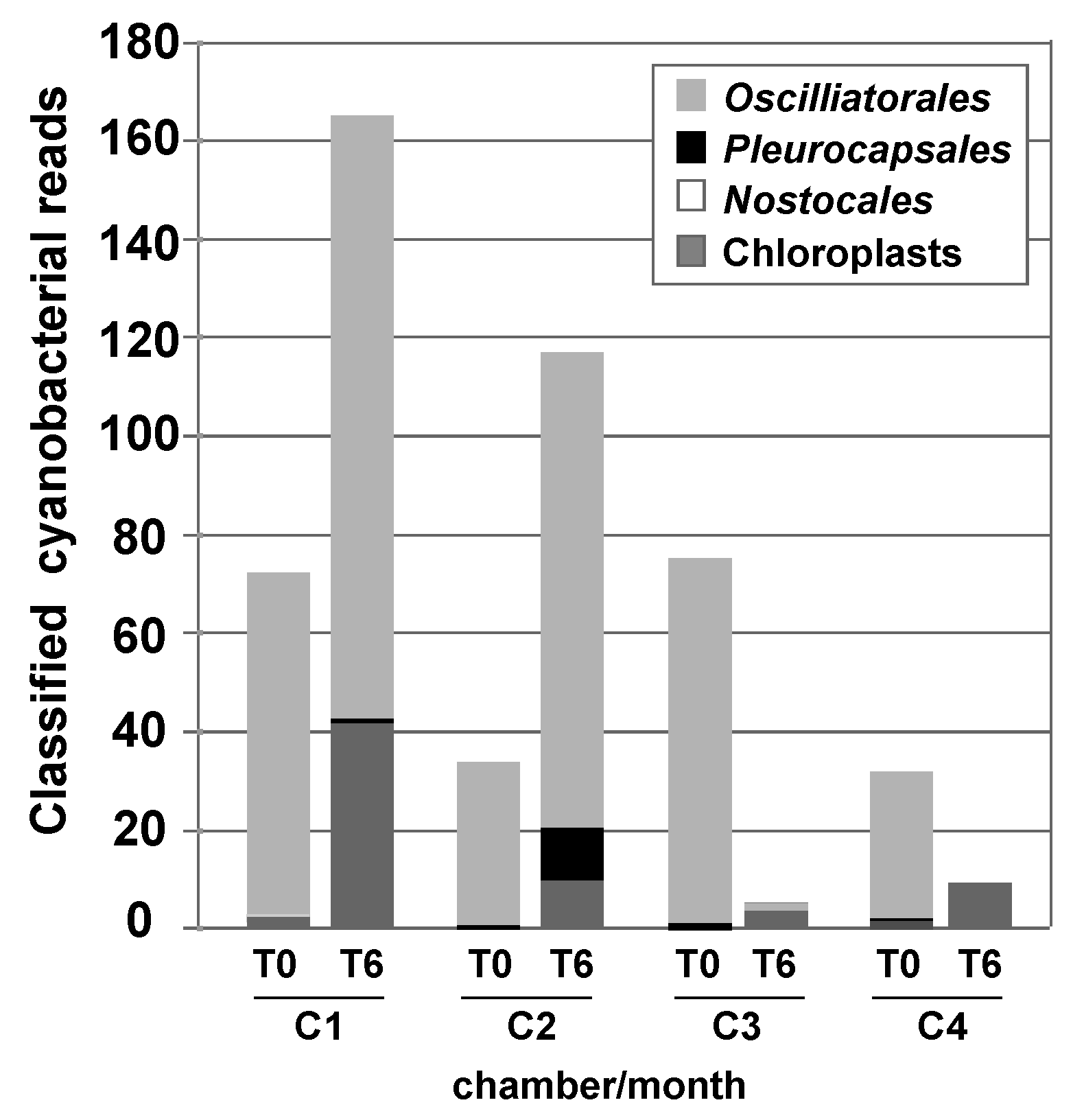
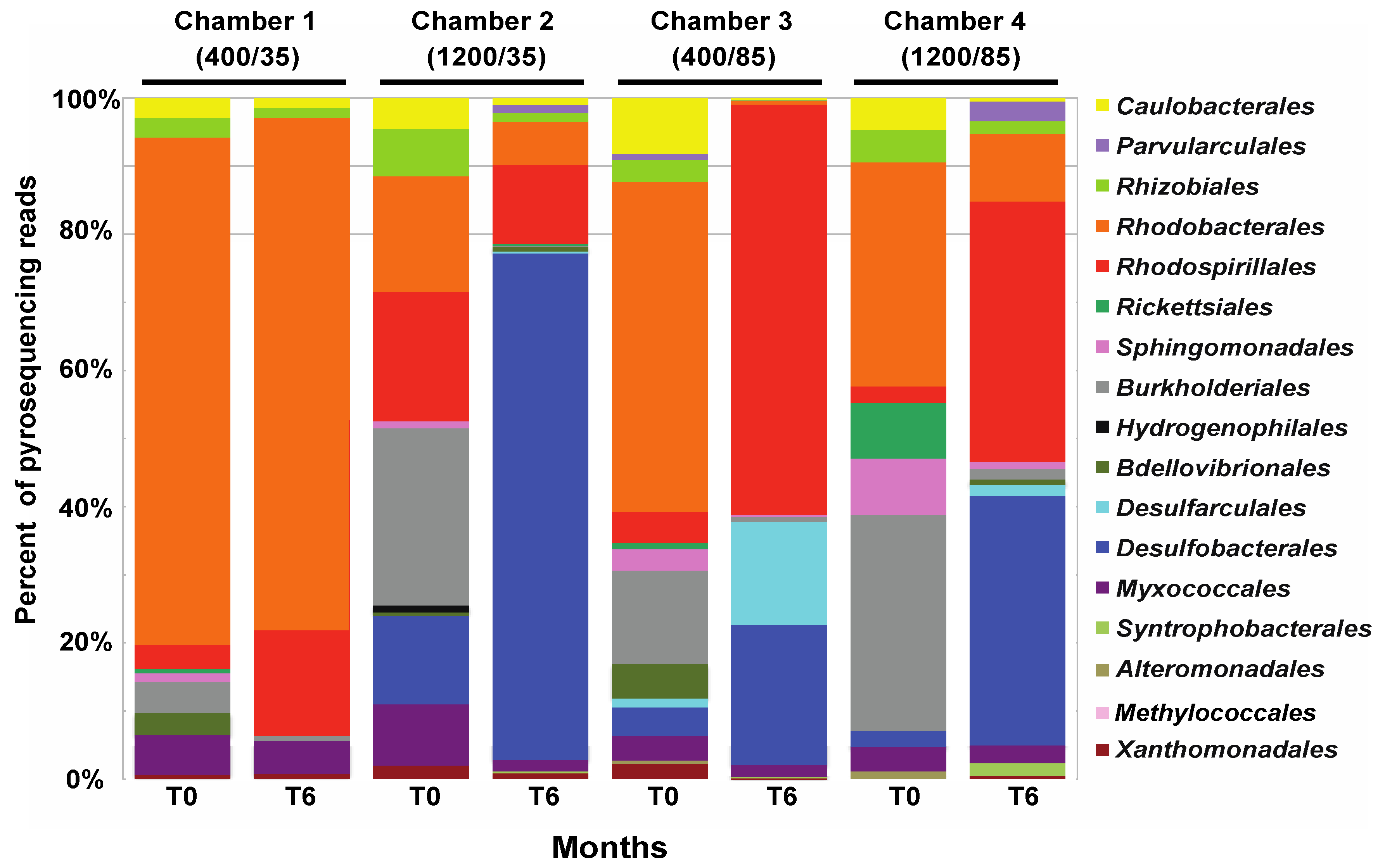
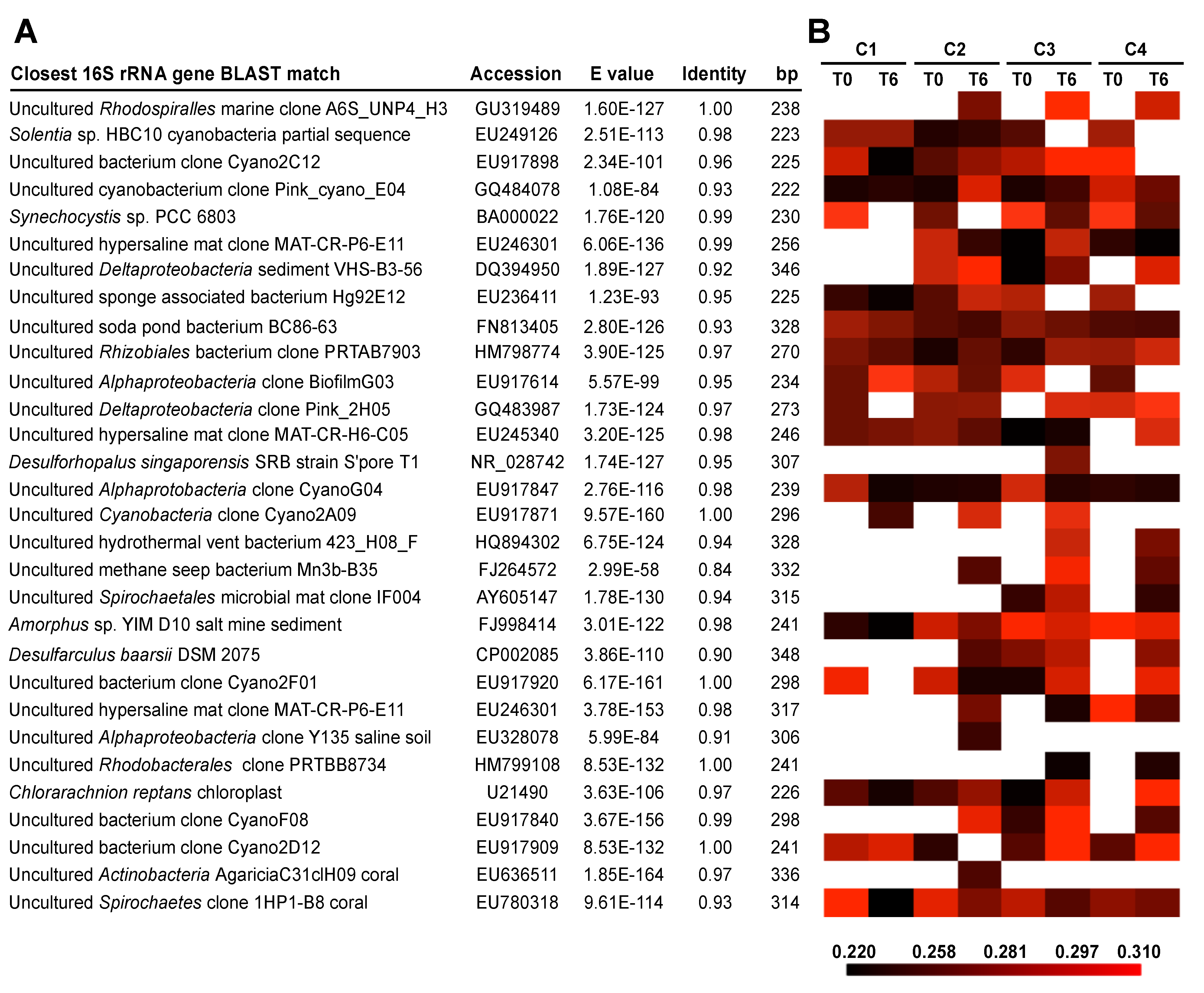
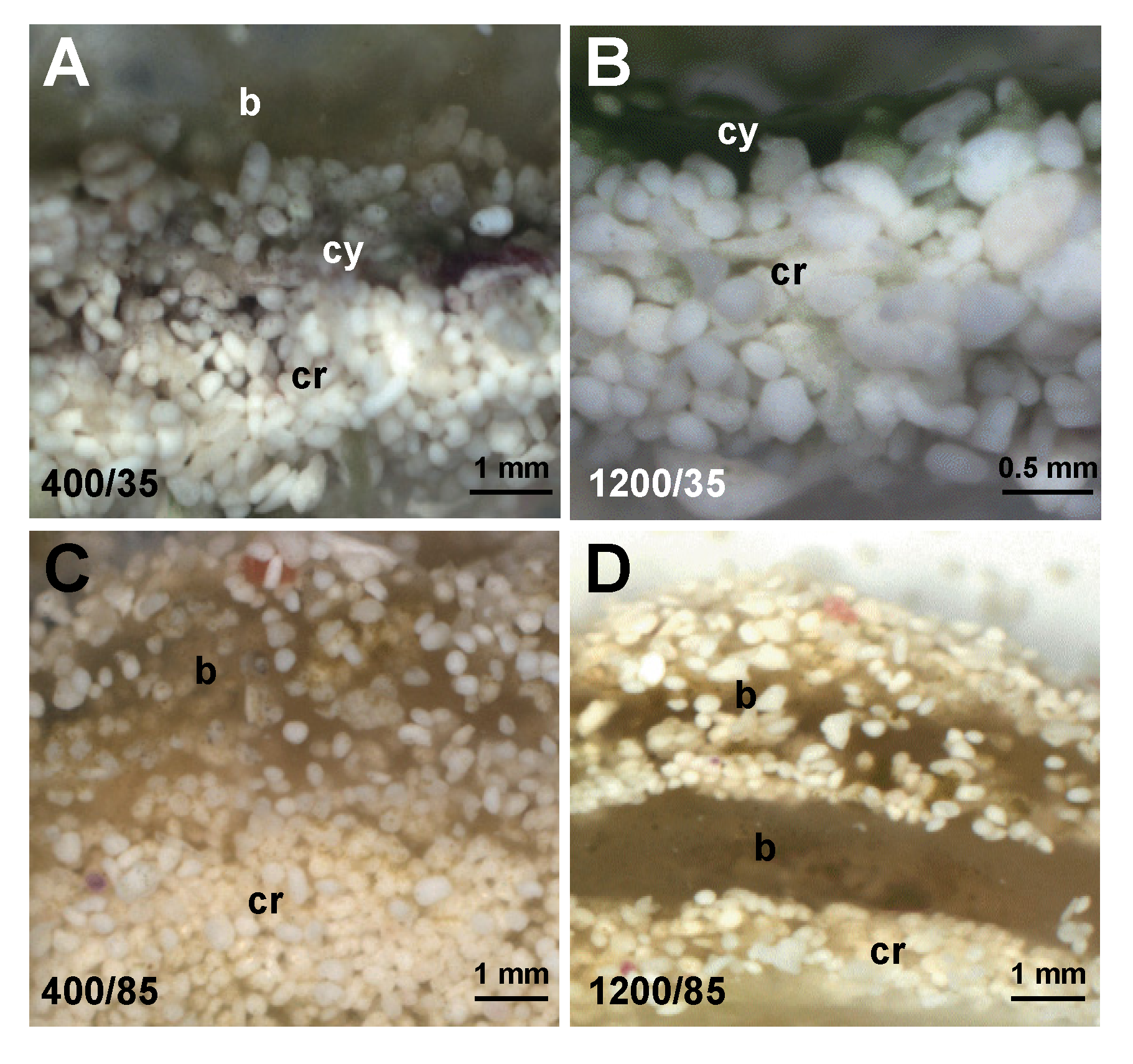
3.3. Effects of Environment on Microbialite Community Structure and Key Taxa
4. Discussion
4.1. Changes in Microbial Diversity in Response to Elevated CO2 and Salinity
4.2. Changes in pH During Environmental Manipulations and Impact on Community
4.3. Impact of Salinity on Microbialite Diversity
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kasting, J.F. Earth’s early atmosphere. Science 1993, 259, 920–926. [Google Scholar] [CrossRef]
- Joint, I.; Doney, S.C.; Karl, D.M. Will ocean acidification affect marine microbes? Int. Soc. Microb. Ecol. J. 2010, 5, 1–7. [Google Scholar]
- Azam, F. Microbial control of oceanic carbon flux: the plot thickens. Science 1998, 280, 694–696. [Google Scholar] [CrossRef]
- Falkowski, P.; Scholes, R.J.; Boyle, E.; Canadell, J.; Canfield, D.; Elser, J.; Gruber, N.; Hibbard, K.; Hogberg, P.; Linder, S.; et al. The global carbon cycle: A test of our knowledge of earth as a system. Science 2000, 290, 291–296. [Google Scholar] [CrossRef]
- Joos, F.; Plattner, G.K.; Stocker, T.F.; Marchal, O.; Schmittner, A. Global warming and marine carbon cycle feedbacks on future atmospheric CO2. Science 1999, 284, 464–467. [Google Scholar] [CrossRef]
- Post, W.M.; Izaurralde, R.C.; Jastrow, J.D.; McCarl, B.A.; Amonette, J.E.; Bailey, V.L.; Jardine, P.M.; West, T.O.; Zhou, J.Z. Enhancement of carbon sequestration in US soils. Bioscience 2004, 54, 895–908. [Google Scholar] [CrossRef]
- Hoegh-Guldberg, O.; Mumby, P.J.; Hooten, A.J.; Steneck, R.S.; Greenfield, P.; Gomez, E.; Harvell, C.D.; Sale, P.F.; Edwards, A.J.; Caldeira, K.; et al. Coral reefs under rapid climate change and ocean acidification. Science 2007, 318, 1737–1742. [Google Scholar] [CrossRef]
- Orr, J.C.; Fabry, V.J.; Aumont, O.; Bopp, L.; Doney, S.C.; Feely, R.A.; Gnanadesikan, A.; Gruber, N.; Ishida, A.; Joos, F.; et al. Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 2005, 437, 681–686. [Google Scholar] [CrossRef] [Green Version]
- Burne, R.V.; Moore, L.S. Microbialites: Organosedimentary deposits of benthic microbial communities. Palaios 1987, 2, 241–254. [Google Scholar] [CrossRef]
- Grotzinger, J.P.; Knoll, A.H. Stromatolites in Precambrian carbonates: Evolutionary mileposts or environmental dipsticks? Annu. Rev. Earth Planet. Sci. 1999, 27, 313–358. [Google Scholar] [CrossRef]
- Dupraz, C.; Visscher, P.T. Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 2005, 13, 429–438. [Google Scholar] [CrossRef]
- Dupraz, C.; Reid, R.P.; Braissant, O.; Decho, A.W.; Norman, R.S.; Visscher, P.T. Processes of carbonate precipitation in modern microbial mats. Earth Sci. Rev. 2009, 96, 141–162. [Google Scholar] [CrossRef]
- Visscher, P.T.; Van Gemerden, H. Sulfur Cycling in Laminated Marine Microbial Ecosystems. In Biogeochemistry of Global Change; Oremland, R.S., Ed.; Chapman and Hall: London, UK, 1993; pp. 672–690. [Google Scholar]
- Van Gemerden, H. Microbial mats: A joint venture. Mar. Geol. 1993, 113, 3–25. [Google Scholar] [CrossRef]
- Visscher, P.T.; Reid, R.P.; Bebout, B.M.; Hoeft, S.E.; Macintyre, I.G.; Thompson, J.A. Formation of lithified micritic laminae in modern marine stromatolites (Bahamas): The role of sulfur cycling. Am. Mineral. 1998, 83, 1482–1493. [Google Scholar]
- Visscher, P.T.; Stolz, J.F. Microbial mats as bioreactors: Populations, processes and products. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2005, 219, 87–100. [Google Scholar] [CrossRef]
- Baumgartner, L.K.; Spear, J.R.; Buckley, D.H.; Pace, N.R.; Reid, R.P.; Visscher, P.T. Microbial diversity in modern marine stromatolites, Highborne Cay, Bahamas. Environ. Microbiol. 2009, 11, 2710–2719. [Google Scholar] [CrossRef]
- Foster, J.S.; Green, S.J.; Ahrendt, S.R.; Hetherington, K.L.; Golubic, S.; Reid, R.P.; Bebout, L. Molecular and morphological characterization of cyanobacterial diversity in the marine stromatolites of Highborne Cay, Bahamas. Int. Soc. Microb. Ecol. J. 2009, 3, 573–587. [Google Scholar]
- Mobberley, J.M.; Ortega, M.C.; Foster, J.S. Comparative microbial diversity analyses of modern marine thrombolitic mats by barcoded pyrosequencing. Environ. Microbiol. 2012, 14, 82–100. [Google Scholar] [CrossRef]
- Warthmann, R.; Vasconcelos, C.; Bittermann, A.G.; McKenzie, J.A. The Role of Purple Sulphur Bacteria in Carbonate Precipitation of Modeern and Possibly Early Precambian Stromatolites. In Lecture Notes in Earth Sciences: Advances in Stromatolites Geobiology; Reitner, J., Quéric, N.V., Arp, G., Eds.; Springer: Berlin, Germany, 2011; Volume 131, pp. 141–149. [Google Scholar]
- Havemann, S.A.; Foster, J.S. Comparative characterization of the microbial diversities of an artificial microbialite model and a natural stromatolite. Appl. Environ. Microbiol. 2008, 74, 7410–7421. [Google Scholar] [CrossRef]
- Foster, J.S.; Green, S.J. Microbial Diversity in Modern Stromatolites. In Cellular Origin, Life in Extreme Habitats and Astrobiology: Interactions with Sediments; Seckbach, J., Tewari, V., Eds.; Springer: Berlin, Germany, 2011; pp. 385–405. [Google Scholar]
- Pinckney, J.; Paerl, H.W.; Bebout, B.M. Salinity control of benthic microbial mat community production in a Bahamian hypersaline lagoon. J. Exp. Mar. Biol. Ecol. 1995, 187, 223–237. [Google Scholar] [CrossRef]
- Pinckney, J.; Paerl, H.W.; Reid, R.P.; Bebout, B.M. Ecophysiology of stromatolitic microbial mats, Lee Stocking Island, Exuma Cays, Bahamas. Microb. Ecol. 1995, 29, 19–37. [Google Scholar]
- Abed, R.M.; Kohls, K.; de Beer, D. Effect of salinity changes on the bacterial diversity, photosynthesis and oxygen consumption of cyanobacterial mats from an intertidal flat of the Arabian Gulf. Environ. Microbiol. 2007, 9, 1384–1392. [Google Scholar] [CrossRef]
- Green, S.J.; Blackford, C.; Bucki, P.; Jahnke, L.L.; Prufert-Bebout, L. A salinity and sulfate manipulation of hypersaline microbial mats reveals stasis in the cyanobacterial community structure. Int. Soc. Microb. Ecol. J. 2008, 2, 457–470. [Google Scholar]
- Andres, M.S.; Reid, R.P. Growth morphologies of modern marine stromatolites: A case study from Highborne Cay, Bahamas. Sediment. Geol. 2006, 185, 319–328. [Google Scholar] [CrossRef]
- Paterson, D.M.; Aspden, R.J.; Visscher, P.T.; Consalvey, M.; Andres, M.S.; Decho, A.W.; Stolz, J.; Reid, R.P. Light-dependant biostabilisation of sediments by stromatolite assemblages. PLoS One 2008, 3, e3176. [Google Scholar] [CrossRef] [Green Version]
- Reid, R.P.; Visscher, P.T.; Decho, A.W.; Stolz, J.F.; Bebout, B.M.; Dupraz, C.; Macintyre, I.G.; Paerl, H.W.; Pinckney, J.L.; Prufert-Bebout, L.; et al. The role of microbes in accretion, lamination and early lithification of modern marine stromatolites. Nature 2000, 406, 989–992. [Google Scholar] [CrossRef]
- Bowlin, E.M.; Klaus, J.; Foster, J.S.; Andres, M.; Custals, L.; Reid, R.P. Environmental controls on microbial community cycling in modern marine stromatolites. Sediment. Geol. 2012, 263–264, 45–55. [Google Scholar]
- Visscher, P.T.; Beukema, J.; van Gemerden, H. In situ characterization of sediments: Measurement of oxygen and sulfide profiles with a novel combined needle electrode. Limnol. Oceanogr. 1991, 36, 1476–1480. [Google Scholar] [CrossRef]
- Myshrall, K.; Mobberley, J.M.; Green, S.J.; Visscher, P.T.; Havemann, S.A.; Reid, R.P.; Foster, J.S. Biogeochemical cycling and microbial diversity in the modern marine thrombolites of Highborne Cay, Bahamas. Geobiology 2010, 8, 337–354. [Google Scholar] [CrossRef]
- Hamady, M.; Walker, J.J.; Harris, J.K.; Gold, N.J.; Knight, R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 2008, 5, 235–237. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Kunin, V.; Engelbrektson, A.; Ochman, H.; Hugenholtz, P. Wrinkles in the rare biosphere: Pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 2010, 12, 118–123. [Google Scholar] [CrossRef]
- Huse, S.M.; Huber, J.A.; Morrison, H.G.; Sogin, M.L.; Welch, D.M. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007, 8, R143. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Schloss, P.D. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput. Biol. 2010, 6, e1000844. [Google Scholar] [CrossRef]
- Huse, S.M.; Welch, D.M.; Morrison, H.G.; Sogin, M.L. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 2010, 12, 1889–1898. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005, 71, 1501–1506. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Lozupone, C.; Hamady, M.; Knight, R. UniFrac: An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 2006, 7, 371. [Google Scholar] [CrossRef]
- Hamady, M.; Lozupone, C.; Knight, R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. Int. Soc. Microb. Ecol. J. 2010, 4, 17–27. [Google Scholar]
- Badger, M.R.; Bek, E.J. Multiple Rubisco forms in proteobacteria: Their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 2008, 59, 1525–1541. [Google Scholar] [CrossRef]
- Burns, B.P.; Goh, F.; Allen, M.; Neilan, B.A. Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ. Microbiol. 2004, 6, 1096–1101. [Google Scholar] [CrossRef]
- Dupraz, C.; Visscher, P.T.; Baumgartner, L.K.; Reid, R.P. Microbe-mineral interactions: Early carbonate precipitation in a hypersaline lake (Eleuthera Island, Bahamas). Sedimentology 2004, 51, 1–21. [Google Scholar] [CrossRef]
- Macintyre, I.G.; Prufert-Bebout, L.; Reid, R.P. The role of endolithic cyanobacteria in the formation of lithified lamiae in Bahamian stromatolites. Sedimentology 2000, 47, 915–921. [Google Scholar] [CrossRef]
- Grüter, D.; Schmid, B.; Brandl, H. Influence of plant diversity and elevated atmospheric carbon dioxide levels on belowground bacterial diversity. BMC Microbiol. 2006, 6, 68. [Google Scholar] [CrossRef] [Green Version]
- Sadowsky, M.; Schortemeyer, M. Soil microbial responses to increased concentrations of atmospheric CO2. Glob. Chang. Biol. 1997, 3, 217–224. [Google Scholar]
- Grayston, S.J.; Campbell, C.D.; Lutze, J.L.; Gifford, R.M. Impact of elevated CO2 on the metabolic diversity of microbial communities in N-limited grass wards. Plant Soil 1998, 203, 289–300. [Google Scholar] [CrossRef]
- Khodadad, C.L.; Zimmerman, A.R.; Green, S.J.; Uthandi, S.; Foster, J.S. Taxa-specific changes in soild microbial composition induced by pyrogenic carbon amendments. Soil Biol. Biochem. 2011, 43, 385–392. [Google Scholar] [CrossRef]
- Rothman, D.H. Atmospheric carbon dioxide levels for the last 500 million years. Proc. Natl. Acad. Sci. USA 2002, 99, 4167–4171. [Google Scholar] [CrossRef]
- Riding, R. Cyanobacterial calcification, carbon dioxide concentration mechanisms, and Proterozoic-Cambrian changes in atmospheric composition. Geobiology 2006, 4, 299–316. [Google Scholar] [CrossRef]
- Badger, M.R.; Price, G.D.; Long, B.M.; Woodger, F.J. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J. Exp. Bot. 2006, 57, 249–265. [Google Scholar]
- Myshrall, K.L.; Dupraz, C.; Visscher, P.T. Patterns in Microbialites Throughout Geolotic Time: Is the Present Really the Key to the Past? In Experimental Approaches to Understanding Fossil Organisms; Hembree, D.I., Platt, B.F., Smith, J.J., Eds.; Springer: Berlin, Germany, 2014; Volume 41. [Google Scholar]
- Berner, R.A. GEOCARBSULF: A combined model for Phanerozoic atmospheric O2 and CO2. Geochim. Cosmochim. Acta 2006, 70, 5653–5664. [Google Scholar] [CrossRef]
- Visscher, P.T.; Reid, R.P.; Bebout, B.M. Microscale observations of sulfate reduction: Correlation of microbial activity with lithified micritic laminae in modern marine stromatolites. Geology 2000, 28, 919–922. [Google Scholar] [CrossRef]
- Baumgartner, L.K.; Reid, R.P.; Dupraz, C.; Decho, A.W.; Buckley, D.H.; Spear, J.R.; Przekop, K.M.; Visscher, P.T. Sulfate reducing bacteria in microbial mats: Changing paradigms, new discoveries. Sediment. Geol. 2006, 185, 131–145. [Google Scholar] [CrossRef]
- Lee, J.Z.; Burow, L.C.; Woebken, D.; Everroad, R.C.; Kubo, M.D.; Spormann, A.M.; Weber, P.K.; Pett-Ridge, J.; Bebout, B.M.; Hoehler, T.M. Fermentation couples Chloroflexi and sulfate-reducing bacteria to Cyanobacteria in hypersaline microbial mats. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Gallagher, K.L.; Dupraz, C.; Visscher, P.T. Two opposing effects of sulfate reduction on carbonate precipitation in normal marine, hypersaline, and alkaline environment. Geology 2014, 42, 313–314. [Google Scholar] [CrossRef]
- Gallagher, K.L.; Kading, T.J.; Braissant, O.; Dupraz, C.; Visscher, P.T. Inside the alkalinity engine: The role of electron donors in the organomineralization potential of sulfate-reducing bacteria. Geobiology 2012, 10, 518–530. [Google Scholar] [CrossRef]
- Stal, L.J.; Moezelaar, R. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 1997, 21, 179–211. [Google Scholar] [CrossRef]
- Meister, P. Two opposing effects of sulfate reduction on carbonate precipitation in normal marine, hypersaline, and alkaline environments: Comment. Geology 2013, 41, 499–502. [Google Scholar] [CrossRef]
- Meister, P. Two opposing effects of sulfate reduction on carbonate precipitation in normal marine, hypersaline, and alkaline environments: Reply. Geology 2014, 42, e315. [Google Scholar] [CrossRef]
- Castro, H.F.; Classen, A.T.; Austin, E.E.; Norby, R.J.; Schadt, C.W. Soil microbial community responses to multiple experimental climate change drivers. Appl. Environ. Microbiol. 2010, 76, 999–1007. [Google Scholar] [CrossRef]
- DeAngelis, K.M.; Silver, W.L.; Thompson, A.W.; Firestone, M.K. Microbial communities acclimate to recurring changes in soil redox potential status. Environ. Microbiol. 2010, 12, 3137–3149. [Google Scholar] [CrossRef]
- Aloisi, G.; Gloter, A.; Kruger, M.; Wallmann, K.; Guyot, F.; Zuddas, P. Nucleation of calcium carbonate on bacterial nanoglobules. Geology 2006, 34, 1017–1020. [Google Scholar] [CrossRef]
- Bosak, T.; Newman, D.K. Microbial nucleation of calcium carbonate in the Precambrian. Geology 2003, 31, 577–580. [Google Scholar] [CrossRef]
- Feely, R.A.; Sabine, C.L.; Lee, K.; Berelson, W.; Kleypas, J.; Fabry, V.J.; Millero, F.J. Impact of anthropogenic CO2 on the CaCO3 system in the oceans. Science 2004, 305, 362–366. [Google Scholar] [CrossRef]
- Feely, R.A.; Sabine, C.L.; Hernandez-Ayon, J.M.; Ianson, D.; Hales, B. Evidence for upwelling of corrosive “acidified” water onto the continental shelf. Science 2008, 320, 1490–1492. [Google Scholar]
- Takahashi, T.; Sutherland, S.C.; Feely, R.A.; Wanninkhof, R. Decadel change of surface water pCO2 in the North Pacific: A synthesis of 35 years of observation. J. Geophys. Res. 2006, 111. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Field, D.; Huang, Y.; Edwards, R.; Li, W.; Gilna, P.; Joint, I. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 2008, 3, e3042. [Google Scholar] [CrossRef] [Green Version]
- Shibca, S.V.; Hedgpeth, D.W.; Park, P.K. Dissolved Oxygen and pH Increases by Primary Production in the Surface Water of Arthur Harboe, Antarctica 1970–1971. In Adaptation within Antarctic Ecosystems: Proceedings of the Third SCAR Symposium on Antactic Biology; Smithsonian Institution: Washington, DC, USA, 1977; pp. 83–97. [Google Scholar]
- Felix, E.A.; Rushforth, S.R. The alga flora of the Great Salt Lake. Nova Hedwig. 1979, 31, 163–194. [Google Scholar]
- Potts, M. Blue-green algae (Cyanophyta) in marine coastal environments of the Sinai peninusla: Distribution, zonation, stratification and taxonomic diversity. Phycologia 1980, 19, 60–73. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Kühl, M.; Nübel, U.; Muyzer, G. Salinity-dependent limitation of photosynthesis and oxygen exchange in microbial mats. J. Phycol. 1999, 35, 227–238. [Google Scholar] [CrossRef]
- Casamayor, E.O.; Massana, R.; Benlloch, S.; Ovreas, L.; Diez, B.; Goddard, V.J.; Gasol, J.M.; Joint, I.; Rodriguez-Valera, F.; Pedros-Alio, C. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 2002, 4, 338–348. [Google Scholar] [CrossRef]
- Caumette, P.; Matheron, R.; Raymond, N.; Relexans, J.C. Microbial mats in the hypersaline ponds of Meditteranean salterns (Salins-de-Giraud, France). FEMS Microbiol. Ecol. 1994, 13, 273–286. [Google Scholar]
- Fourçans, A.; de Oteyza, T.G.; Wieland, A.; Solé, A.; Diestra, E.; van Bleijswijk, J.; Grimalt, J.O.; Kühl, M.; Esteve, I.; Muyzer, G.; et al. Characterization of functional bacterial groups in a hypersaline microbial mat community (Salins-de-Giraud, Camargue, France). FEMS Microbiol. Ecol. 2004, 51, 55–70. [Google Scholar]
- Visscher, P.T.; Dupraz, C.; Braissant, O.; Gallagher, K.L.; Glunk, C.; Casillas, L.; Reed, R.E.S. Biogeochemistry of Carbon Cycling in Hypersaline Mats: Linking the Present to the Past Through Biosignatures. In Cellular Origin, Life in Extreme Habitats and Astrobiology; Seckbach, J., Oren, A., Eds.; Springer: Berlin, Germany, 2010; Volume 14, pp. 443–468. [Google Scholar]
- Paerl, H.W.; Steppe, T.F.; Buchan, K.C.; Potts, M. Hypersaline cyanobacterial mats as indicators of elevated tropical hurricane activity and associated climate change. Ambio 2003, 32, 87–90. [Google Scholar]
- Yannarell, A.C.; Paerl, H.W. Effects of salinity and light on organic carbon and nitrogen uptake in a hypersaline microbial mat. FEMS Microbiol. Ecol. 2007, 62, 345–353. [Google Scholar] [CrossRef]
- Liu, H.B.; Buskey, E.J. Hypersalinity enhances the production of extracellular polymeric substances (EPS) in the Texas brown tide alga, Aureoumbra lagunensis (Pelagophyceae). J. Phycol. 2000, 36, 71–77. [Google Scholar] [CrossRef]
- Abdullahi, A.S.; Underwood, G.J.C.; Gretz, M.R. Extracellular matric assembly in diatoms (Bacillariophyceae). V. Environmental effects on polysaccharide synthesis in the model diatom, Phaeodactylum tricornutum. J. Phycol. 2006, 42, 363–378. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Decho, A.W. Isolation and biochemical characterization of extracellular polymeric secretions (EPS) from modern soft marine stromatolites (Bahamas) and its inhibitory effect on CaCO3 precipitation. Prep. Biochem. Biotechnol. 2002, 32, 51–63. [Google Scholar] [CrossRef]
- Braissant, O.; Decho, A.W.; Dupraz, C.; Glunk, C.; Przekop, K.M.; Visscher, P.T. Exopolymeric substances of sulfate-reducing bacteria: Interactions with calcium at alkaline pH and implication for formation of carbonate minerals. Geobiology 2007, 5, 401–411. [Google Scholar] [CrossRef]
- Bissett, A.; Reimer, A.; de Beer, D.; Shiraishi, F.; Arp, G. Metabolic microenvironmental control by photosynthetic biofilms under changing macroenvironmental temperature and pH conditions. Appl. Environ. Microbiol. 2008, 74, 6306–6312. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ahrendt, S.R.; Mobberley, J.M.; Visscher, P.T.; Koss, L.L.; Foster, J.S. Effects of Elevated Carbon Dioxide and Salinity on the Microbial Diversity in Lithifying Microbial Mats. Minerals 2014, 4, 145-169. https://doi.org/10.3390/min4010145
Ahrendt SR, Mobberley JM, Visscher PT, Koss LL, Foster JS. Effects of Elevated Carbon Dioxide and Salinity on the Microbial Diversity in Lithifying Microbial Mats. Minerals. 2014; 4(1):145-169. https://doi.org/10.3390/min4010145
Chicago/Turabian StyleAhrendt, Steven R., Jennifer M. Mobberley, Pieter T. Visscher, Lawrence L. Koss, and Jamie S. Foster. 2014. "Effects of Elevated Carbon Dioxide and Salinity on the Microbial Diversity in Lithifying Microbial Mats" Minerals 4, no. 1: 145-169. https://doi.org/10.3390/min4010145






