Genome-Wide Characterization of DNA Demethylase Genes and Their Association with Salt Response in Pyrus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Chromosomal Mapping
2.2. Protein Properties and Sequence Analyses
2.3. Multiple Sequences Alignment and Phylogenetic Analysis
2.4. Ks Calculation and Divergence Time Estimation of Homologous Gene Pairs
2.5. Plant Materials and Stress Treatments
2.6. RNA Isolation and Reverse Transcription-Quantitative PCR
2.7. Bisulfite Conversion of Genomic DNA and Methylation Sequencing
3. Results
3.1. Genome-Wide Identification of DMEs in Pear
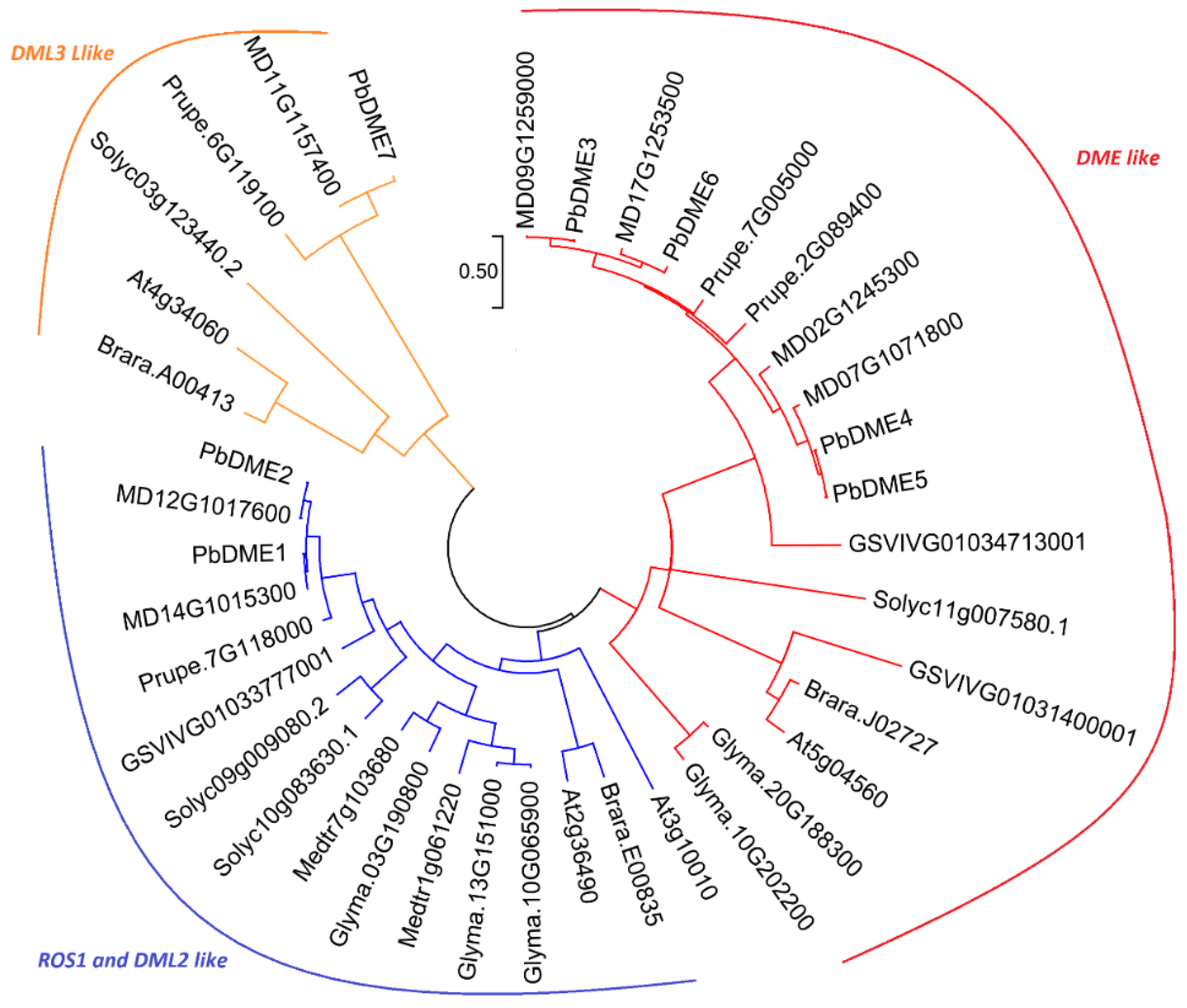
3.2. Classification and Phylogeny Analysis of the PbDMEs
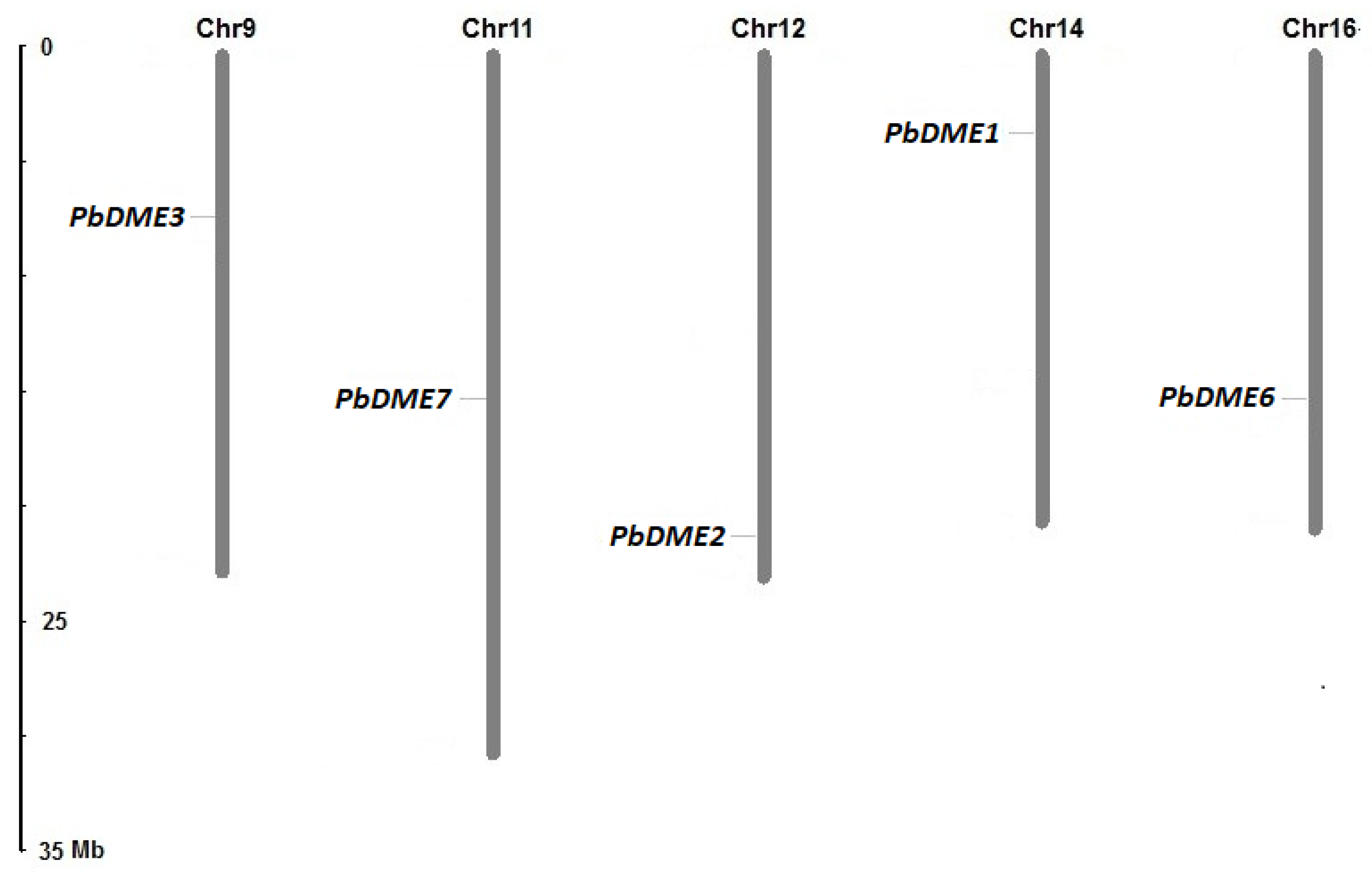
3.3. Characteristics and Chromosomal Location of Pear DME Genes
3.4. Evolution of DME Family Genes in the Pomoideae
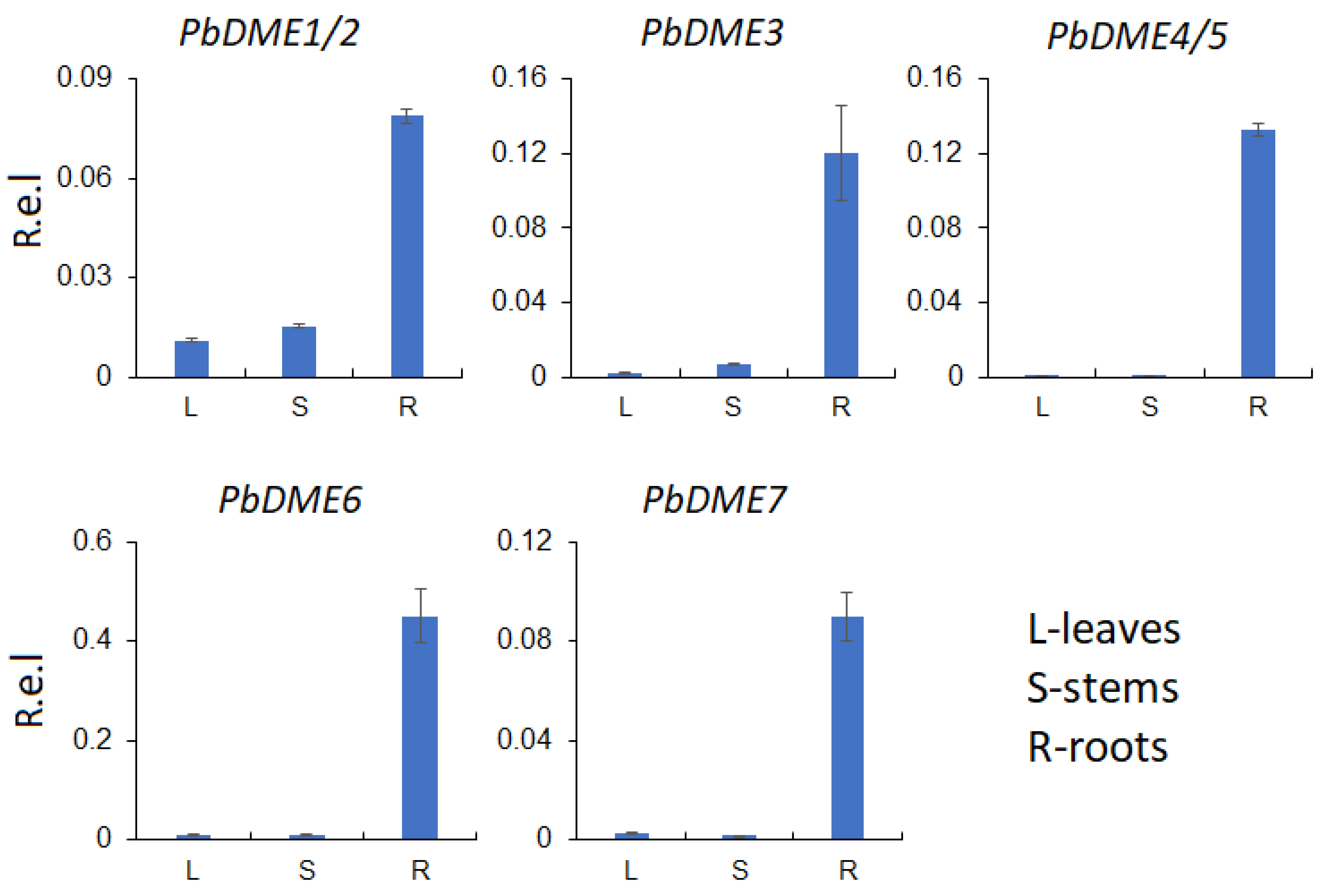
3.5. Expression of PbDMEs and Salt-Related Genes in Response to Salt Stress
3.6. Changes in the Level of Methylation of Some Salt-Responsive Genes in Response to Salt Stress
4. Discussion
4.1. Complex Gene Structures May Indicate Various Functions
4.2. DME Genes in Other Plants
4.3. PbDME Genes are Evolutionarily Conserved in Rosaceae Plants
4.4. Alteration of DNA Methylation Level is Essential to Salt Response in P. betulaefolia
4.5. Excellent Candidates for Pear Improvement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vanyushin, B.F.; Ashapkin, V.V. DNA methylation in higher plants: Past, present and future. Biochim. Biophys. Acta 2011, 1809, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Du, J.; Hale, C.J.; Gallego-Bartolome, J.; Feng, S.; Vashisht, A.A.; Chory, J.; Wohlschlegel, J.A.; Patel, D.J.; Jacobsen, S.E. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 2014, 157, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. Canalization of development and the inheritance of acquired characters. Nature 1942, 150, 563–565. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.J.; Chen, T.; Zhu, J.K. Regulation and function of DNA methylation in plants and animals. Cell. Res. 2011, 21, 442–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, D.; Ju, Z.; Gao, C.; Mei, X.; Fu, D.; Zhu, H.; Luo, Y.; Zhu, B. Genomewide identification of cytosine-5 DNA methyltransferases and demethylases in Solanum lycopersicum. Gene 2014, 550, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Morales-Ruiz, T.; Ortega-Galisteo, A.P.; Ponferrada-Marin, M.I.; Martinez-Macias, M.I.; Ariza, R.R.; Roldan-Arjona, T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Nat. Acad. Sci. USA 2006, 103, 6853–6858. [Google Scholar] [CrossRef] [PubMed]
- Penterman, J.; Zilberman, D.; Huh, J.H.; Ballinger, T.; Henikoff, S.; Fischer, R.L. DNA demethylation in the Arabidopsis genome. Proc. Nat. Acad. Sci. USA 2007, 104, 6752–6757. [Google Scholar] [CrossRef] [PubMed]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Nat. Acad. Sci. USA 2017, 114, E4511–E4519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega-Galisteo, A.P.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008, 67, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T.; David, L.; Zhu, J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 2002, 111, 803–814. [Google Scholar] [CrossRef]
- Martínez-Macías, M.I.; Qian, W.; Miki, D.; Pontes, O.; Liu, Y.; Tang, K.; Liu, R.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T.; Zhu, J.K. A DNA 3’ phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell. 2012, 45, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Duan, C.G.; Zhu, X.; Qian, W.; Zhu, J.K. A DNA ligase required for active DNA demethylation and genomic imprinting in Arabidopsis. Cell. Res. 2015, 25, 757–760. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Frost, J.M.; Park, K.; Ohr, H.; Park, G.T.; Kim, S.; Eom, H.; Lee, I.; Brooks, J.S.; Fischer, R.L.; Choi, Y. Control of DEMETER DNA demethylase gene transcription in male and female gamete companion cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 2078–2083. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.J.; Fischer, R.L. Genome demethylation and imprinting in the endosperm. Curr. Opin. Plant. Biol. 2011, 14, 162–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Gao, C.; Bian, X.; Zhao, S.; Zhao, C.; Xia, H.; Song, H.; Hou, L.; Wan, S.; Wang, X. Genome-Wide identification and comparative analysis of cytosine-5 DNA methyltransferase and demethylase families in wild and cultivated peanut. Front. Plant. Sci. 2016, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Eroglu, S.; Aksoy, E. Genome-wide analysis of gene expression profiling revealed that COP9 signalosome is essential for correct expression of Fe homeostasis genes in Arabidopsis. Biometals 2017, 30, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gho, Y.S.; Jung, K.H.; Kim, S.R. Genome-Wide identification and analysis of genes, conserved between japonica and indica rice cultivars, that respond to low-temperature stress at the vegetative growth stage. Front. Plant Sci. 2017, 8, 1120. [Google Scholar] [CrossRef] [PubMed]
- Di, F.; Jian, H.; Wang, T.; Chen, X.; Ding, Y.; Du, H.; Lu, K.; Li, J.; Liu, L. Genome-Wide analysis of the PYL gene family and identification of PYL genes that respond to abiotic stress in Brassica napus. Genes 2018, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Kimatu, J.N.; Xu, K.; Liu, B. DNA cytosine methylation in plant development. J. Genet. Genom. 2010, 37, 1–12. [Google Scholar] [CrossRef]
- Ding, H.; Gao, J.; Qin, C.; Ma, H.; Huang, H.; Song, P.; Luo, X.; Lin, H.; Shen, Y.; Pan, G.; Zhang, Z. The dynamics of DNA methylation in maize roots under Pb stress. Int. J. Mol. Sci. 2014, 15, 23537–23554. [Google Scholar] [CrossRef] [PubMed]
- Plitta, B.P.; Michalak, M.; Bujarska-Borkowska, B.; Barciszewska, M.Z.; Barciszewski, J.; Chmielarz, P. Effect of desiccation on the dynamics of genome-wide DNA methylation in orthodox seeds of Acer platanoides L. Plant Physiol. Biochem. 2014, 85, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Feng, L.L.; Li, J.M.; He, Z.H. Genetic and epigenetic control of plant heat responses. Front. Plant. Sci. 2015, 6, 267. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, Y.; Zheng, H.; Lu, W.; Wu, C.; Huang, J.; Yan, K.; Yang, G.; Zheng, C. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 2015, 66, 5997–6008. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Chevala, V.N.; Shankar, R.; Jain, M. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 2015, 5, 144922. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.J.; Azevedo, V.; Maroco, J.; Oliveira, M.M.; Santos, A.P. Salt tolerant and sensitive rice varieties display differential methylome flexibility under salt stress. PLoS ONE 2015, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Cheng, S.F.; Liu, X.Y.; Du, H.; Dai, M.Q.; Zhou, D.X.; Yang, W.J.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.H.; Zhang, M.; Fu, R.; Qian, X.W.; Rong, P.; Zhang, Y.; Jiang, P.; Wang, J.J.; Lu, X.K.; Wang, D.L.; Ye, W.W.; Zhu, X.Y. Epigenetic mechanisms of salt tolerance and heterosis in upland cotton (Gossypium hirsutum L.) revealed by methylation-sensitive amplified polymorphism analysis. Euphytica 2016, 208, 477–491. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Q.; Zhang, M.; Yang, C.; Sha, W.; Liu, B. Alteration of DNA methylation level and pattern in sorghum (Sorghum bicolor L.) pure-lines and inter-line F1 hybrids following low-dose laser irradiation. J. Photochem. Photobiol. 2010, 99, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ji, D.; Li, S.; Wang, P.; Li, Q.; Xiang, F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 2012, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Xian, Z.; Hu, G.; Li, Z. SlAGO4A, a core factor of RNA-directed DNA methylation (RdDM) pathway, plays an important role under salt and drought stress in tomato. Mol. Breed. 2016, 36, 1–13. [Google Scholar] [CrossRef]
- Liang, D.; Zhang, Z.; Wu, H.; Huang, C.; Shuai, P.; Ye, C.; Tang, S.; Wang, Y.; Yang, L.; Wang, J.; Yin, W.; Xia, X. Singlebase-resolution methylomes of Populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet. 2014, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef] [PubMed]
- Le, T.N.; Schumann, U.; Smith, N.A.; Tiwari, S.; Au, P.C.; Zhu, Q.H.; Taylor, J.M.; Kazan, K.; Llewellyn, D.J.; Zhang, R.; Dennis, E.S.; Wang, M.B. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014, 15, 458. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002, 14, 465. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.S.; Sano, H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genom. 2007, 277, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Alina, R.; Sgorbati, S.; Santagostino, A.; Labra, M.; Ghiani, A.; Citterio, S. Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol. Plant 2004, 121, 472–480. [Google Scholar]
- Labra, M.; Ghiani, A.; Citterio, S.; Sgorbati, S.; Sala, F.; Vannini, C.; Ruffini-Castiglione, M.; Bracale, M. Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol. 2002, 4, 694–699. [Google Scholar] [CrossRef]
- Filek, M.; Keskinen, R.; Hartikainen, H.; Szarejko, I.; Janiak, A.; Miszalski, Z.; Golda, A. The protective role of selenium in rape seedlings subjected to cadmium stress. J. Plant Physiol. 2008, 165, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.H.; Choi, J.; Nino, M.; Cho, Y.; Kang, K.; Jung, Y. Regulation of abiotic stress response through NtROS2a-mediated demethylation in tobacco. Plant Breed. Biotech. 2015, 3, 108–118. [Google Scholar] [CrossRef]
- Demeulemeester, M.A.C.; Van Stallen, N.M.; De Profit, M.P. Degree of DNA methylation in chicory (Cichorium intybus L.): Influence of plant age and vernalization. Plant Sci. 1999, 142, 101–108. [Google Scholar] [CrossRef]
- Yong, W.; Xu, Y.; Xu, W.; Wang, X.; Li, N.; Wu, J.; Liang, T.; Chong, K.; Xu, Z.; Tan, K.; et al. Vernalization-induced flowering in wheat is mediated by a lectin-like gene VER2. Planta 2003, 217, 261–270. [Google Scholar] [PubMed]
- Kumar, M.; Choi, J.; An, G.; Kim, S.R. Ectopic expression of OsSta2 enhances salt stress tolerance in rice. Front. Plant Sci. 2017, 8, 316. [Google Scholar] [CrossRef] [PubMed]
- Murshed, R.; Lopez-Lauri, F.; Sallanon, H. Effect of salt stress on tomato fruit antioxidant systems depends on fruit development stage. Physiol. Mol. Biol. Plants 2014, 20, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Gharsallah, C.; Fakhfakh, H.; Grubb, D.; Gorsane, F. Effect of salt stress on ion concentration, proline content, antioxidant enzyme activities and gene expression in tomato cultivars. AoB Plants 2016, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.H.; Li, H.; Cong, Y.; Lin, J.; Sheng, B.L. Characterization and expression of a phytochelatin synthase gene in birchleaf pear (Pyrus betulaefolia Bunge). Plant Mol. Biol. Rep. 2012, 30, 1329–1337. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Awais Khan, A.; Tao, S.; Korban, S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 3, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Okubo, M.; Furukawa, Y.; Sakuratani, T. Growth, flowering and leaf properties of pear cultivars grafted on two Asian pear rootstock seedlings under NaCl irrigation. Sci. Hortic-Amsterda 2000, 85, 91–101. [Google Scholar] [CrossRef]
- Matsumoto, K.; Tamura, F.; Chun, J.P.; Ikeda, T.; Imanishi, K.; Tanabe, K. Enhancement in salt tolerance of Japanese pear by using Pyrus betulaefolia rootstock (soil management, fertilization & irrigation). Hortic. Res. 2007, 6, 47–52. [Google Scholar]
- Okubo, M.; Sakuratani, T. Effects of sodium chloride on survival and stem elongation of two Asian pear rootstock seedlings. Sci. Hortic-Amsterda 2000, 85, 85–90. [Google Scholar] [CrossRef]
- Expression Atlas. Available online: https://www.ebi.ac.uk/gxa/plant/experiments/ (accessed on 27 July 2018).
- AtGenExpress Visualization Tool (VAT). Available online: http://jsp.weigelworld.org/expviz/expviz.jsp/ (accessed on 27 July 2018).
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- ExPASy. Available online: https://web.expasy.org/compute_pi/ (accessed on 1 June 2018).
- The MEME Suite. Available online: http://meme-suite.org/tools/meme/ (accessed on 20 May 2018).
- GSDS2.0 Gene Structure Display Server. Available online: http://gsds.cbi.pku.edu.cn/ (accessed on 21 May 2018).
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Li, H.; Cong, Y.; Chang, Y.; Lin, J. Comparison of two CBL genes on stress tolerance functions from Pyrus betulaefolia. J. Fruit. Sci. 2014, 31, 529–535. [Google Scholar]
- Tang, J.; Lin, J.; Li, H.; Li, X.; Yang, Q.; Cheng, Z.M.; Chang, Y. Characterization of CIPK family in asian pear (Pyrus bretschneideri Rehd) and co-expression analysis related to salt and osmotic stress responses. Front. Plant Sci. 2016, 7, 1361. [Google Scholar] [CrossRef] [PubMed]
- Qüesta, J.I.; Fina, J.P.; Casati, P. DDM1 and ROS1 have a role in UV-B induced- and oxidative DNA damage in A. thaliana. Front. Plant Sci. 2013, 4, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López Sánchez, A.; Stassen, J.H.; Furci, L.; Smith, L.M.; Ton, J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 2016, 88, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Gehring, M.; Johnson, L.; Hannon, M.; Harada, J.J.; Goldberg, R.B.; Jacobsen, S.E.; Fischer, R.L. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 2002, 110, 33–42. [Google Scholar] [CrossRef]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- International Peach Genome Initiative. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, L.; Li, M.; Lou, Q.; Xia, H.; Wang, P.; Li, T.; Liu, H.; Luo, L. Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished difference to drought resistance. PLoS ONE 2013, 8, e80253. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Miao, X.; Cui, J.; Deng, J.; Wang, X.; Wang, Y.; Zhang, Y.; Gao, S.; Yang, K. Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across different salt stress in maize (Zea mays L.). Euphytica 2018, 214, 25. [Google Scholar] [CrossRef]
- Yaish, M.W.; Al-Lawati, A.; Al-Harrasi, I.; Patankar, H.V. Genome-wide DNA Methylation analysis in response to salinity in the model plant caliph medic (Medicago truncatula). BMC Genom. 2018, 19, 78. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Wang, X.; Ynag, G.; Wu, Y.; Su, S.; Li, S.; Liu, H.; Yuan, Y. Analysis of the DNA methylation of maize (Zea mays L.) in response to cold stress based on methylation-sensitive amplified polymorphisms. J. Plant Biol. 2013, 56, 32–38. [Google Scholar] [CrossRef]
- Lira-Medeiros, C.F.; Parisod, C.; Fernandes, R.A.; Mata, C.S.; Cardoso, M.A.; Ferreira, P.C.G. Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE 2010, 5, e10326. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Gene ID | Position | No. of Intron | CDS (bp) | Size (aa) | MW (kDa) | pI | Arabidopsis Ortholog |
|---|---|---|---|---|---|---|---|---|
| PbDME1 | Pbr016811.1 | Chr14:3306106–3315557(−) | 18 | 5433 | 1810 | 202.43 | 5.89 | AT2G36490/AT3G10010 |
| PbDME2 | Pbr005494.1 | Chr12:20839378–20849041(+) | 18 | 5406 | 1801 | 201.51 | 5.95 | AT2G36490/AT3G10010 |
| PbDME3 | Pbr014401.1 | Chr9:6995060–7006208(−) | 20 | 6048 | 2015 | 225.75 | 6.38 | AT5G04560 |
| PbDME4 | Pbr025523.1 | scaffold404.0:407609–416687(−) | 19 | 5778 | 1925 | 216.71 | 8.52 | AT5G04560 |
| PbDME5 | Pbr025515.1 | scaffold404.0:259775–268853(−) | 19 | 5778 | 1925 | 216.71 | 8.52 | AT5G04560 |
| PbDME6 | Pbr038018.2 | Chr16:14869003–14879562(−) | 19 | 5880 | 1959 | 220.49 | 7.20 | AT5G04560 |
| PbDME7 | Pbr027983.1 | Chr11:19457199–19465105(+) | 19 | 3702 | 1233 | 138.88 | 9.01 | AT4G34060 |
| PbCIPK1 | Length of Target Genome Sequence (204 bp) | Number of Bisulfite Sequences (Used/Excluded/Total) | ||||||||||||||||||||
| Number of CpGs (7) | 7/0/7 | |||||||||||||||||||||
| Number of Sites | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||
| CpG Positio | 11 | 37 | 48 | 63 | 74 | 80 | 85 | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | Total | |
| Controls Me-CpG | 0/10 | 0/10 | 0/10 | 1/10 | 0/10 | 0/10 | 0/10 | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | 1/70 | |
| 0.0% | 0.0% | 0.0% | 10.0% | 0.0% | 0.0% | 0.0% | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | 1.4% | ||
| NaCl Me-CpG | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | 0/70 | |
| 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | – – | 0.0% | ||
| PbCIPK20 | Length of Target Genome Sequence (225 bp) | |||||||||||||||||||||
| Number of CpGs (20) | 20/0/20 | |||||||||||||||||||||
| CpG Position | 4 | 10 | 15 | 44 | 53 | 67 | 76 | 86 | 97 | 100 | 110 | 122 | 129 | 141 | 144 | 164 | 179 | 207 | 222 | 224 | Total | |
| Controls Me-CpG | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 1/200 | |
| 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 10.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.5% | ||
| NaCl Me-CpG | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/200 | |
| 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | ||
| PbCIPK22 | Length of Target Genome Sequence (215 bp) | |||||||||||||||||||||
| Number of CpGs (11) | 11/0/11 | |||||||||||||||||||||
| CpG Position | 1 | 5 | 27 | 29 | 108 | 140 | 159 | 170 | 179 | 198 | 214 | – – | – – | – – | – – | – – | – – | – – | – – | – – | Total | |
| Controls Me-CpG | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | – – | – – | – – | – – | – – | – – | – – | – – | – – | 1/110 | |
| 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 10.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | – – | – – | – – | – – | – – | – – | – – | – – | – – | 0.9% | ||
| NaCl Me-CpG | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | – – | – – | – – | – – | – – | – – | – – | – – | – – | 0/110 | |
| 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | – – | – – | – – | – – | – – | – – | – – | – – | – – | 0.0% | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Li, H.; Lin, J.; Wang, Y.; Xu, X.; Cheng, Z.-M.; Chang, Y. Genome-Wide Characterization of DNA Demethylase Genes and Their Association with Salt Response in Pyrus. Genes 2018, 9, 398. https://doi.org/10.3390/genes9080398
Liu C, Li H, Lin J, Wang Y, Xu X, Cheng Z-M, Chang Y. Genome-Wide Characterization of DNA Demethylase Genes and Their Association with Salt Response in Pyrus. Genes. 2018; 9(8):398. https://doi.org/10.3390/genes9080398
Chicago/Turabian StyleLiu, Chunxiao, Hui Li, Jing Lin, Ying Wang, Xiaoyang Xu, Zong-Ming (Max) Cheng, and Yonghong Chang. 2018. "Genome-Wide Characterization of DNA Demethylase Genes and Their Association with Salt Response in Pyrus" Genes 9, no. 8: 398. https://doi.org/10.3390/genes9080398




