Phenotype- and SSR-Based Estimates of Genetic Variation between and within Two Important Elymus Species in Western and Northern China
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Phenotype Observation
2.2. DNA Extraction and Polymerase Chain Reaction Amplification
2.3. Data Analysis
3. Results
3.1. Phenotypic Differentiation between Two Elymus Species
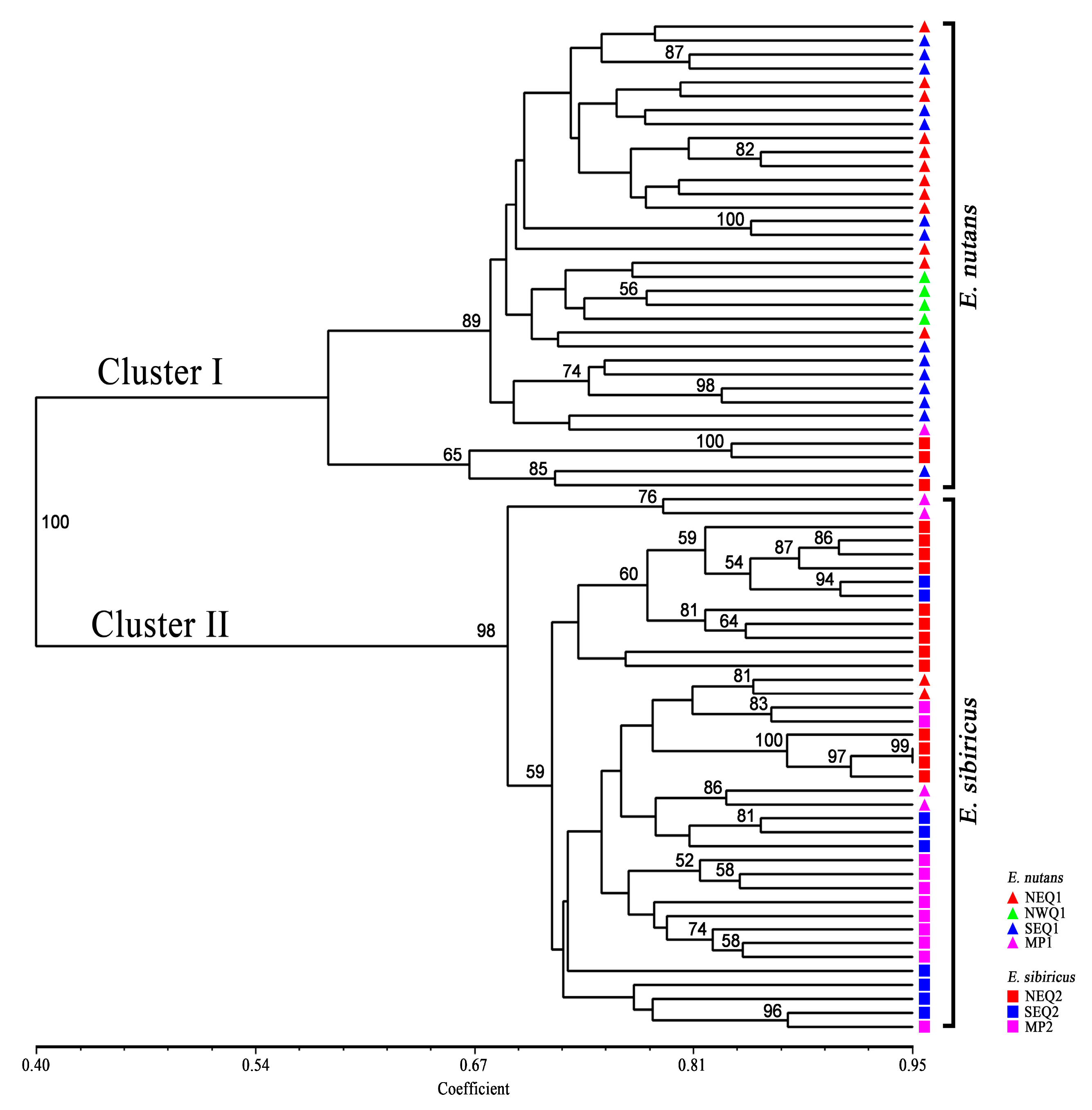
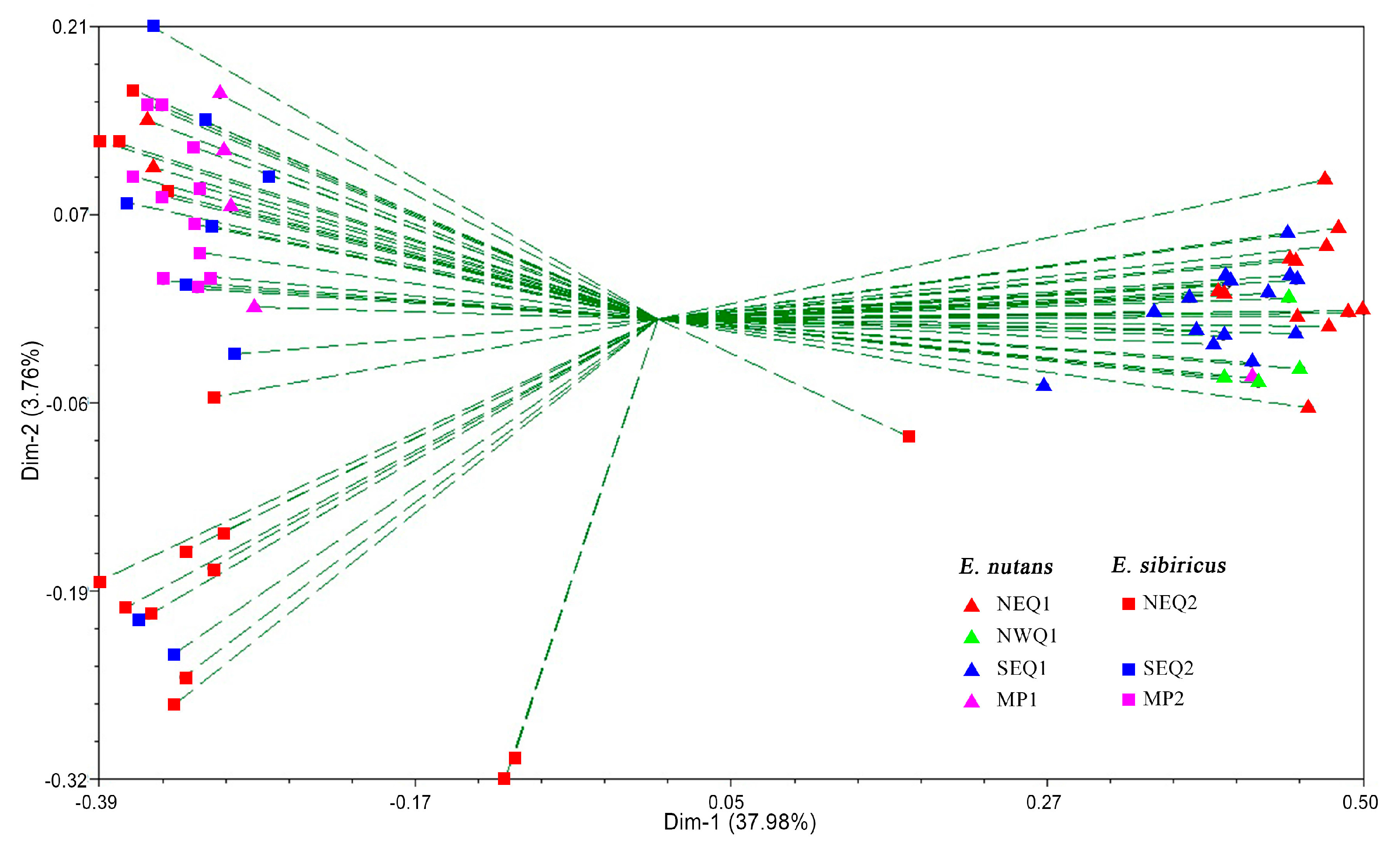
3.2. Genetic Diversity and Genetic Relationship between Two Elymus Species
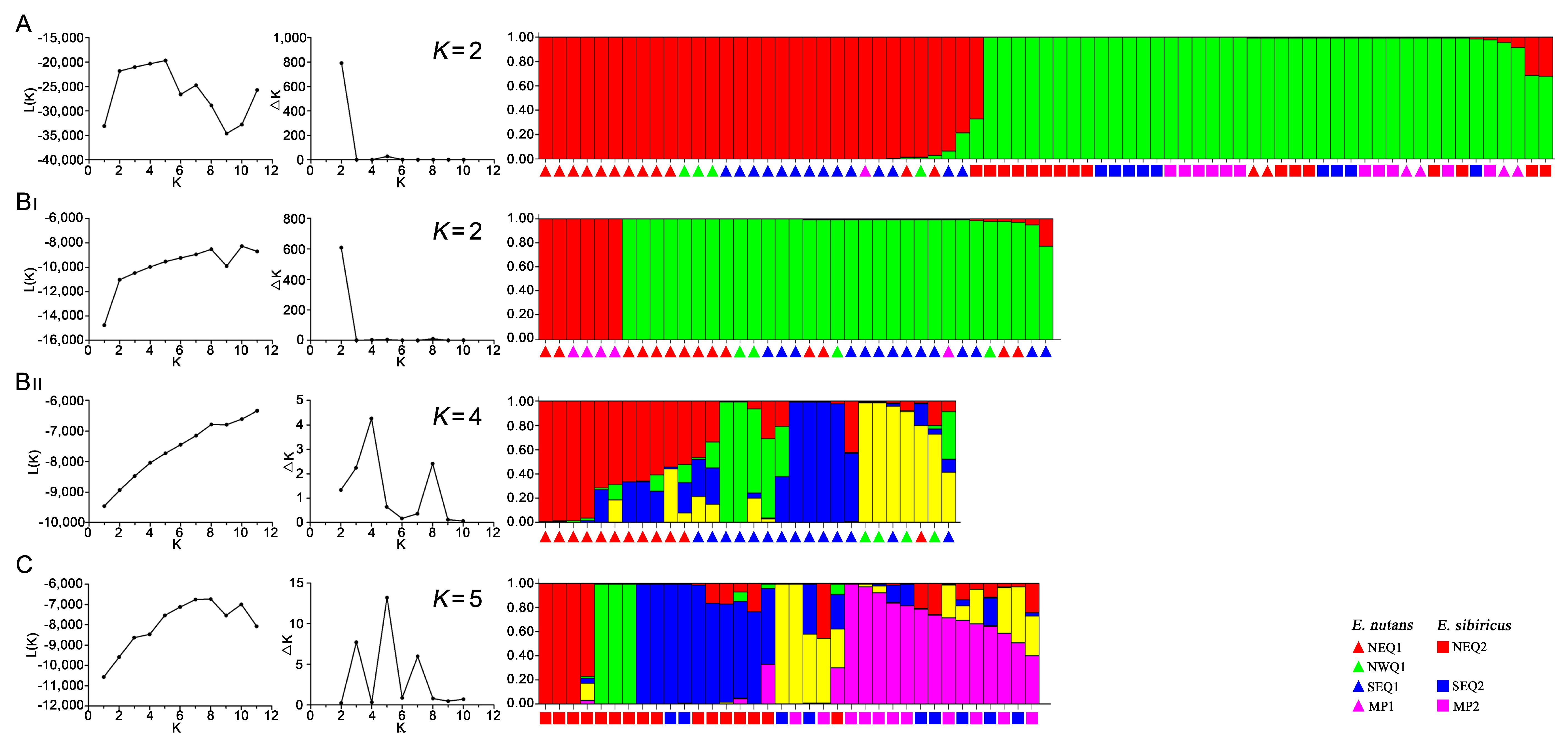
3.3. Genetic Structure between and within the Two Elymus Species
3.4. Genetic Variation among and within Geographic Groups
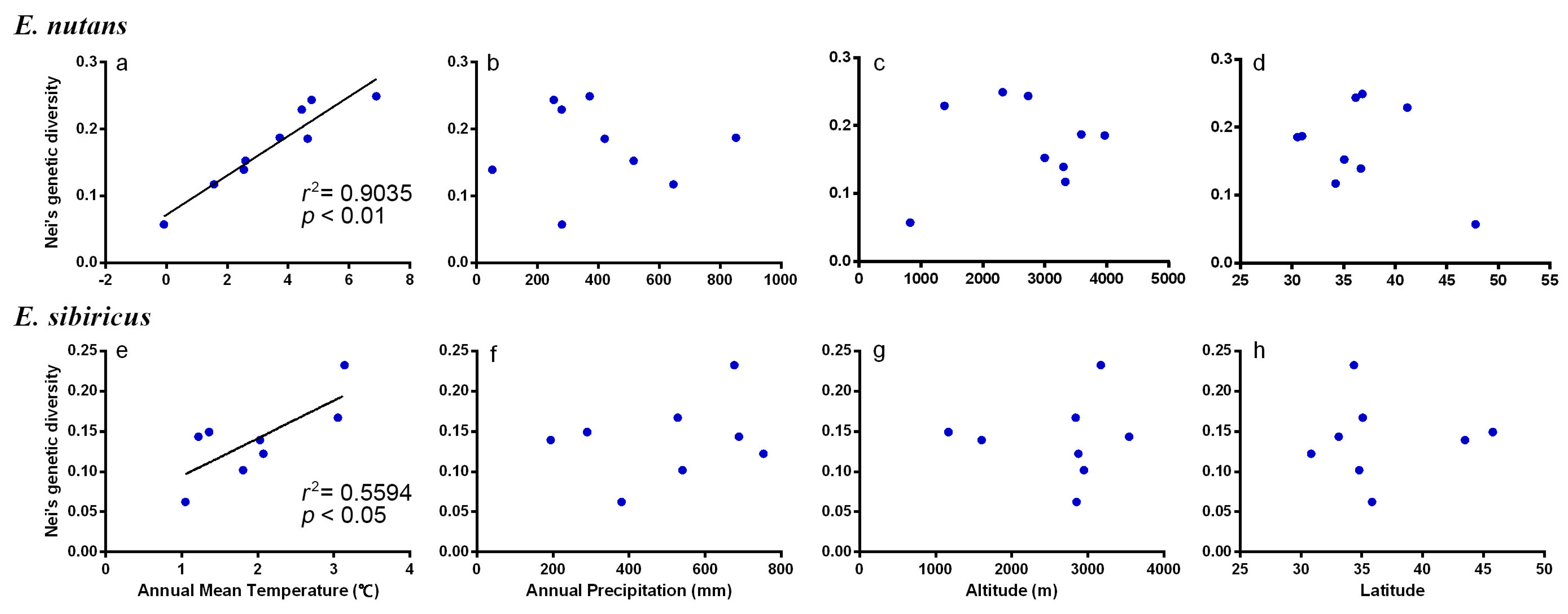
3.5. Association between Habitat Parameters and Genetic Variation
4. Discussion
4.1. Phenotypic and Genetic Diversity between the Two Elymus Species
4.2. Genetic Variation of Geographic Groups and Its Correlation to Environment Factors
4.3. Conservation Implications
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dewey, D.R. The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. Taxon 1984, 35, 202. [Google Scholar]
- Chen, Z.H.; Miao, J.M.; Zhong, J.C.; Ma, X.; Chen, S.Y.; Zhang, X.Q. Genetic diversity of wild Elymus nutans germplasm detected by SRAP markers. Acta Pratacult. Sin. 2009, 18, 192–200. [Google Scholar]
- Ma, X.; Chen, S.Y.; Zhang, X.Q.; Bai, S.Q.; Zhang, C.B. Assessment of worldwide genetic diversity of Siberian wild rye (Elymus sibiricus L.) germplasm based on gliadin analysis. Molecules 2012, 17, 4424–4434. [Google Scholar] [CrossRef] [PubMed]
- You, M.H.; Liu, J.P.; Bai, S.Q.; Li, D.X.; Yan, J.J.; Zhang, C.B. Influence of row spaces and planting years on yield of fresh grass and seeds of Elymus sibiricus. Pratacult. Sci. 2012, 29, 1278–1284. [Google Scholar]
- Lu, B.R. Meiotic studies of Elymus nutans and E. jacquemontii (Poaceae, Triticeae) and their hybrids with Pseudoroegneria spicata and seventeen Elymus species. Plant Syst. Evol. 1993, 186, 193–212. [Google Scholar] [CrossRef]
- Knapp, E.E.; Rice, K.J. Genetic structure and gene flow in Elymus glaucus (blue wildrye): Implications for native grassland restoration. Restor. Ecol. 1996, 4, 1–10. [Google Scholar] [CrossRef]
- Xie, W.G.; Zhao, X.H.; Zhang, J.Q.; Wang, Y.R.; Liu, W.X. Assessment of genetic diversity of Siberian wild rye (Elymus sibiricus L.) germplasms with variation of seed shattering and implication for future genetic improvement. Biochem. Syst. Ecol. 2015, 58, 211–218. [Google Scholar] [CrossRef]
- Lanwrenee, T. Inheritance of a dwarf character in Russian wild ryegrass, Elymus junceus. Can. J. Genet. Cytol. 1967, 9, 126–128. [Google Scholar]
- Sharma, H.C.; Gill, B.S. New hybrids between Agropyron and wheat. Theor. Appl. Genet. 1983, 66, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.B.; Guo, Y.X.; Zhou, H.; Wang, K. Isozyme variability among Elymus species indigenous to the Tibetan and Inner Mongolian Plateaus. Grassl. Sci. 2007, 53, 91–96. [Google Scholar] [CrossRef]
- Díaz, O.; Salomon, B.; Bothmer, R.V. Genetic diversity and structure in populations of Elymus caninus (L.) L. (Poaceae). Hereditas 1999, 131, 63–74. [Google Scholar] [CrossRef]
- Last, L.; Lüscher, G.; Widmer, F.; Boller, B.; Kölliker, R. Indicators for genetic and phenotypic diversity of Dactylis glomerata in Swiss permanent grassland. Ecol. Ind. 2014, 38, 181–191. [Google Scholar] [CrossRef]
- Xie, W.G.; Zhang, X.Q.; Cai, H.W.; Liu, W.; Peng, Y. Genetic diversity analysis and transferability of cereal EST-SSR markers to orchardgrass (Dactylis glomerata L.). Biochem. Syst. Ecol. 2010, 38, 740–749. [Google Scholar] [CrossRef]
- Madesis, P.; Abraham, E.M.; Kalivas, A.; Ganopoulos, I.; Tsaftaris, A. Genetic diversity and structure of natural Dactylis glomerata L. populations revealed by morphological and microsatellite-based (SSR/ISSR) markers. Genet. Mol. Res. 2014, 13, 4226–4240. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Luo, D.; Ma, L.C.; Xie, W.G.; Wang, Y.; Wang, Y.R.; Liu, Z.P. Development and cross-species transferability of EST-SSR markers in Siberian wildrye (Elymus sibiricus L.) using illumina sequencing. Sci. Rep. 2016, 6, 20549. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Geethanjali, S.; Sameeyafarheen, F.; Maheswaran, M. Molecular marker based genetic diversity analysis in rice (Oryza sativa L.) using RAPD and SSR markers. Euphytica 2003, 133, 243–252. [Google Scholar] [CrossRef]
- Huang, H.; Lu, J.; Ren, Z.B.; Hunter, W.; Dowd, S.E.; Dang, P. Mining and validating grape (Vitis L.) ESTs to develop EST-SSR markers for genotyping and mapping. Mol. Breed. 2011, 28, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.K.; Huang, X.; Yan, H.D.; Yin, G.H.; Zhang, X.Q.; Tian, Y.; Zhang, Y.; Jiang, X.M.; Yan, Y.H.; Ma, X.; et al. Constructing DNA fingerprinting of Hemarthria cultivars using EST-SSR and SCoT markers. Genet. Resour. Crop Evol. 2014, 61, 1047–1055. [Google Scholar] [CrossRef]
- Ma, X.; Zhou, Y.H.; Yu, H.Q.; Zhang, H.Q. Genetic diversity of Gliadin in wild germplasm of Elymus nutans Griseb. Hereditas 2006, 28, 699–706. [Google Scholar] [PubMed]
- Miao, J.M.; Zhang, X.Q.; Chen, S.Y.; Ma, X.; Chen, Z.H.; Zhong, J.C.; Bai, S.Q. Gliadin analysis of Elymus nutans Griseb. from the Qinghai-Tibetan Plateau and Xinjiang, China. Grassl. Sci. 2011, 57, 127–134. [Google Scholar] [CrossRef]
- Yan, J.J.; Bai, S.Q.; Zhang, X.Q.; You, M.H.; Zhang, C.B.; Li, D.X.; Zeng, Y. Genetic diversity of wild Elymus sibiricus germplasm from the Qinghai-Tibetan Plateau in China detected by SRAP markers. Acta Pratacult. Sin. 2010, 19, 173–183. [Google Scholar]
- Zhao, X.H.; Jiang, X.; Zhao, K.; Zhao, X.H.; Yin, J.; Xie, W.G. Screening of germplasm with low seed shattering rate and evaluation on agronomic traits in Elymus sibiricus L. J. Plant Gen. Resour. 2015, 16, 691–699. [Google Scholar]
- Shan, Z.; Wu, H.L.; Li, C.L.; Chen, H.; Wu, Q. Improved SDS method for general plant genomic DNA extraction. Guangdong Agric. Sci. 2011, 38, 113–115. [Google Scholar]
- Bushman, B.S.; Larson, S.R.; Mott, I.W.; Cliften, P.F.; Wang, R.R.; Chatterton, N.J.; Hernandez, A.G.; Ali, S.; Kim, R.W.; Thimmapuram, J.; et al. Development and annotation of perennial Triticeae ESTs and SSR markers. Genome 2008, 51, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana/Chicago, IL, USA, 1949. [Google Scholar]
- Zhang, M.Q.; Wang, Y.R.; Zhang, J.Y.; Liu, Z.P.; Zhang, L.; Nie, B.; Zhou, J. A study on genetic diversity of reproductive characters in Elymus nutans germplasm resources. Acta Pratacult. Sin. 2011, 20, 182–191. [Google Scholar]
- Deng, W.K.; Wang, Y.B.; Liu, Z.X.; Cheng, H.; Xue, Y. HemI: A Toolkit for Illustrating Heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Wang, M.X.; Chen, Y.W. An analysis of population genetic structure of masson pine by isozyme technique. Sci. Silvae Scin. 1988, 24, 399–409. [Google Scholar]
- Ghislain, M.; Zhang, D.P.; Fajardo, D.; Huamán, Z.; Hijmans, R.J. Marker-assisted sampling of the cultivated Andean potato Solanum phureja collection using RAPD markers. Genet. Resour. Crop Evol. 1999, 46, 547–555. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.C.; Boyle, T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 157. [Google Scholar]
- Rohlf, F.J. NTSYS-pc-Numerical Taxonomy and Multivariate Analysis System; Exeter Software: Setauket, NY, USA, 1988. [Google Scholar]
- Yap, I.V.; Nelson, R.J. Winboot: A Program for Performing Bootstrap Analysis of Binary Data to Determine the Confidence Limits of UPGMA-based Dendrograms; International Rice Research Institute: Manila, Philippines, 1996. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [PubMed]
- Zhang, C.L.; Zhang, J.B.; Fan, Y.; Sun, M.; Wu, W.D.; Zhao, W.D.; Yang, X. P; Huang, L.K.; Peng, Y.; Ma, X.; et al. Genetic structure and eco-geographical differentiation of wild sheep fescue (Festuca ovina L.) in Xinjiang, Northwest China. Molecules 2017, 22, 1316. [Google Scholar] [CrossRef] [PubMed]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Shehzad, T.; Okuizumi, H.; Kawase, M.; Okuno, K. Development of SSR-based sorghum (Sorghum bicolor (L.) Moench) diversity research set of germplasm and its evaluation by morphological traits. Genet. Resour. Crop Evol. 2009, 56, 809–827. [Google Scholar] [CrossRef]
- Chen, S.Y.; Chen, Z.H.; Zhou, Q.P.; Li, S.D.; Ma, X.; Zhang, X.Q. Study on morphological diversity of Elymus nutans germplasm resources from Qinghai-Tibetan Plateau. Chin. J. Grassl. 2016, 38, 27–33. [Google Scholar]
- Zhang, J.B.; Bai, S.Q.; Zhang, X.Q.; Ma, X.; Yan, J.J.; Zhang, C.B.; You, M.H. Study on ear characters of Elymus nutans Griseb. in the northwestern plateau of Sichuan province. J. Sichuan Univ. 2009, 46, 1505–1509. [Google Scholar]
- Zhang, J.C.; Xie, W.G.; Wang, Y.R.; Zhao, X.H. Potential of Start Codon Targeted (SCoT) Markers to Estimate Genetic Diversity and Relationships among Chinese Elymus sibiricus Accessions. Molecules 2015, 20, 5987–6001. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Zhang, X.Q.; Ma, X.; Huang, L.K. Assessment of genetic diversity and differentiation of Elymus nutans indigenous to Qinghai-Tibet Plateau using simple sequence repeats markers. Can. J. Plant Sci. 2013, 93, 1089–1096. [Google Scholar] [CrossRef]
- Xie, W.G.; Zhang, X.Q.; Ma, X.; Cai, H.W.; Huang, L.K.; Peng, Y.; Zeng, B. Diversity comparison and phylogenetic relationships of cocksfoot (Dactylis glomerata L.) germplasm as revealed by SSR markers. Can. J. Plant Sci. 2010, 90, 13–21. [Google Scholar] [CrossRef]
- Stevens, L.; Salomon, B.; Sun, G. Microsatellite variability and heterozygote excess in Elymus trachycaulus population from British Columbia in Canada. Biochem. Syst. Ecol. 2007, 35, 725–736. [Google Scholar] [CrossRef]
- Wilson, B.L.; Kitzmiller, J.; Rolle, W.; Hipkins, V.D. Isozyme variation and its environmental correlates in Elymus glaucus from California Floristic Province. Can. J. Bot. 2001, 79, 139–153. [Google Scholar]
- Yan, X.B.; Guo, Y.X.; Liu, F.Y.; Zhao, C.; Liu, Q.L.; Lu, B.R. Population structure affected by excess gene flow in self-pollinating Elymus nutans and E. burchan-buddae (Triticeae: Poaceae). Popul. Ecol. 2010, 52, 233–241. [Google Scholar] [CrossRef]
- Huang, H.; Liu, Y. Natural hybridization, introgression breeding, and cultivar improvement in the genus actinidia. Tree Genet. Genomes 2014, 10, 1113–1122. [Google Scholar] [CrossRef]
- Jensen, K.B.; Mott, I.W.; Robins, J.G.; Waldron, B.L.; Nelson, M. Genetic improvement and diversity in Snake River wheatgrass (Elymus wawawaiensis) (Poaceae: Triticeae). Rangel. Ecol. Manag. 2012, 65, 76–84. [Google Scholar] [CrossRef]
- Robins, J.G.; Bushman, B.S.; Jensen, K.B. Dry matter yield combing ability among nine sources of cocksfoot germplasm. Euphytica 2012, 188, 419–428. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Zhang, J.C.; Zhao, X.H.; Xie, W.G.; Wang, Y.R. Assessing and broadening genetic diversity of Elymus sibiricus germplasm for the improvement of seed shattering. Molecules 2016, 21, 869. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.H.; Zhang, J.C.; Zhang, Z.Y.; Wang, Y.R.; Xie, W.G. Hybrid identification and genetic variation of Elymus sibiricus hybrid populations using EST-SSR markers. Hereditas 2017, 154, 15. [Google Scholar] [CrossRef] [PubMed]
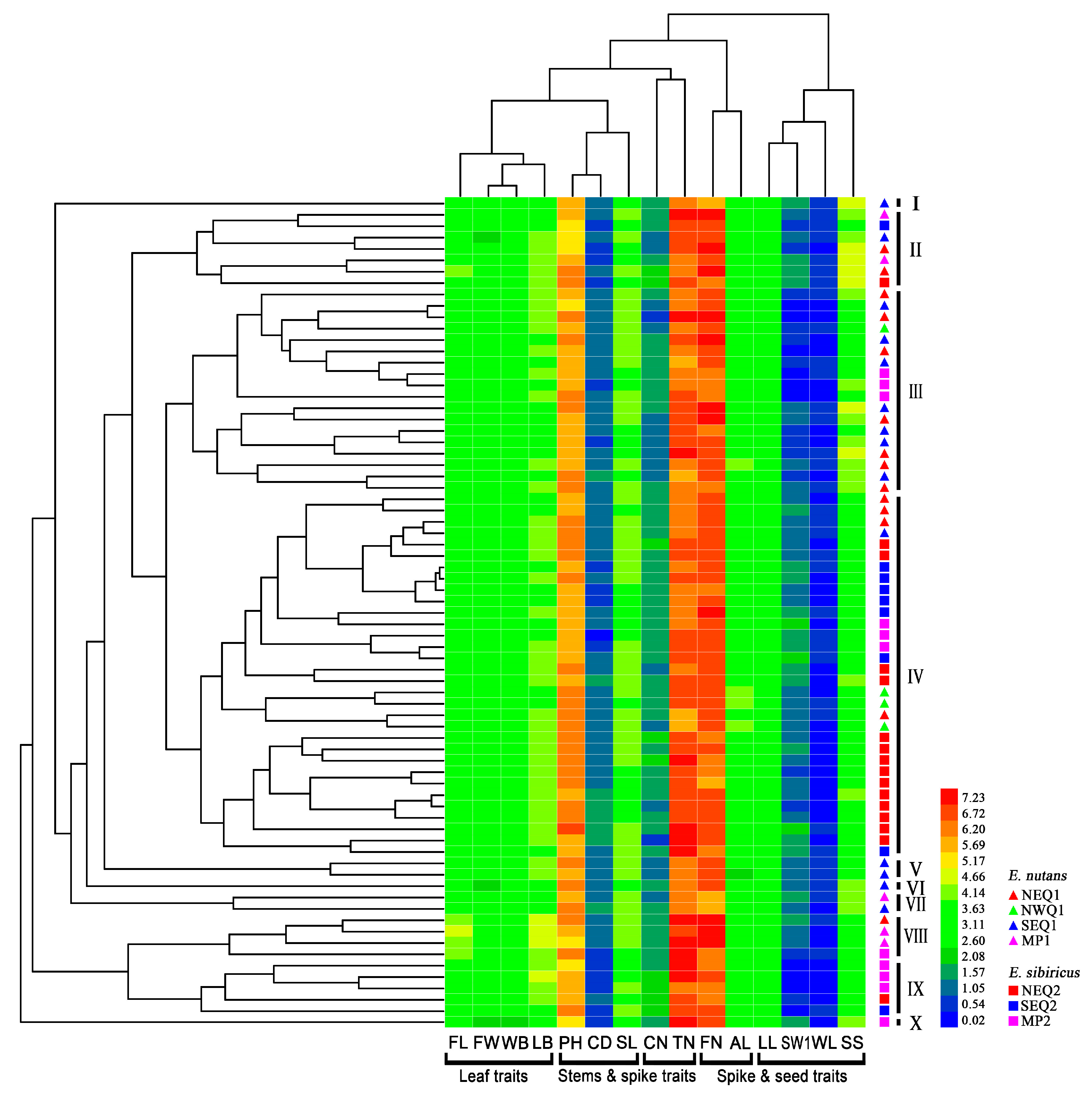
| Traits | E. nutans | E. sibiricus | Pst (S) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variance Portion | Percent of Variance Portion (%) | Pst | Variance Portion | Percent of Variance Portion (%) | Pst | ||||||
| AP (σ2t/s) | WP (σ2s) | AP (σ2t/s) | WP (σ2s) | AP (σ2t/s) | WP (σ2s) | AP (σ2t/s) | WP (σ2s) | ||||
| FL | 6.23 | 14.43 | 21.34 | 49.40 | 30.17% | 2.14 | 8.67 | 10.99 | 44.53 | 19.80% | 5.02% |
| FW | 1.19 | 2.94 | 18.52 | 45.93 | 28.74% | 0.01 | 0.05 | 15.41 | 60.29 | 20.37% | 1.86% |
| LB | 3.46 | 15.08 | 11.77 | 51.34 | 18.65% | 2.09 | 5.92 | 11.91 | 33.71 | 26.10% | 6.60% |
| WB | 0.88 | 2.84 | 14.83 | 47.59 | 23.76% | 0.02 | 0.04 | 23.20 | 49.94 | 31.73% | 3.93% |
| PH | 29.18 | 138.09 | 11.57 | 54.78 | 17.44% | 210.93 | 155.07 | 39.67 | 29.16 | 57.63% | 0.00% |
| CN | 0.02 | 0.34 | 3.59 | 53.45 | 6.29% | 0.02 | 0.48 | 0.00 | 55.21 | 0.00% | 16.56% |
| TN | 22.41 | 1175.19 | 0.91 | 47.91 | 1.87% | 81.55 | 823.07 | 3.49 | 35.24 | 9.02% | 11.72% |
| CD | 0.00 | 0.07 | 0.00 | 29.55 | 0.00% | 0.35 | 0.23 | 0.00 | 0.25 | 0.00% | 8.94% |
| SL | 0.65 | 5.69 | 0.00 | 48.67 | 0.00% | 0.69 | 7.88 | 5.13 | 58.23 | 8.09% | 5.10% |
| FN | 128.25 | 797.29 | 7.04 | 43.75 | 13.86% | 170.48 | 432.22 | 15.25 | 38.67 | 28.29% | 0.00% |
| LL | 0.01 | 0.01 | 10.95 | 46.86 | 18.95% | 0.01 | 0.01 | 18.78 | 35.90 | 34.35% | 3.79% |
| WL | 0.01 | 0.01 | 0.21 | 47.04 | 0.44% | 0.00 | 0.00 | 0.00 | 0.91 | 0.00% | 0.00% |
| AL | 0.01 | 0.08 | 11.60 | 65.66 | 15.02% | 0.01 | 0.03 | 3.53 | 60.37 | 5.53% | 18.44% |
| SW1 | 0.04 | 0.42 | 6.78 | 77.78 | 8.02% | 0.06 | 1.17 | 4.61 | 87.56 | 5.00% | 3.10% |
| SS | 1.50 | 20.64 | 1.76 | 24.15 | 6.78% | 1.08 | 9.09 | 5.09 | 43.14 | 10.57% | 23.43% |
| Mean | 12.92 | 144.87 | 8.06 | 48.92 | 12.67% | 31.29 | 96.26 | 10.47 | 42.21 | 17.10% | 7.23% |
| Traits | Range | Mean Value | Standard Deviation | CV | H’ | PCA of E. nutans | PCA of E. sibiricus | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EN | ES | EN | ES | EN | ES | EN | ES | EN | ES | PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | ||
| Leaf traits | FL (cm) | 7.20–25.60 | 7.50–23.40 | 12.38 | 12.29 | 1.93 | 1.64 | 15.66% | 13.61% | 2.15 | 2.08 | 0.78 | −0.37 | 0.32 | 0.71 | −0.42 | −0.11 |
| FW (mm) | 4.90–13.80 | 5.30–12.90 | 9.31 | 9.12 | 1.01 | 0.99 | 11.28% | 11.14% | 2.11 | 2.17 | 0.86 | 0.15 | −0.17 | 0.88 | −0.26 | 0.20 | |
| LB (cm) | 12.60–30.80 | 13.30–25.60 | 19.00 | 19.03 | 2.18 | 1.63 | 11.62% | 8.54% | 1.90 | 2.08 | 0.78 | −0.41 | 0.19 | 0.81 | −0.29 | −0.11 | |
| WB (mm) | 6.10–15.10 | 5.90–13.80 | 10.91 | 10.44 | 1.04 | 1.04 | 9.88% | 10.26% | 2.23 | 2.07 | 0.84 | 0.09 | −0.33 | 0.82 | −0.18 | 0.30 | |
| Stems traits | PH (cm) | 44.30–99.60 | 46.70–114.90 | 71.09 | 72.12 | 6.56 | 7.55 | 9.54% | 10.70% | 1.98 | 2.08 | 0.14 | 0.66 | −0.49 | 0.50 | 0.34 | −0.33 |
| CN (No.) | 1.00–4.50 | 1.00–5.30 | 2.97 | 3.75 | 0.39 | 0.49 | 13.25% | 13.69% | 2.07 | 2.22 | 0.60 | 0.27 | 0.21 | 0.18 | −0.34 | 0.43 | |
| TN (No.) | 65.60–214.5 | 79.30–198.60 | 110.95 | 130.76 | 23.10 | 25.92 | 20.46% | 19.97% | 1.93 | 2.09 | 0.36 | −0.49 | 0.15 | 0.15 | −0.11 | −0.36 | |
| CD (mm) | 1.60–3.10 | 1.40–3.40 | 2.43 | 2.30 | 0.29 | 0.29 | 11.89% | 12.33% | 1.87 | 1.98 | 0.26 | 0.25 | −0.65 | 0.57 | 0.46 | −0.51 | |
| Spike and seed traits | SL (cm) | 12.30–24.70 | 11.10–24.70 | 18.63 | 17.57 | 1.57 | 1.46 | 8.52% | 8.47% | 2.10 | 2.21 | 0.55 | −0.03 | −0.13 | 0.33 | 0.69 | −0.04 |
| FN (No.) | 56.10–211.3 | 68.20–155.00 | 128.57 | 113.55 | 24.17 | 17.79 | 19.88% | 15.47% | 2.11 | 2.12 | 0.48 | −0.54 | 0.17 | 0.12 | 0.30 | 0.50 | |
| LL (mm) | 7.30–10.60 | 7.30–12.30 | 9.21 | 9.59 | 0.46 | 0.69 | 4.99% | 7.12% | 2.15 | 2.05 | 0.35 | 0.68 | 0.26 | 0.25 | 0.66 | −0.15 | |
| WL (mm) | 1.00–1.80 | 1.10–1.70 | 1.49 | 1.41 | 0.14 | 0.13 | 9.55% | 9.43% | 2.22 | 1.90 | 0.17 | 0.64 | 0.54 | 0.09 | −0.05 | 0.69 | |
| AL (mm) | 5.00–19.00 | 8.90–15.60 | 13.45 | 11.52 | 1.14 | 1.02 | 8.67% | 9.03% | 2.13 | 2.26 | 0.20 | 0.19 | −0.45 | 0.15 | 0.26 | 0.49 | |
| SW1 (g) | 1.10–4.00 | 1.00–5.00 | 2.36 | 2.71 | 0.19 | 0.21 | 8.51% | 8.64% | 1.81 | 1.51 | 0.37 | 0.65 | 0.47 | 0.12 | 0.71 | 0.30 | |
| SS (gf) | 11.70–33.00 | 9.30–28.20 | 19.54 | 14.74 | 5.52 | 2.21 | 28.11% | 15.45% | 1.64 | 2.07 | −0.43 | 0.21 | 0.48 | 0.02 | 0.51 | 0.29 | |
| Mean | 12.79% | 11.59% | 2.03 | 2.06 | |||||||||||||
| EV | 4.27 | 2.80 | 2.05 | 3.47 | 2.67 | 2.00 | |||||||||||
| %V | 28.44 | 18.64 | 13.70 | 23.13 | 17.77 | 13.34 | |||||||||||
| C% | 28.44 | 47.08 | 60.77 | 23.13 | 40.90 | 54.24 | |||||||||||
| Species | Pop | AMT (°C) | AP (mm) | A (m) | L (N) | N | NPL | PPL (%) | NPB | Na | Ne | I | H |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. nutans | NEQ1 | 4.00 | 446.20 | 2819.30 | 35°63′ | 14 | 359 | 88.64 | 4 | 1.82 ± 0.03 | 1.45 ± 0.02 | 0.42 ± 0.01 | 0.27 ± 0.01 |
| NWQ1 | 2.50 | 51.30 | 3300.00 | 36°67′ | 4 | 156 | 38.52 | 0 | 1.02 ± 0.04 | 1.24 ± 0.02 | 0.21 ± 0.01 | 0.14 ± 0.01 | |
| SEQ1 | 4.20 | 636.50 | 3779.30 | 30°75′ | 14 | 306 | 75.56 | 5 | 1.63 ± 0.03 | 1.36 ± 0.02 | 0.34 ± 0.01 | 0.22 ± 0.01 | |
| MP1 | 2.60 | 279.70 | 1158.00 | 43°82′ | 5 | 283 | 69.88 | 11 | 1.50 ± 0.04 | 1.38 ± 0.02 | 0.35 ± 0.01 | 0.23 ± 0.01 | |
| Mean | 276.0 | 68.15 | 5.0 | 1.49 ± 0.02 | 1.36 ± 0.01 | 0.33 ± 0.01 | 0.22 ± 0.01 | ||||||
| 47 | |||||||||||||
| E. sibiricus | NEQ2 | 2.30 | 521.80 | 2930.90 | 35°06′ | 16 | 311 | 81.84 | 62 | 1.78 ± 0.03 | 1.37 ± 0.02 | 0.36 ± 0.01 | 0.23 ± 0.01 |
| SEQ2 | 1.60 | 717.30 | 3244.40 | 32°10′ | 9 | 204 | 53.68 | 3 | 1.29 ± 0.04 | 1.29 ± 0.02 | 0.27 ± 0.01 | 0.18 ± 0.01 | |
| MP2 | 1.70 | 237.60 | 1405.80 | 44°52′ | 11 | 205 | 53.95 | 5 | 1.31 ± 0.04 | 1.28 ± 0.02 | 0.26 ± 0.01 | 0.17 ± 0.01 | |
| Mean | 240.0 | 63.16 | 23.30 | 1.46 ± 0.02 | 1.31 ± 0.01 | 0.29 ± 0.01 | 0.19 ± 0.01 | ||||||
| 40 |
| E. nutans | NEQ1 | NWQ1 | SEQ1 | MP1 |
| NEQ1 | 0.11 | 0.05 | 0.14 | |
| NWQ1 | 0.17 | 0.09 | 0.31 | |
| SEQ1 | 0.07 | 0.17 | 0.23 | |
| MP1 | 0.17 | 0.37 | 0.26 | |
| E. sibiricus | NEQ2 | SEQ2 | MP2 | |
| NEQ2 | 0.04 | 0.05 | ||
| SEQ2 | 0.08 | 0.04 | ||
| MP2 | 0.09 | 0.09 |
| Species | Source of Variance | Degrees of Freedom | Sum of Squares | Mean Square | Variance Components | Total Variance | p-Value |
|---|---|---|---|---|---|---|---|
| E. nutans | Among geographic groups | 3 | 503.15 | 167.72 | 13.07 | 18.53% | <0.001 |
| Within geographic groups | 33 | 1897.06 | 57.49 | 56.49 | 81.47% | <0.001 | |
| Total | 36 | 2400.21 | 69.56 | ||||
| E. sibiricus | Among geographic groups | 2 | 218.15 | 109.08 | 5.45 | 10.68% | <0.001 |
| Within geographic groups | 33 | 1505.43 | 45.62 | 45.62 | 89.32% | <0.001 | |
| Total | 35 | 1723.58 | 51.07 |
| Species | Pearson Coefficient | Annual Mean Temperature | Annual Precipitation | Altitude | Latitude |
|---|---|---|---|---|---|
| E. nutans | r | 0.95 | −0.03 | 0.16 | −0.39 |
| p | <0.01 ** | 0.95 ns | 0.68 ns | 0.29 ns | |
| E. sibiricus | r | 0.78 | 0.28 | 0.04 | 0.03 |
| p | 0.03 * | 0.51 ns | 0.92 ns | 0.95 ns |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Xie, W.; Zhang, J.; Zhao, X.; Zhao, Y.; Wang, Y. Phenotype- and SSR-Based Estimates of Genetic Variation between and within Two Important Elymus Species in Western and Northern China. Genes 2018, 9, 147. https://doi.org/10.3390/genes9030147
Zhang Z, Xie W, Zhang J, Zhao X, Zhao Y, Wang Y. Phenotype- and SSR-Based Estimates of Genetic Variation between and within Two Important Elymus Species in Western and Northern China. Genes. 2018; 9(3):147. https://doi.org/10.3390/genes9030147
Chicago/Turabian StyleZhang, Zongyu, Wengang Xie, Junchao Zhang, Xuhong Zhao, Yongqiang Zhao, and Yanrong Wang. 2018. "Phenotype- and SSR-Based Estimates of Genetic Variation between and within Two Important Elymus Species in Western and Northern China" Genes 9, no. 3: 147. https://doi.org/10.3390/genes9030147
APA StyleZhang, Z., Xie, W., Zhang, J., Zhao, X., Zhao, Y., & Wang, Y. (2018). Phenotype- and SSR-Based Estimates of Genetic Variation between and within Two Important Elymus Species in Western and Northern China. Genes, 9(3), 147. https://doi.org/10.3390/genes9030147




