Mcm10: A Dynamic Scaffold at Eukaryotic Replication Forks
Abstract
:1. Efficient Replication of Large Eukaryotic Genomes
2. Discovery and Biochemical Characterization of Mcm10
3. The Multifaceted Regulation of Mcm10 Function
4. Mcm10 is a Central Player in Multiple Steps of DNA Replication
5. Mcm10 Promotes Assembly of the Replicative Helicase
6. Activation of the CMG Helicase Relies on Mcm10
7. Mcm10-Dependent Polymerase Loading
8. Replication Fork Progression and Stability Relies on Mcm10
9. Emerging Connections between Mcm10 and Cancer Development
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sun, J.; Kong, D. DNA replication origins, ORC/DNA interaction, and assembly of pre-replication complex in eukaryotes. Acta Biochim. Biophys. Sin. 2010, 42, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Edenberg, H.J.; Huberman, J.A. Eukaryotic chromosome replication. Annu. Rev. Genet. 1975, 9, 245–284. [Google Scholar] [CrossRef] [PubMed]
- Heichinger, C.; Penkett, C.J.; Bahler, J.; Nurse, P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006, 25, 5171–5179. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, M.K.; Winzeler, E.A.; Collingwood, D.; Hunt, S.; Wodicka, L.; Conway, A.; Lockhart, D.J.; Davis, R.W.; Brewer, B.J.; Fangman, W.L. Replication dynamics of the yeast genome. Science 2001, 294, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Wyrick, J.J.; Aparicio, J.G.; Chen, T.; Barnett, J.D.; Jennings, E.G.; Young, R.A.; Bell, S.P.; Aparicio, O.M. Genome-wide distribution of ORC and Mcm proteins in S. cerevisiae: High-resolution mapping of replication origins. Science 2001, 294, 2357–2360. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Carrington, J.T.; Albergante, L.; Al Mamun, M.; Haagensen, E.J.; Komseli, E.S.; Gorgoulis, V.G.; Newman, T.J.; Blow, J.J. Unreplicated DNA remaining from unperturbed s phases passes through mitosis for resolution in daughter cells. Proc. Natl. Acad. Sci. USA 2016, 113, E5757–E5764. [Google Scholar] [CrossRef] [PubMed]
- Picard, F.; Cadoret, J.C.; Audit, B.; Arneodo, A.; Alberti, A.; Battail, C.; Duret, L.; Prioleau, M.N. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet. 2014, 10, e1004282. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Singh, S.; Pradhan, S.; Narayan, G. Mcm paradox: Abundance of eukaryotic replicative helicases and genomic integrity. Mol. Biol. Int. 2014, 2014, 574850. [Google Scholar] [CrossRef] [PubMed]
- Donovan, S.; Harwood, J.; Drury, L.S.; Diffley, J.F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 1997, 94, 5611–5616. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.K.; MacAlpine, H.K.; Prinz, J.A.; Li, Y.; Belsky, J.A.; MacAlpine, D.M. Dynamic loading and redistribution of the Mcm2-7 helicase complex through the cell cycle. EMBO J. 2015, 34, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Alver, R.C.; Chadha, G.S.; Blow, J.J. The contribution of dormant origins to genome stability: From cell biology to human genetics. DNA Repair 2014, 19, 182–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blow, J.J.; Ge, X.Q.; Jackson, D.A. How dormant origins promote complete genome replication. Trends Biochem. Sci. 2011, 36, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Al Mamun, M.; Albergante, L.; Moreno, A.; Carrington, J.T.; Blow, J.J.; Newman, T.J. Inevitability and containment of replication errors for eukaryotic genome lengths spanning megabase to gigabase. Proc. Natl. Acad. Sci. USA 2016, 113, E5765–E5774. [Google Scholar] [CrossRef] [PubMed]
- Besnard, E.; Babled, A.; Lapasset, L.; Milhavet, O.; Parrinello, H.; Dantec, C.; Marin, J.M.; Lemaitre, J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with g-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012, 19, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Newman, T.J.; Mamun, M.A.; Nieduszynski, C.A.; Blow, J.J. Replisome stall events have shaped the distribution of replication origins in the genomes of yeasts. Nucleic Acids Res. 2013, 41, 9705–9718. [Google Scholar] [CrossRef] [PubMed]
- Takeda, D.Y.; Dutta, A. DNA replication and progression through s phase. Oncogene 2005, 24, 2827–2843. [Google Scholar] [CrossRef] [PubMed]
- Evrin, C.; Clarke, P.; Zech, J.; Lurz, R.; Sun, J.; Uhle, S.; Li, H.; Stillman, B.; Speck, C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA 2009, 106, 20240–20245. [Google Scholar] [CrossRef]
- Masai, H.; Matsumoto, S.; You, Z.; Yoshizawa-Sugata, N.; Oda, M. Eukaryotic chromosome DNA replication: Where, when, and how? Annu. Rev. Biochem. 2010, 79, 89–130. [Google Scholar] [CrossRef] [PubMed]
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.; On, K.F.; Diffley, J.F. Regulating DNA replication in eukarya. Cold Spring Harbor Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Bleichert, F.; Botchan, M.R.; Berger, J.M. Crystal structure of the eukaryotic origin recognition complex. Nature 2015, 519, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Ticau, S.; Friedman, L.J.; Ivica, N.A.; Gelles, J.; Bell, S.P. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell 2015, 161, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Bielinsky, A.K. Replication origins: Why do we need so many? Cell Cycle 2003, 2, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P.; Dutta, A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002, 71, 333–374. [Google Scholar] [CrossRef] [PubMed]
- Deegan, T.D.; Diffley, J.F. Mcm: One ring to rule them all. Curr. Opin. Struct. Biol. 2016, 37, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Mulia, J.C.; Gilbert, D.M. Replicating large genomes: Divide and conquer. Mol. Cell 2016, 62, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Schwacha, A. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell 2008, 31, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Moyer, S.E.; Lewis, P.W.; Botchan, M.R. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 2006, 103, 10236–10241. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; van Deursen, F.; Polychronopoulos, D.; Foltman, M.; Jones, R.C.; Edmondson, R.D.; Calzada, A.; Labib, K. A key role for Ctf4 in coupling the Mcm2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009, 28, 2992–3004. [Google Scholar] [CrossRef] [PubMed]
- Ricke, R.M.; Bielinsky, A.K. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell 2004, 16, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Wohlschlegel, J.A.; Dhar, S.K.; Prokhorova, T.A.; Dutta, A.; Walter, J.C. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol. Cell 2002, 9, 233–240. [Google Scholar] [CrossRef]
- Douglas, M.E.; Diffley, J.F. Recruitment of Mcm10 to sites of replication initiation requires direct binding to the minichromosome maintenance (Mcm) complex. J. Biol. Chem. 2016, 291, 5879–5888. [Google Scholar] [CrossRef] [PubMed]
- Perez-Arnaiz, P.; Bruck, I.; Kaplan, D.L. Mcm10 coordinates the timely assembly and activation of the replication fork helicase. Nucleic Acids Res. 2016, 44, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Xia, Y.; Liu, L.; Cui, J.; Li, Z.; Cao, Q.; Chen, X.S.; Campbell, J.L.; Lou, H. Cell-cycle-regulated interaction between Mcm10 and double hexameric Mcm2-7 is required for helicase splitting and activation during S phase. Cell Rep. 2015, 13, 2576–2586. [Google Scholar] [CrossRef] [PubMed]
- Alabert, C.; Bukowski-Wills, J.C.; Lee, S.B.; Kustatscher, G.; Nakamura, K.; de Lima Alves, F.; Menard, P.; Mejlvang, J.; Rappsilber, J.; Groth, A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014, 16, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Pacek, M.; Tutter, A.V.; Kubota, Y.; Takisawa, H.; Walter, J.C. Localization of Mcm2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 2006, 21, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Gan, H.; Han, J.; Zhou, Z.X.; Jia, S.; Chabes, A.; Farrugia, G.; Ordog, T.; Zhang, Z. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol. Cell 2014, 56, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Stauffer, M.E.; Eichman, B.F. Structural biology of replication initiation factor Mcm10. Sub-Cell. Biochem. 2012, 62, 197–216. [Google Scholar]
- Thu, Y.M.; Bielinsky, A.K. Enigmatic roles of Mcm10 in DNA replication. Trends Biochem. Sci. 2013, 38, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Dumas, L.B.; Lussky, J.P.; McFarland, E.J.; Shampay, J. New temperature-sensitive mutants of Saccharomyces cerevisiae affecting DNA replication. Mol. Gen. Genet. 1982, 187, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Solomon, N.A.; Wright, M.B.; Chang, S.; Buckley, A.M.; Dumas, L.B.; Gaber, R.F. Genetic and molecular analysis of DNA43 and DNA52: Two new cell-cycle genes in Saccharomyces cerevisiae. Yeast 1992, 8, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Maine, G.T.; Sinha, P.; Tye, B.K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 1984, 106, 365–385. [Google Scholar] [PubMed]
- Merchant, A.M.; Kawasaki, Y.; Chen, Y.; Lei, M.; Tye, B.K. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 3261–3271. [Google Scholar] [CrossRef] [PubMed]
- Aves, S.J.; Tongue, N.; Foster, A.J.; Hart, E.A. The essential schizosaccharomyces pombe CDC23 DNA replication gene shares structural and functional homology with the Saccharomyces cerevisiae DNA43 (MCM10) gene. Curr. Genet. 1998, 34, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Christensen, T.W.; Tye, B.K. Drosophila Mcm10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol. Biol. Cell 2003, 14, 2206–2215. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Yanagi, K.; Mizuno, T.; Yokoi, M.; Kawasaki, Y.; Moon, K.Y.; Hurwitz, J.; Yatagai, F.; Hanaoka, F. The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G2 phase. Nucleic Acids Res. 2000, 28, 4769–4777. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Jeon, Y.; Jeon, C.H.; Kim, J.H.; Lee, H. Targeted disruption of Mcm10 causes defective embryonic cell proliferation and early embryo lethality. Biochim. Biophys. Acta 2011, 1813, 1777–1783. [Google Scholar] [CrossRef] [PubMed]
- Shultz, R.W.; Tatineni, V.M.; Hanley-Bowdoin, L.; Thompson, W.F. Genome-wide analysis of the core DNA replication machinery in the higher plants arabidopsis and rice. Plant Physiol. 2007, 144, 1697–1714. [Google Scholar] [CrossRef] [PubMed]
- Kurth, I.; O’Donnell, M. New insights into replisome fluidity during chromosome replication. Trends Biochem. Sci. 2013, 38, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Thu, Y.M.; Bielinsky, A.K. Mcm10: One tool for all-integrity, maintenance and damage control. Semin. Cell Dev. Biol. 2014, 30, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Robertson, P.D.; Warren, E.M.; Zhang, H.; Friedman, D.B.; Lary, J.W.; Cole, J.L.; Tutter, A.V.; Walter, J.C.; Fanning, E.; Eichman, B.F. Domain architecture and biochemical characterization of vertebrate Mcm10. J. Biol. Chem. 2008, 283, 3338–3348. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, O.M.; Weinstein, D.M.; Bell, S.P. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of Mcm proteins and Cdc45p during S phase. Cell 1997, 91, 59–69. [Google Scholar] [CrossRef]
- Dungrawala, H.; Rose, K.L.; Bhat, K.P.; Mohni, K.N.; Glick, G.G.; Couch, F.B.; Cortez, D. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 2015, 59, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Moore, K.; Murray, J.; Aves, S.J.; Price, C. Mcm10 interacts with Rad4/Cut5(TopBP1) and its association with origins of DNA replication is dependent on Rad4/Cut5(TopBP1). DNA Repair 2011, 10, 1154–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Perna, R.; Aria, V.; De Falco, M.; Sannino, V.; Okorokov, A.L.; Pisani, F.M.; De Felice, M. The physical interaction of Mcm10 with Cdc45 modulates their DNA-binding properties. Biochem. J. 2013, 454, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, S.; Korza, G.; Carson, J.; Liachko, I.; Tye, B.K. Novel DNA binding properties of the Mcm10 protein from Saccharomyces cerevisiae. J. Biol. Chem. 2009, 284, 25412–25420. [Google Scholar] [CrossRef] [PubMed]
- Warren, E.M.; Huang, H.; Fanning, E.; Chazin, W.J.; Eichman, B.F. Physical interactions between Mcm10, DNA, and DNA polymerase-alpha. J. Biol. Chem. 2009, 284, 24662–24672. [Google Scholar] [CrossRef] [PubMed]
- Hart, E.A.; Bryant, J.A.; Moore, K.; Aves, S.J. Fission yeast Cdc23 interactions with DNA replication initiation proteins. Curr. Genet. 2002, 41, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Homesley, L.; Lei, M.; Kawasaki, Y.; Sawyer, S.; Christensen, T.; Tye, B.K. Mcm10 and the Mcm2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000, 14, 913–926. [Google Scholar] [PubMed]
- Lee, J.K.; Seo, Y.S.; Hurwitz, J. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc. Natl. Acad. Sci. USA 2003, 100, 2334–2339. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, S.L.; Cheng, I.H.; Chai, W.; Tye, B.K. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J. Mol. Biol. 2004, 340, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Fatoba, S.T.; Tognetti, S.; Berto, M.; Leo, E.; Mulvey, C.M.; Godovac-Zimmermann, J.; Pommier, Y.; Okorokov, A.L. Human SIRT1 regulates DNA binding and stability of the Mcm10 DNA replication factor via deacetylation. Nucleic Acids Res. 2013, 41, 4065–4079. [Google Scholar] [PubMed]
- Ricke, R.M.; Bielinsky, A.K. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-alpha in budding yeast. J. Biol. Chem. 2006, 281, 18414–18425. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Gregan, J.; Lindner, K.; Young, H.; Kearsey, S.E. Nuclear distribution and chromatin association of DNA polymerase-alpha/primase is affected by tev protease cleavage of Cdc23 (Mcm10) in fission yeast. BMC Mol. Biol. 2005, 6, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Ukomadu, C.; Jha, S.; Senga, T.; Dhar, S.K.; Wohlschlegel, J.A.; Nutt, L.K.; Kornbluth, S.; Dutta, A. Mcm10 and And-1/Ctf4 recruit DNA polymerase-alpha to chromatin for initiation of DNA replication. Genes Dev. 2007, 21, 2288–2299. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, Y.; Hiraga, S.; Sugino, A. Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells 2000, 5, 975–989. [Google Scholar] [CrossRef] [PubMed]
- Das-Bradoo, S.; Ricke, R.M.; Bielinsky, A.K. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell. Biol. 2006, 26, 4806–4817. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, R.; Lu, Y.; Liang, C. Ctf4p facilitates Mcm10p to promote DNA replication in budding yeast. Biochem. Biophys. Res. Commun. 2010, 395, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Rochette, P.J.; Feyissa, E.A.; Su, T.V.; Liu, Y. Mcm10 mediates Recq4 association with Mcm2-7 helicase complex during DNA replication. EMBO J. 2009, 28, 3005–3014. [Google Scholar] [CrossRef] [PubMed]
- Warren, E.M.; Vaithiyalingam, S.; Haworth, J.; Greer, B.; Bielinsky, A.K.; Chazin, W.J.; Eichman, B.F. Structural basis for DNA binding by replication initiator Mcm10. Structure 2008, 16, 1892–1901. [Google Scholar] [CrossRef] [PubMed]
- Perez-Arnaiz, P.; Kaplan, D.L. An Mcm10 mutant defective in ssDNA binding shows defects in DNA replication initiation. J. Mol. Biol. 2016, 428, 4608–4625. [Google Scholar] [CrossRef] [PubMed]
- Okorokov, A.L.; Waugh, A.; Hodgkinson, J.; Murthy, A.; Hong, H.K.; Leo, E.; Sherman, M.B.; Stoeber, K.; Orlova, E.V.; Williams, G.H. Hexameric ring structure of human Mcm10 DNA replication factor. EMBO Rep. 2007, 8, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Dalrymple, B.P.; Kongsuwan, K.; Wijffels, G.; Dixon, N.E.; Jennings, P.A. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA 2001, 98, 11627–11632. [Google Scholar] [CrossRef] [PubMed]
- Alver, R.C.; Zhang, T.; Josephrajan, A.; Fultz, B.L.; Hendrix, C.J.; Das-Bradoo, S.; Bielinsky, A.K. The N-terminus of Mcm10 is important for interaction with the 9–1-1 clamp and in resistance to DNA damage. Nucleic Acids Res. 2014, 42, 8389–8404. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Josephrajan, A.; Adhikary, S.; Bowles, T.; Bielinsky, A.K.; Eichman, B.F. Mcm10 self-association is mediated by an N-terminal coiled-coil domain. PLoS ONE 2013, 8, e70518. [Google Scholar] [CrossRef] [PubMed]
- Robertson, P.D.; Chagot, B.; Chazin, W.J.; Eichman, B.F. Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved Mcm motif. J. Biol. Chem. 2010, 285, 22942–22949. [Google Scholar] [CrossRef] [PubMed]
- Dalton, S.; Hopwood, B. Characterization of Cdc47p-minichromosome maintenance complexes in Saccharomyces cerevisiae: Identification of Cdc45p as a subunit. Mol. Cell. Biol. 1997, 17, 5867–5875. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.J.; Shen, J.; Gomez-Llorente, Y.; Martin, C.S.; Carazo, J.M.; Chen, X.S. Double hexamer disruption and biochemical activities of methanobacterium thermoautotrophicum Mcm. J. Biol. Chem. 2005, 280, 42405–42410. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Gibson, S.; Tye, B.K. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes Dev. 1991, 5, 944–957. [Google Scholar] [CrossRef] [PubMed]
- Vo, N.; Anh Suong, D.N.; Yoshino, N.; Yoshida, H.; Cotterill, S.; Yamaguchi, M. Novel roles of HP1a and Mcm10 in DNA replication, genome maintenance and photoreceptor cell differentiation. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kaur, M.; Kar, A.; Ranade, S.M.; Saxena, S. Ultraviolet radiation stress triggers the down-regulation of essential replication factor Mcm10. J. Biol. Chem. 2010, 285, 8352–8362. [Google Scholar] [CrossRef] [PubMed]
- Burich, R.; Lei, M. Two bipartite NLSs mediate constitutive nuclear localization of Mcm10. Curr. Genet. 2003, 44, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Nevins, J.R. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 2001, 10, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Inoue, I. Expression of Mcm10 and TopBP1 is regulated by cell proliferation and UV irradiation via the E2F transcription factor. Oncogene 2004, 23, 6250–6260. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Deng, B.; Xing, G.; Teng, Y.; Tian, C.; Cheng, X.; Yin, X.; Yang, J.; Gao, X.; Zhu, Y.; et al. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc. Natl. Acad. Sci. USA 2007, 104, 7951–7956. [Google Scholar] [CrossRef] [PubMed]
- Miotto, B.; Chibi, M.; Xie, P.; Koundrioukoff, S.; Moolman-Smook, H.; Pugh, D.; Debatisse, M.; He, F.; Zhang, L.; Defossez, P.A. The Rbbp6/ZBTB38/Mcm10 axis regulates DNA replication and common fragile site stability. Cell Rep. 2014, 7, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Sakai, Y.; Saijo, M.; Coelho, K.; Kishino, T.; Niikawa, N.; Taya, Y. cDNA sequence and chromosomal localization of a novel human protein, Rbq-1 (Rbbp6), that binds to the retinoblastoma gene product. Genomics 1995, 30, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Simons, A.; Melamed-Bessudo, C.; Wolkowicz, R.; Sperling, J.; Sperling, R.; Eisenbach, L.; Rotter, V. Pact: Cloning and characterization of a cellular p53 binding protein that interacts with Rb. Oncogene 1997, 14, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.B.; Zhang, Y.V.; Tumbar, T. Gata6 promotes hair follicle progenitor cell renewal by genome maintenance during proliferation. EMBO J. 2017, 36, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.; Chen, S.; Tong, L. MicroRNA-215 regulates fibroblast function: Insights from a human fibrotic disease. Cell Cycle 2015, 14, 1973–1984. [Google Scholar] [CrossRef] [PubMed]
- Wotschofsky, Z.; Gummlich, L.; Liep, J.; Stephan, C.; Kilic, E.; Jung, K.; Billaud, J.N.; Meyer, H.A. Integrated microRNA and mRNA signature associated with the transition from the locally confined to the metastasized clear cell renal cell carcinoma exemplified by mir-146-5p. PLoS ONE 2016, 11, e0148746. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Yatagai, F.; Hanaoka, F. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J. Biol. Chem. 2001, 276, 48526–48531. [Google Scholar] [PubMed]
- Izumi, M.; Yatagai, F.; Hanaoka, F. Localization of human Mcm10 is spatially and temporally regulated during the S phase. J. Biol. Chem. 2004, 279, 32569–32577. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Sharma, A.; Khan, M.; Kar, A.; Saxena, S. Mcm10 proteolysis initiates before the onset of M-phase. BMC Cell Biol. 2010, 11, 84. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, S.; Gorbsky, G.J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015, 16, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Khan, M.M.; Kar, A.; Sharma, A.; Saxena, S. CRL4-DDB1-VprBP ubiquitin ligase mediates the stress triggered proteolysis of Mcm10. Nucleic Acids Res. 2012, 40, 7332–7346. [Google Scholar] [CrossRef] [PubMed]
- Romani, B.; Shaykh Baygloo, N.; Aghasadeghi, M.R.; Allahbakhshi, E. HIV-1 Vpr protein enhances proteasomal degradation of Mcm10 DNA replication factor through the Cul4-DDB1[VprBP] E3 ubiquitin ligase to induce G2/M cell cycle arrest. J. Biol. Chem. 2015, 290, 17380–17389. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.; Xiong, Y. Crl4s: The Cul4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009, 34, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Fultz, B.L.; Das-Bradoo, S.; Bielinsky, A.-K. Mapping ubiquitination sites of S. cerevisiae Mcm10. Biochem. Biophys. Rep. 2016, 8, 212–218. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Chabra, I.; Kornhauser, J.M.; Skrzypek, E.; Zhang, B. Phosphosite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 2004, 4, 1551–1561. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Qiao, J.W.; Patel, J.; Udeshi, N.D.; Clauser, K.R.; Mani, D.R.; Burgess, M.W.; Gillette, M.A.; Jaffe, J.D.; Carr, S.A. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 2013, 10, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Yang, F.; Liu, T.; Mani, D.R.; Petyuk, V.A.; Gillette, M.A.; Clauser, K.R.; Qiao, J.W.; Gritsenko, M.A.; Moore, R.J.; et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteom. 2014, 13, 1690–1704. [Google Scholar] [CrossRef] [PubMed]
- Dephoure, N.; Zhou, C.; Villen, J.; Beausoleil, S.A.; Bakalarski, C.E.; Elledge, S.J.; Gygi, S.P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 10762–10767. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., 3rd; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; D’Souza, R.C.; Tyanova, S.; Schaab, C.; Wisniewski, J.R.; Cox, J.; Mann, M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014, 8, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Ruse, C.I.; McClatchy, D.B.; Lu, B.; Cociorva, D.; Motoyama, A.; Park, S.K.; Yates, J.R., 3rd. Motif-specific sampling of phosphoproteomes. J. Proteome Res. 2008, 7, 2140–2150. [Google Scholar] [CrossRef] [PubMed]
- Mayya, V.; Lundgren, D.H.; Hwang, S.I.; Rezaul, K.; Wu, L.; Eng, J.K.; Rodionov, V.; Han, D.K. Quantitative phosphoproteomic analysis of t cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2009, 2, ra46. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Vermeulen, M.; Santamaria, A.; Kumar, C.; Miller, M.L.; Jensen, L.J.; Gnad, F.; Cox, J.; Jensen, T.S.; Nigg, E.A.; et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010, 3, ra3. [Google Scholar] [CrossRef] [PubMed]
- Kettenbach, A.N.; Schweppe, D.K.; Faherty, B.K.; Pechenick, D.; Pletnev, A.A.; Gerber, S.A. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 2011, 4, rs5. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Schreiber, T.B.; Daub, H. Dual phosphoproteomics and chemical proteomics analysis of erlotinib and gefitinib interference in acute myeloid leukemia cells. J. Proteom. 2012, 75, 1343–1356. [Google Scholar] [CrossRef] [PubMed]
- Klammer, M.; Kaminski, M.; Zedler, A.; Oppermann, F.; Blencke, S.; Marx, S.; Muller, S.; Tebbe, A.; Godl, K.; Schaab, C. Phosphosignature predicts dasatinib response in non-small cell lung cancer. Mol. Cell. Proteom. 2012, 11, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Chadha, G.S.; Gambus, A.; Gillespie, P.J.; Blow, J.J. Xenopus Mcm10 is a CDK-substrate required for replication fork stability. Cell Cycle 2016, 15, 2183–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watase, G.; Takisawa, H.; Kanemaki, M.T. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr. Biol. 2012, 22, 343–349. [Google Scholar] [CrossRef] [PubMed]
- van Deursen, F.; Sengupta, S.; De Piccoli, G.; Sanchez-Diaz, A.; Labib, K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012, 31, 2195–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilves, I.; Petojevic, T.; Pesavento, J.J.; Botchan, M.R. Activation of the Mcm2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 2010, 37, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Sheu, Y.J.; Stillman, B. Cdc7-Dbf4 phosphorylates Mcm proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell 2006, 24, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Umemori, T.; Hirai, K.; Muramatsu, S.; Kamimura, Y.; Araki, H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007, 445, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Diffley, J.F. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Nakato, R.; Katou, Y.; Shirahige, K.; Araki, H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 2011, 21, 2055–2063. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Umemori, T.; Endo, S.; Muramatsu, S.; Kanemaki, M.; Kamimura, Y.; Obuse, C.; Araki, H. Sld7, an Sld3-associated protein required for efficient chromosomal DNA replication in budding yeast. EMBO J. 2011, 30, 2019–2030. [Google Scholar] [CrossRef] [PubMed]
- Lujan, S.A.; Williams, J.S.; Kunkel, T.A. DNA polymerases divide the labor of genome replication. Trends Cell Biol. 2016, 26, 640–654. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 1998, 107, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wold, M.S. Replication protein A: Single-stranded DNA’s first responder: Dynamic DNA-interactions allow Replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. BioEssays 2014, 36, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Okazaki, T. Discontinuous DNA replication. Annu. Rev. Biochem. 1980, 49, 421–457. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, L.; Bambara, R.A. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J. Biol. Chem. 2011, 286, 6865–6870. [Google Scholar] [CrossRef] [PubMed]
- Strzalka, W.; Ziemienowicz, A. Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation. Ann. Bot. 2011, 107, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Gregan, J.; Lindner, K.; Brimage, L.; Franklin, R.; Namdar, M.; Hart, E.A.; Aves, S.J.; Kearsey, S.E. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol. Biol. Cell 2003, 14, 3876–3887. [Google Scholar] [CrossRef] [PubMed]
- Im, J.S.; Ki, S.H.; Farina, A.; Jung, D.S.; Hurwitz, J.; Lee, J.K. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, Recql4, and Mcm10 proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 15628–15632. [Google Scholar] [CrossRef] [PubMed]
- Heller, R.C.; Kang, S.; Lam, W.M.; Chen, S.; Chan, C.S.; Bell, S.P. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 2011, 146, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Jares, P.; Blow, J.J. Xenopus Cdc7 function is dependent on licensing but not on xORC, xCdc6, or CDK activity and is required for xCdc45 loading. Genes Dev. 2000, 14, 1528–1540. [Google Scholar] [PubMed]
- Walter, J.C. Evidence for sequential action of Cdc7 and Cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 2000, 275, 39773–39778. [Google Scholar] [CrossRef] [PubMed]
- On, K.F.; Beuron, F.; Frith, D.; Snijders, A.P.; Morris, E.P.; Diffley, J.F. Prereplicative complexes assembled in vitro support origin-dependent and independent DNA replication. EMBO J. 2014, 33, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Schwacha, A. The Mcm complex: Unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 2009, 73, 652–683. [Google Scholar] [CrossRef] [PubMed]
- Kanke, M.; Kodama, Y.; Takahashi, T.S.; Nakagawa, T.; Masukata, H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012, 31, 2182–2194. [Google Scholar] [CrossRef] [PubMed]
- Im, J.S.; Park, S.Y.; Cho, W.H.; Bae, S.H.; Hurwitz, J.; Lee, J.K. Recql4 is required for the association of Mcm10 and Ctf4 with replication origins in human cells. Cell Cycle 2015, 14, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Kliszczak, M.; Sedlackova, H.; Pitchai, G.P.; Streicher, W.W.; Krejci, L.; Hickson, I.D. Interaction of Recq4 and Mcm10 is important for efficient DNA replication origin firing in human cells. Oncotarget 2015, 6, 40464–40479. [Google Scholar] [PubMed]
- Wang, J.T.; Xu, X.; Alontaga, A.Y.; Chen, Y.; Liu, Y. Impaired p32 regulation caused by the lymphoma-prone Recq4 mutation drives mitochondrial dysfunction. Cell Rep. 2014, 7, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Hardy, C.F.; Dryga, O.; Seematter, S.; Pahl, P.M.; Sclafani, R.A. Mcm5/Cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl. Acad. Sci. USA 1997, 94, 3151–3155. [Google Scholar] [CrossRef] [PubMed]
- Bruck, I.; Kaplan, D.L. The Dbf4-Cdc7 kinase promotes Mcm2-7 ring opening to allow for single-stranded DNA extrusion and helicase assembly. J. Biol. Chem. 2015, 290, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Bruck, I.; Kaplan, D. Dbf4-Cdc7 phosphorylation of Mcm2 is required for cell growth. J. Biol. Chem. 2009, 284, 28823–28831. [Google Scholar] [CrossRef] [PubMed]
- Fien, K.; Cho, Y.S.; Lee, J.K.; Raychaudhuri, S.; Tappin, I.; Hurwitz, J. Primer utilization by DNA polymerase-alpha/primase is influenced by its interaction with Mcm10p. J. Biol. Chem. 2004, 279, 16144–16153. [Google Scholar] [CrossRef] [PubMed]
- Langston, L.D.; Zhang, D.; Yurieva, O.; Georgescu, R.E.; Finkelstein, J.; Yao, N.Y.; Indiani, C.; O’Donnell, M.E. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc. Natl. Acad. Sci. USA 2014, 111, 15390–15395. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, S.; Hirai, K.; Tak, Y.S.; Kamimura, Y.; Araki, H. Cdk-dependent complex formation between replication proteins Dpb11, Sld2, Pol-e, and GINS in budding yeast. Genes Dev. 2010, 24, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Nasheuer, H.P.; Grosse, F. Immunoaffinity-purified DNA polymerase-alpha displays novel properties. Biochemistry 1987, 26, 8458–8466. [Google Scholar] [CrossRef] [PubMed]
- Kouprina, N.; Kroll, E.; Bannikov, V.; Bliskovsky, V.; Gizatullin, R.; Kirillov, A.; Shestopalov, B.; Zakharyev, V.; Hieter, P.; Spencer, F.; et al. Ctf4 (Chl15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 5736–5747. [Google Scholar] [CrossRef] [PubMed]
- Boehm, E.M.; Washington, M.T.R.I.P. To the PIP: PCNA-binding motif no longer considered specific: PIP motifs and other related sequences are not distinct entities and can bind multiple proteins involved in genome maintenance. BioEssays 2016, 38, 1117–1122. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Kawasaki, Y.; Sasanuma, H.; Tye, B.K.; Sugino, A. Budding yeast Mcm10/Dna43 mutant requires a novel repair pathway for viability. Genes Cells 2003, 8, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Liachko, I.; Bouten, R.; Kelman, Z.; Tye, B.K. Alternative mechanisms for coordinating polymerase-alpha and Mcm helicase. Mol. Cell. Biol. 2010, 30, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Tittel-Elmer, M.; Alabert, C.; Pasero, P.; Cobb, J.A. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J. 2009, 28, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Lukas, C.; Savic, V.; Bekker-Jensen, S.; Doil, C.; Neumann, B.; Pedersen, R.S.; Grofte, M.; Chan, K.L.; Hickson, I.D.; Bartek, J.; et al. 53bp1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011, 13, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, R.D.; Soni, D.V.; Wollman, R.; Hahn, A.T.; Yee, M.C.; Guan, A.; Hesley, J.A.; Miller, S.C.; Cromwell, E.F.; Solow-Cordero, D.E.; et al. A genome-wide sirna screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell 2009, 35, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Santocanale, C.; Diffley, J.F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 1998, 395, 615–618. [Google Scholar] [PubMed]
- Tercero, J.A.; Diffley, J.F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 2001, 412, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.R.; Nguyen, H.D.; Wang, X.; Bielinsky, A.K. Mcm10 deficiency causes defective-replisome-induced mutagenesis and a dependency on error-free postreplicative repair. Cell Cycle 2014, 13, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Thu, Y.M.; Van Riper, S.K.; Higgins, L.; Zhang, T.; Becker, J.R.; Markowski, T.W.; Nguyen, H.D.; Griffin, T.J.; Bielinsky, A.K. Slx5/Slx8 promotes replication stress tolerance by facilitating mitotic progression. Cell Rep. 2016, 15, 1254–1265. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Bielinsky, A.K. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol. Biol. Cell 2007, 18, 4085–4095. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Bang, S.W.; Jeon, Y.; Kang, S.; Hwang, D.S. Knockdown of human Mcm10 exhibits delayed and incomplete chromosome replication. Biochem. Biophys. Res. Commun. 2008, 365, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Bang, S.W.; Kim, S.H.; Hwang, D.S. Knockdown of human Mcm10 activates G2 checkpoint pathway. Biochem. Biophys. Res. Commun. 2008, 365, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Bielinsky, A.K. Mcm10: The glue at replication forks. Cell Cycle 2016, 15, 3024–3025. [Google Scholar] [CrossRef] [PubMed]
- Koppen, A.; Ait-Aissa, R.; Koster, J.; van Sluis, P.G.; Ora, I.; Caron, H.N.; Volckmann, R.; Versteeg, R.; Valentijn, L.J. Direct regulation of the minichromosome maintenance complex by MYCN in neuroblastoma. Eur. J. Cancer 2007, 43, 2413–2422. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Aragoncillo, E.; Carrillo, J.; Lalli, E.; Agra, N.; Gomez-Lopez, G.; Pestana, A.; Alonso, J. Dax1, a direct target of EWS/Fli1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing’s tumor cells. Oncogene 2008, 27, 6034–6043. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.X.; Zeng, J.M.; Zhou, J.J.; Yang, G.H.; Ding, K.; Zhang, X.J. Silencing of RTKN2 by siRNA suppresses proliferation, and induces G1 arrest and apoptosis in human bladder cancer cells. Mol. Med. Rep. 2016, 13, 4872–4878. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Prasad, S.B.; Yadav, S.S.; Govardhan, H.B.; Pandey, L.K.; Singh, S.; Pradhan, S.; Narayan, G. Over expression of minichromosome maintenance genes is clinically correlated to cervical carcinogenesis. PLoS ONE 2013, 8, e69607. [Google Scholar] [CrossRef] [PubMed]
- Li, W.M.; Huang, C.N.; Ke, H.L.; Li, C.C.; Wei, Y.C.; Yeh, H.C.; Chang, L.L.; Huang, C.H.; Liang, P.I.; Yeh, B.W.; et al. Mcm10 overexpression implicates adverse prognosis in urothelial carcinoma. Oncotarget 2016, 7, 77777–77792. [Google Scholar] [PubMed]
- Wu, C.; Zhu, J.; Zhang, X. Integrating gene expression and protein-protein interaction network to prioritize cancer-associated genes. BMC Bioinform. 2012, 13, 182. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, G.; Hwang, W.C.; Do, I.G.; Wang, K.; Kang, S.Y.; Lee, J.; Park, S.H.; Park, J.O.; Kang, W.K.; Jang, J.; et al. Exome sequencing identifies early gastric carcinoma as an early stage of advanced gastric cancer. PLoS ONE 2013, 8, e82770. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Ragland, R.L.; Patel, S.; Rivard, R.S.; Smith, K.; Peters, A.A.; Bielinsky, A.K.; Brown, E.J. RNF4 and Plk1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013, 27, 2259–2273. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
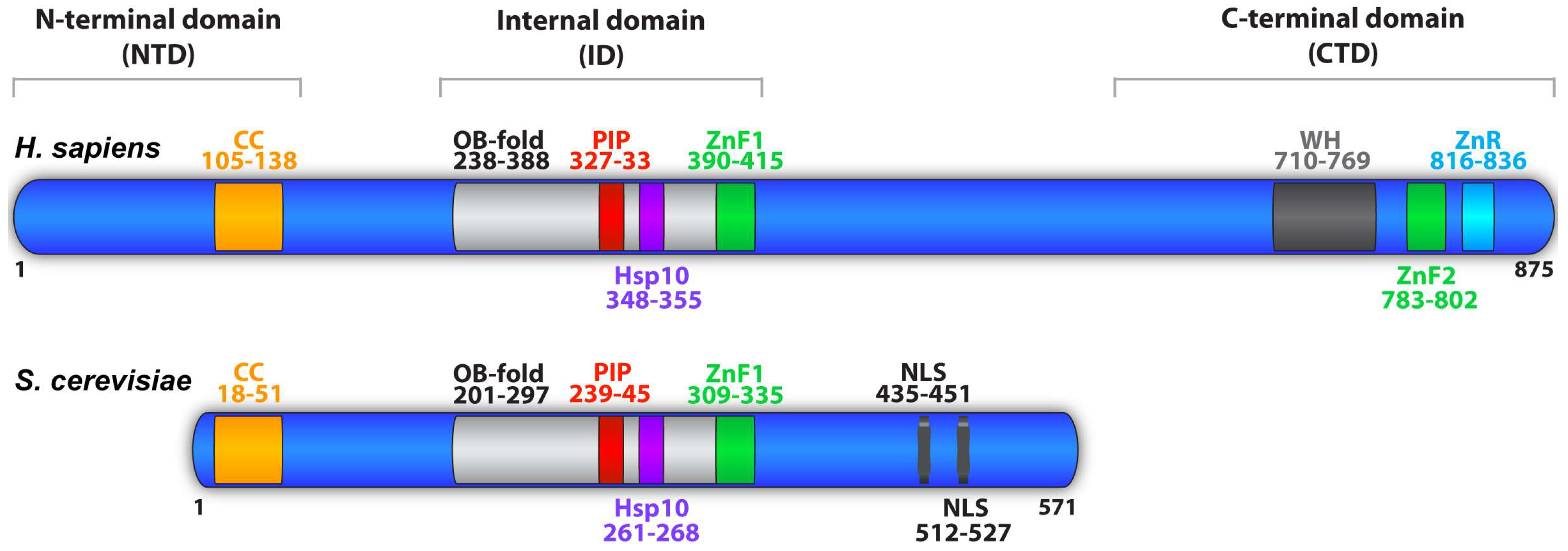
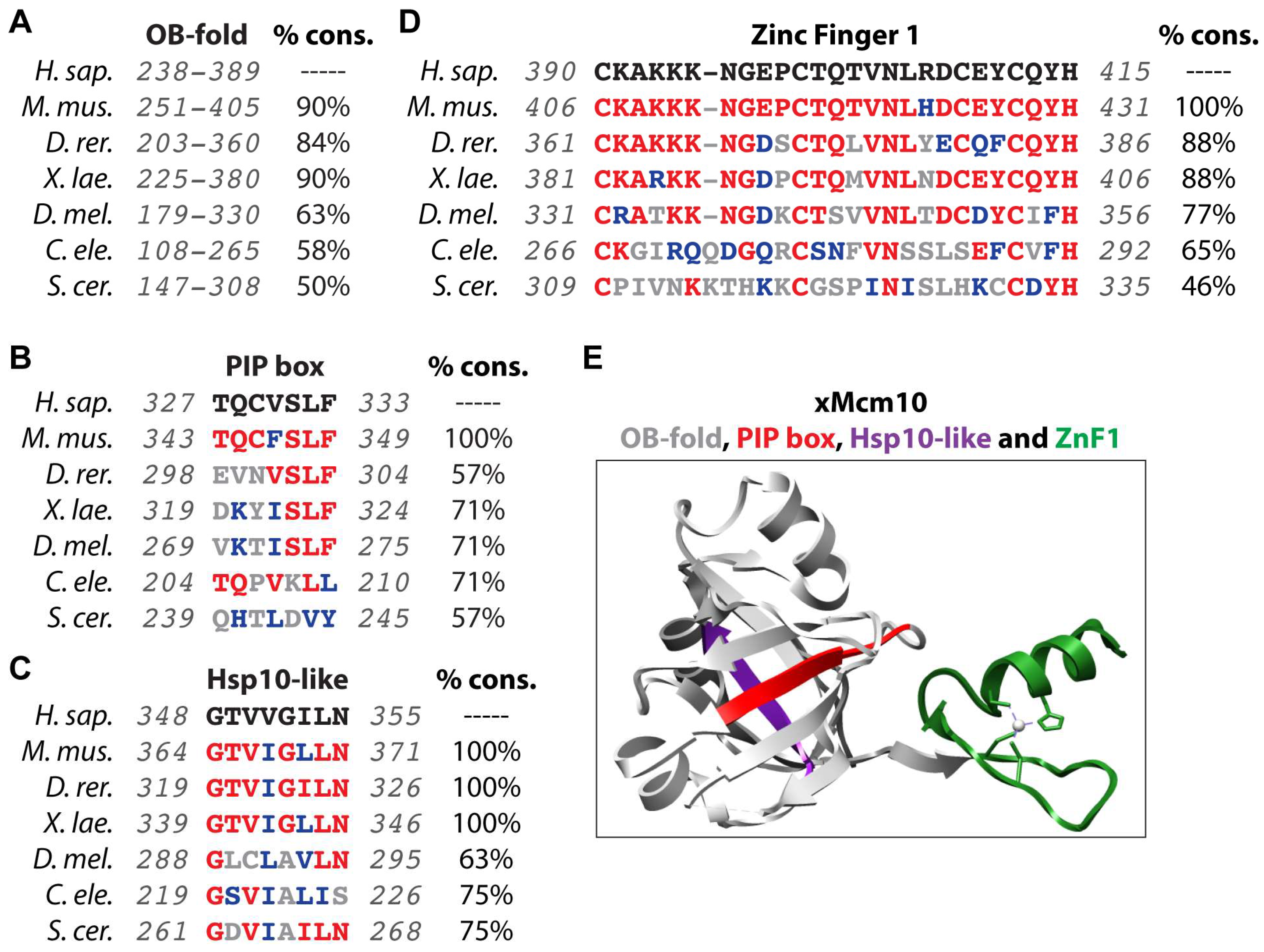

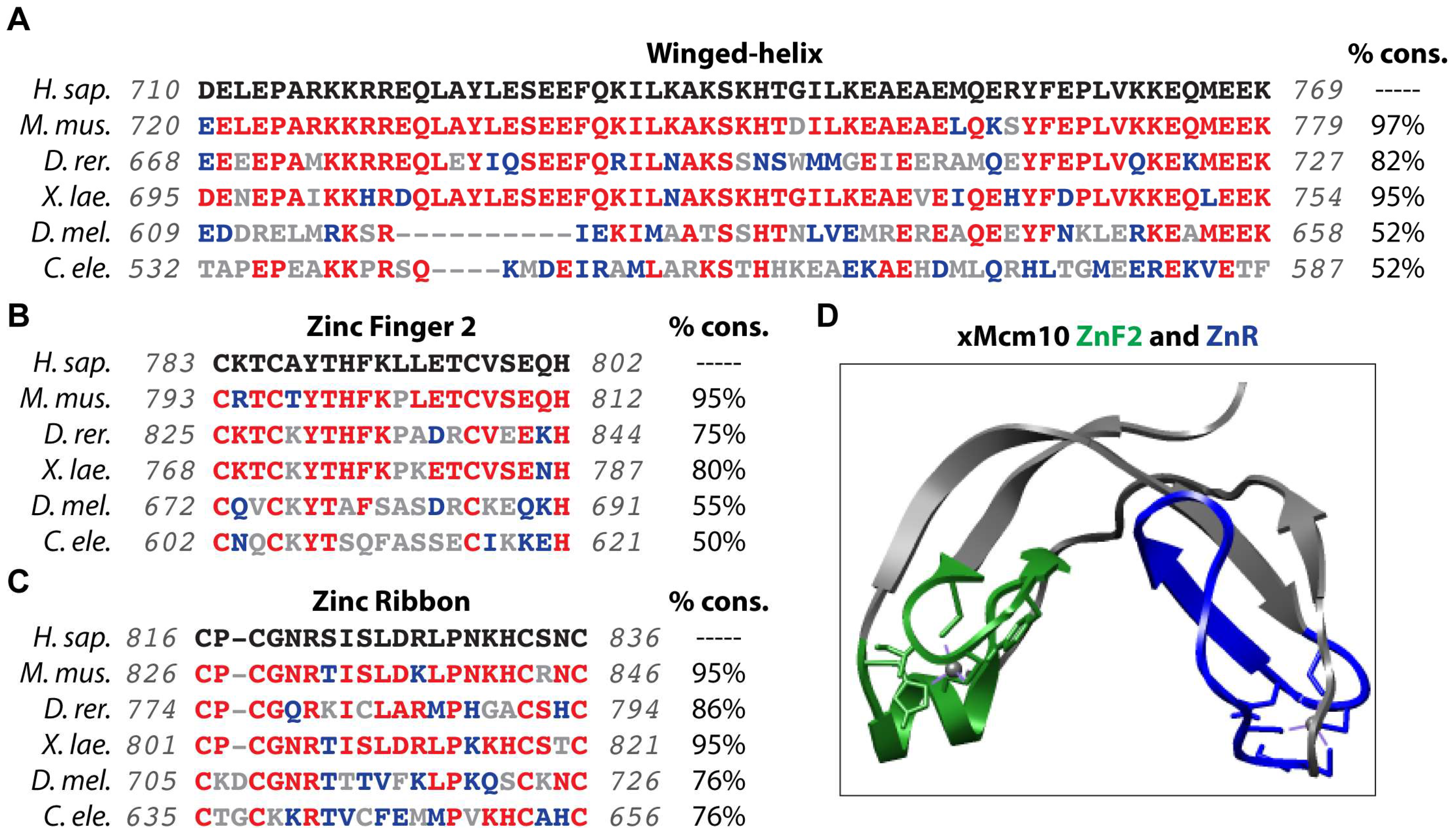
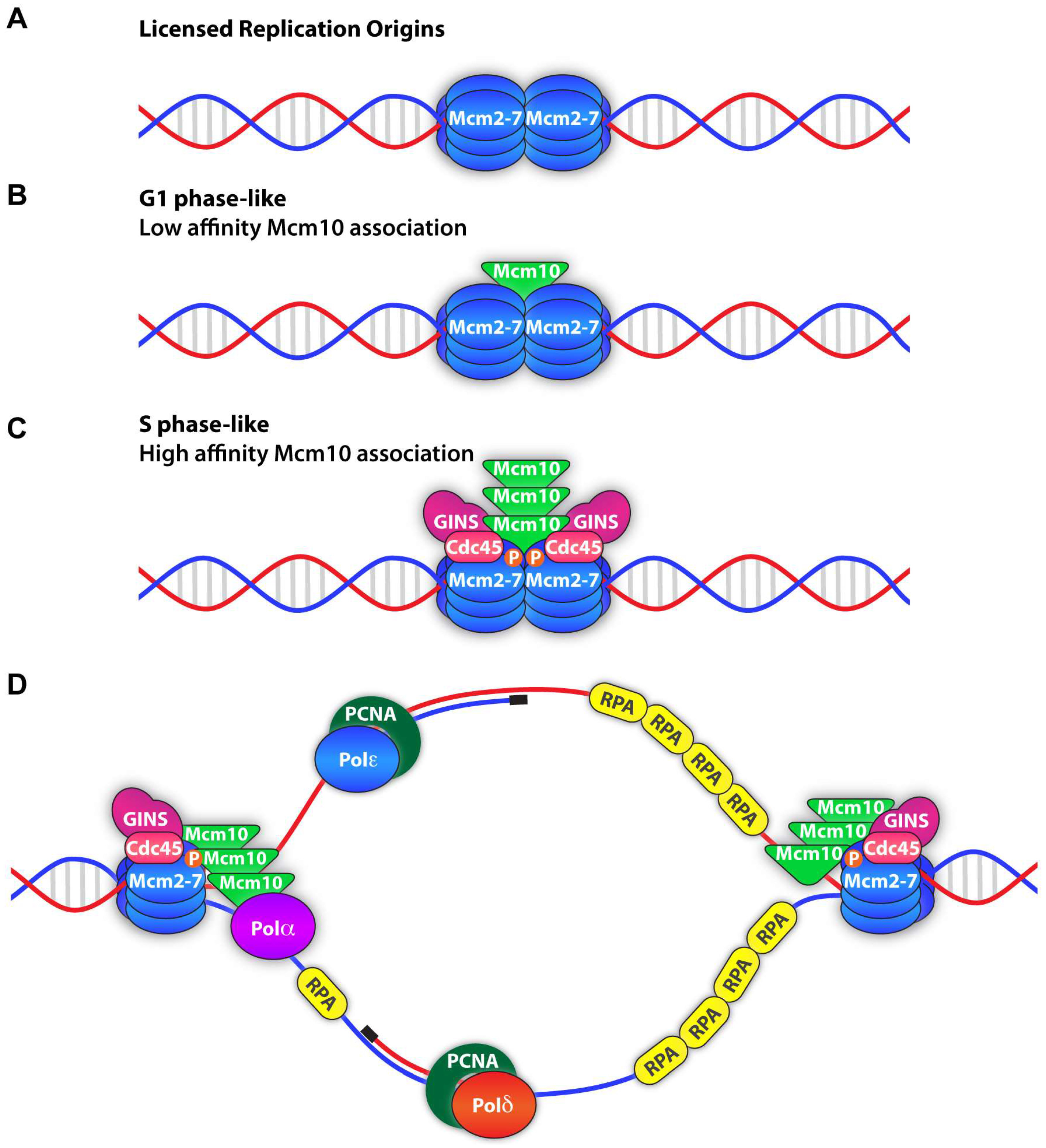

| Modification | Role | Species/System | Region/Residue(s) | Enzyme | Reference(s) |
|---|---|---|---|---|---|
| Ubiquitination | Target for proteasome dependent degradation | Human Mcm10 (HeLa, U2OS) in vivo | 440–525 783–803 843–875 (regions that can mediate degradation) | Cul4-DDB1-VprBP | [93,95,97,98] |
| Ubiquitination | Functional regulation during S-phase | Yeast Mcm10 (Saccharomyces cerevisiae) | K85, K122, K319, K372, K414, K436 | Not identified | [67,100] |
| Phosphorylation | Unknown function | Human Mcm10 (HeLa) | T85, S93, S150, S155, A182, S203, S204, A210, S212, T217, R286, T296, S488, S548, S555, S559, S577, S593, Y641, S644, T663, S706, S824 (* only sites identified in more than 2 datasets are listed) | Not identified, except T85 which is ATR or ATM dependent. | [93,101,102,103,104,105,106,107,108,109,110,111,112] |
| Phosphorylation | Replisome stability | Xenopus extract | S154, S173, S206, S596, S630, S690, S693 | S-CDK | [113] |
| Acetylation | Protein stability and DNA binding | Human Mcm10 | K267, K312 *, K318, K390 *, K657, K664, K668, K674 *, K681 *, K682 *, K683 *, K685 *, K737 *, K739 *, K745 *, K761 *, K768 *, K783, K847 *, K849 *, K853, K868, K874 | p300 (acetylase) SIRT1 * deacetylase) * indicates subset of SIRT1 target residues | [62] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baxley, R.M.; Bielinsky, A.-K. Mcm10: A Dynamic Scaffold at Eukaryotic Replication Forks. Genes 2017, 8, 73. https://doi.org/10.3390/genes8020073
Baxley RM, Bielinsky A-K. Mcm10: A Dynamic Scaffold at Eukaryotic Replication Forks. Genes. 2017; 8(2):73. https://doi.org/10.3390/genes8020073
Chicago/Turabian StyleBaxley, Ryan M., and Anja-Katrin Bielinsky. 2017. "Mcm10: A Dynamic Scaffold at Eukaryotic Replication Forks" Genes 8, no. 2: 73. https://doi.org/10.3390/genes8020073






