Complete Mitochondrial Genome Sequencing of a Burial from a Romano–Christian Cemetery in the Dakhleh Oasis, Egypt: Preliminary Indications
Abstract
:1. Introduction
2. Materials and Methods
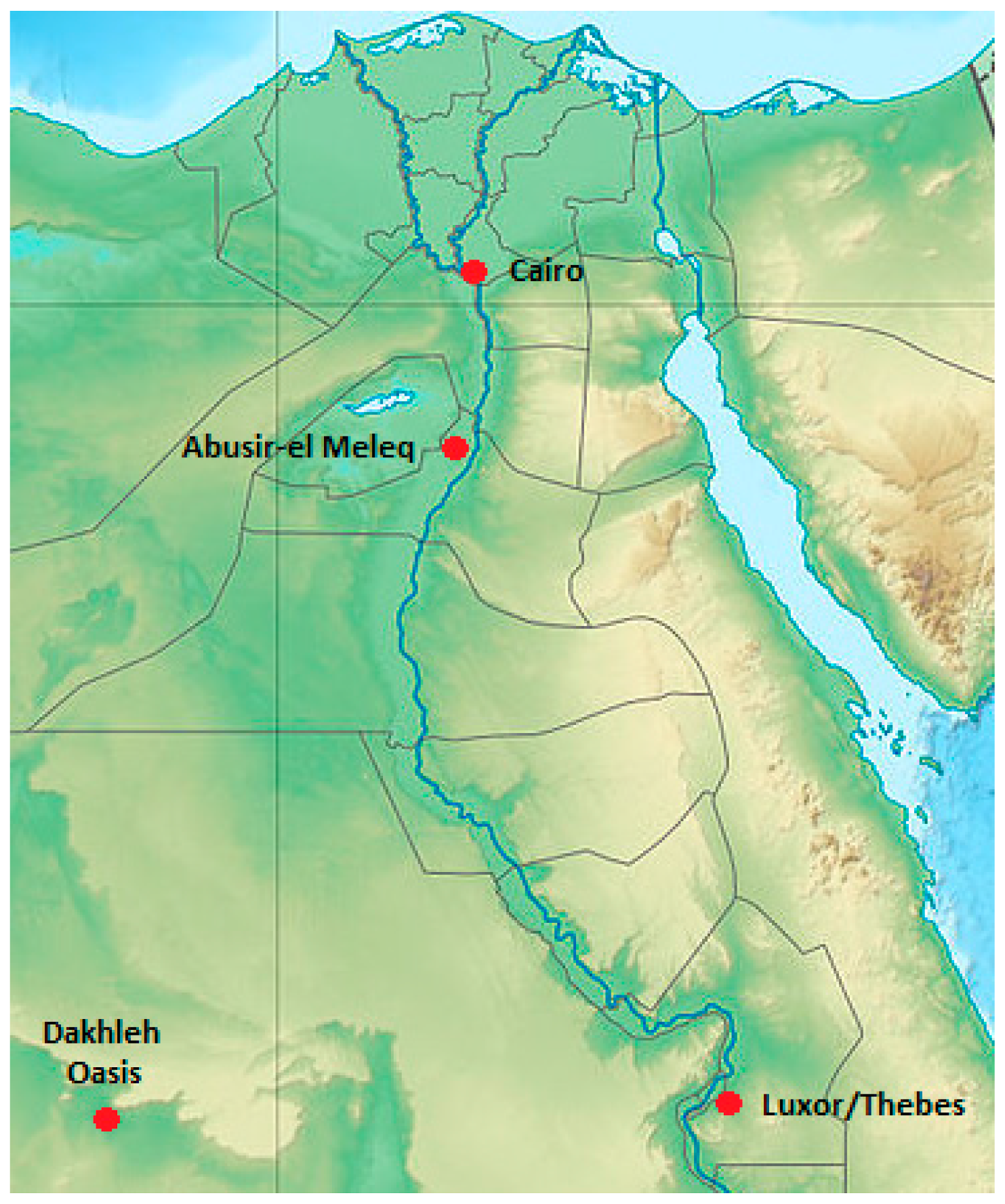
2.1. Material
2.2. Methods
3. Results
3.1. Authenticity of the Results
3.2. Contamination in the mtDNA Data
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Molto, J.E. The comparative skeletal biology and paleoepidemiology of the people from Ein Tirgh and Kellis 2, Dakhleh, Egypt. In The Oasis Papers I: The Proceedings of the First Conference of the Dakhleh Oasis Project; Marlow, C.A., Mills, A.J., Eds.; Oxbow Books: Oxford, UK, 2001. [Google Scholar]
- Schwartz, H.P.; Dupras, T.L.; Fairgrieve, S.I. 15N Enrichment in the Sahara: In Search of a Global Relationship. J. Arch. Sci. 1999, 26, 629–636. [Google Scholar] [CrossRef]
- Mills, A. Pharaonic Egyptians in the Dakhleh Oasis. In Reports from the Survey of the Dakhleh Oasis, Western Desert of Egypt, 1977–1987; Oxbow Books: Oxford, UK, 1999. [Google Scholar]
- Stewart, J.D.; Molto, J.E. The Chronology of Kellis 2: The interpretative significance of radiocarbon dating of human remains. In Dakhleh Oasis Project Monograph; Bowen, G.E., Hope, C.A., Eds.; Oxbow Books: Oxford, UK, 2003; pp. 373–378. [Google Scholar]
- Molto, J.E. Bio-archaeological research of Kellis 2: An overview. In Dakhleh Oasis Project: Preliminary Reports of the 1994–1995 to 1998–1999 Field Seasons; Hope, C.A., Bowen, G.E., Eds.; Oxbow Books: Oxford, UK, 2002; pp. 239–255. [Google Scholar]
- Parr, R.L. Mitochondrial DNA sequence analysis from skeletal remains from the Kellis 2 cemetery. In Dakhleh Oasis Project: Preliminary Reports of the 1994–1995 to 1998–1999 Field Seasons; Hope, C.A., Bowen, G.E., Eds.; Oxbow Books: Oxford, UK, 2002; pp. 257–261. [Google Scholar]
- Graver, A.M.; Molto, J.E.; Parr, R.L.; Walters, S.; Praymack, R.C.; Maki, J.M. Mitochondrial DNA research in the Dakhleh Oasis, Egypt: A preliminary report. Anc. Biomol. 2001, 3, 239–253. [Google Scholar]
- Donoghue, H.D.; Marcsik, A.; Matheson, C.; Vernon, K.; Nuorala, E.; Molto, J.E.; Greenblatt, C.L.; Spigelman, M. Co-infection of Mycobacterium tuberculosis and Mycobacterium leprae in human archaeological samples: A possible explanation for the historical decline of leprosy. Proc. Biol. Sci. 2005, 272, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Henderson, M.A. Craniometrics analysis of samples from Ein Tirchi and Kellis 2, Dakhleh, Egypt. Lakehead University, Thunder Bay, ON, Canada. Unpublished work. 1993. [Google Scholar]
- Fairgrieve, S.I.; Molto, J.E. Cribra orbitalia in two temporally disjunct population samples from the Dakhleh Oasis, Egypt. Am. J. Phys. Anthropol. 2000, 111, 319–331. [Google Scholar] [CrossRef]
- Worp, K.A. Greek papyri from Kellis: I. Dakhleh Oasis Project: Monograph 3; Worp, K.A., Ed.; Oxbow Books: Oxford, UK, 1995. [Google Scholar]
- Gardner, I. Kellis Literary Texts, Volume I. Dakhleh Oasis Project: Monograph 4; Gardner, I., Ed.; Oxbow Books: Oxford, UK, 1995. [Google Scholar]
- Andrews, R.M.; Kubacka, I.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999, 23, 147. [Google Scholar] [PubMed]
- Lindgreen, S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res. Notes 2012, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Wang, W.; Hu, P.; Lyon, G.J.; Hakonarson, H. SNVer: A statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucl. Acids Res. 2011, 39, e132. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, H.; Ginolhac, A.; Schubert, M.; Johnson, P.L.; Orlando, L. MapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 2013, 29, 1682–1684. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Mittnik, A.; Johnson, P.L.F.; Bos, K.; Lari, M.; Bollongino, R.; Sun, C.; Giemsch, L.; Schmitz, R.; Burger, J.; et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 2013, 23, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Llamas, B.; Valverde, G.; Fehren-Schmitz, L.; Weyrich, L.S.; Cooper, A.; Haak, W. From the field to the laboratory: Controlling DNA contamination in human ancient DNA research in the high-throughput sequencing era. Sci. Technol. Archaeol. Res. 2017, 3, 1–14. [Google Scholar] [CrossRef]
- Brandon, M.C.; Ruiz-Pesini, E.; Mishmar, D.; Procaccio, V.; Lott, M.T.; Nguyen, K.C.; Spolim, S.; Patil, U.; Baldi, P.; Wallace, D.C. MITOMASTER: A bioinformatics tool for the analysis of mitochondrial DNA sequences. Hum. Mutat. 2009, 30, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lott, M.T.; Leipzig, J.N.; Derbeneva, O.; Xie, H.M.; Chalkia, D.; Sarmady, M.; Procaccio, V.; Wallace, D.C. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr. Protoc. Bioinform. 2013, 44, 1–26. [Google Scholar]
- Fu, Q.; Rudan, P.; Paabo, S.; Krause, J. Complete mitochondrial genomes reveal neolithic expansion into Europe. PLoS ONE 2012, 7, e32473. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Olivieri, A.; Pala, M.; Metspalu, E.; Fornarino, S.; Battaglia, V.; Accetturo, M.; Kutuev, I.; Khusnutdinova, E.; Pennarun, E.; et al. Mitochondrial DNA variation of modern Tuscans supports the near eastern origin of Etruscans. Am. J. Hum. Genet. 2007, 80, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Hughey, J.R.; Paschou, P.; Drineas, P.; Mastropaolo, D.; Lotakis, D.M.; Navas, P.A.; Michalodimitrakis, M.; Stamatoyannopoulos, J.A.; Stamatoyannopoulos, G. A European population in Minoan Bronze Age Crete. Nat. Commun. 2013, 4, 1861. [Google Scholar] [CrossRef] [PubMed]
- Matisoo-Smith, E.A.; Gosling, A.L.; Boocock, J.; Kardailsky, O.; Kurumilian, Y.; Roudesli-Chebbi, S.; Badre, L.; Morel, J.P.; Sebai, L.L.; Zalloua, P.A. A European Mitochondrial Haplotype Identified in Ancient Phoenician Remains from Carthage, North Africa. PLoS ONE 2016, 11, e0155046. [Google Scholar] [CrossRef] [PubMed]
- Schuenemann, V.J.; Peltzer, A.; Welte, B.; van Pelt, W.P.; Molak, M.; Wang, C.C.; Furtwangler, A.; Urban, C.; Reiter, E.; Nieselt, K.; et al. Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat. Commun. 2017, 8, 15694. [Google Scholar] [CrossRef] [PubMed]
- Behar, D.M.; Metspalu, E.; Kivisild, T.; Rosset, S.; Tzur, S.; Hadid, Y.; Yudkovsky, G.; Rosengarten, D.; Pereira, L.; Amorim, A.; et al. Counting the founders: The matrilineal genetic ancestry of the Jewish Diaspora. PLoS ONE 2008, 3, e2062. [Google Scholar] [CrossRef] [PubMed]
- Schonberg, A.; Theunert, C.; Li, M.; Stoneking, M.; Nasidze, I. High-throughput sequencing of complete human mtDNA genomes from the Caucasus and West Asia: High diversity and demographic inferences. EJHG 2011, 19, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Lippold, S.; Xu, H.; Ko, A.; Li, M.; Renaud, G.; Butthof, A.; Schroder, R.; Stoneking, M. Human paternal and maternal demographic histories: Insights from high-resolution Y chromosome and mtDNA sequences. Investig. Genet. 2014, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Derenko, M.; Malyarchuk, B.; Bahmanimehr, A.; Denisova, G.; Perkova, M.; Farjadian, S.; Yepiskoposyan, L. Complete mitochondrial DNA diversity in Iranians. PLoS ONE 2013, 8, e80673. [Google Scholar] [CrossRef] [PubMed]
- Margaryan, A.; Derenko, M.; Hovhannisyan, H.; Malyarchuk, B.; Heller, R.; Khachatryan, Z.; Avetisyan, P.; Badalyan, R.; Bobokhyan, A.; Melikyan, V.; et al. Eight Millennia of Matrilineal Genetic Continuity in the South Caucasus. Curr. Biol. 2017, 27, 2023–2028. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.P.; Whitchurch, D.M.; Muhlstein, K. Rethinking burial dates at a Graeco-Roman Cemetary: Fag el-Gamous, Fayoum, Egypt. J. Arch. Sci. 2015, 2, 209–214. [Google Scholar]
- Hope, C.A. Three seasons of excavation at Ismant el-Kharab in Dakhleh Oasis, Egypt. Medi. Arch. 1988, 1, 160–178. [Google Scholar]
- Hope, C.A. The find context of the Kellis agricultural account book. In The Kellis Agricultural Account Book; Bagnall, R.S., Ed.; Oxbow Books: Oxford, UK, 1997; pp. 5–16. [Google Scholar]
- Hope, C.A. Objects from the Temple of Tutu. In Egyptian Religion—The Last Thousand Years: Studies Dedicated to the Memory of Jan Quaggebeur; Peeters: Leuven, Belgium, 1998; pp. 803–858. [Google Scholar]
- Hope, C.A. Observation on the dating of the occupation at Ismant el-Kharab. In The Oasis Papers I: The Proceedings of the First Conference of the Dakhleh Oasis Project; Marlow, C.A., Mills, A.J., Eds.; Oxbow Books: Oxford, UK, 2001; pp. 43–59. [Google Scholar]
- Dupras, T.L.; Schwarcz, H.P. Strangers in a strange land: Stable isotope evidencefor human migration in the Dakhleh oasis, Egypt. J. Arch. Sci. 2001, 28, 1199–1208. [Google Scholar] [CrossRef]

| Libraries | AF-Lib1 | AF-Lib2 | UI-Lib |
|---|---|---|---|
| Sequencing method | Single-end, 50 cycles | Paired-end, 2 × 150 cycles | Paired-end, 2 × 80 cycles |
| Raw reads | 3,176,685 | 7,260,250 | 38,178,602 |
| After merging and Trimming | 3,110,635 1,* | 2,697,704 1 | 17,402,064 2 |
| # Reads mapped to rCRS | 161,621 (5.2%) 1 | 166,798 (6.18%) 1 | 1,357,361 (7.8%) 3 |
| % Duplicates | 82.30% | 81.78% | 99.39% |
| # Unique reads | 28,531 1 22,602 4 | 30,397 1 27,290 4 | 8200 3 7987 4 |
| Total # nucleotides | 1,365,862 1 1,088,882 4 | 2,071,569 1 2,028,073 4 | NA 349,012 4 |
| Coverage | 9–145x | 22–239x | 1–128x |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molto, J.E.; Loreille, O.; Mallott, E.K.; Malhi, R.S.; Fast, S.; Daniels-Higginbotham, J.; Marshall, C.; Parr, R. Complete Mitochondrial Genome Sequencing of a Burial from a Romano–Christian Cemetery in the Dakhleh Oasis, Egypt: Preliminary Indications. Genes 2017, 8, 262. https://doi.org/10.3390/genes8100262
Molto JE, Loreille O, Mallott EK, Malhi RS, Fast S, Daniels-Higginbotham J, Marshall C, Parr R. Complete Mitochondrial Genome Sequencing of a Burial from a Romano–Christian Cemetery in the Dakhleh Oasis, Egypt: Preliminary Indications. Genes. 2017; 8(10):262. https://doi.org/10.3390/genes8100262
Chicago/Turabian StyleMolto, J. Eldon, Odile Loreille, Elizabeth K. Mallott, Ripan S. Malhi, Spence Fast, Jennifer Daniels-Higginbotham, Charla Marshall, and Ryan Parr. 2017. "Complete Mitochondrial Genome Sequencing of a Burial from a Romano–Christian Cemetery in the Dakhleh Oasis, Egypt: Preliminary Indications" Genes 8, no. 10: 262. https://doi.org/10.3390/genes8100262
APA StyleMolto, J. E., Loreille, O., Mallott, E. K., Malhi, R. S., Fast, S., Daniels-Higginbotham, J., Marshall, C., & Parr, R. (2017). Complete Mitochondrial Genome Sequencing of a Burial from a Romano–Christian Cemetery in the Dakhleh Oasis, Egypt: Preliminary Indications. Genes, 8(10), 262. https://doi.org/10.3390/genes8100262






