The History of Tree and Shrub Taxa on Bol'shoy Lyakhovsky Island (New Siberian Archipelago) since the Last Interglacial Uncovered by Sedimentary Ancient DNA and Pollen Data
Abstract
:1. Introduction
2. Materials and Methods
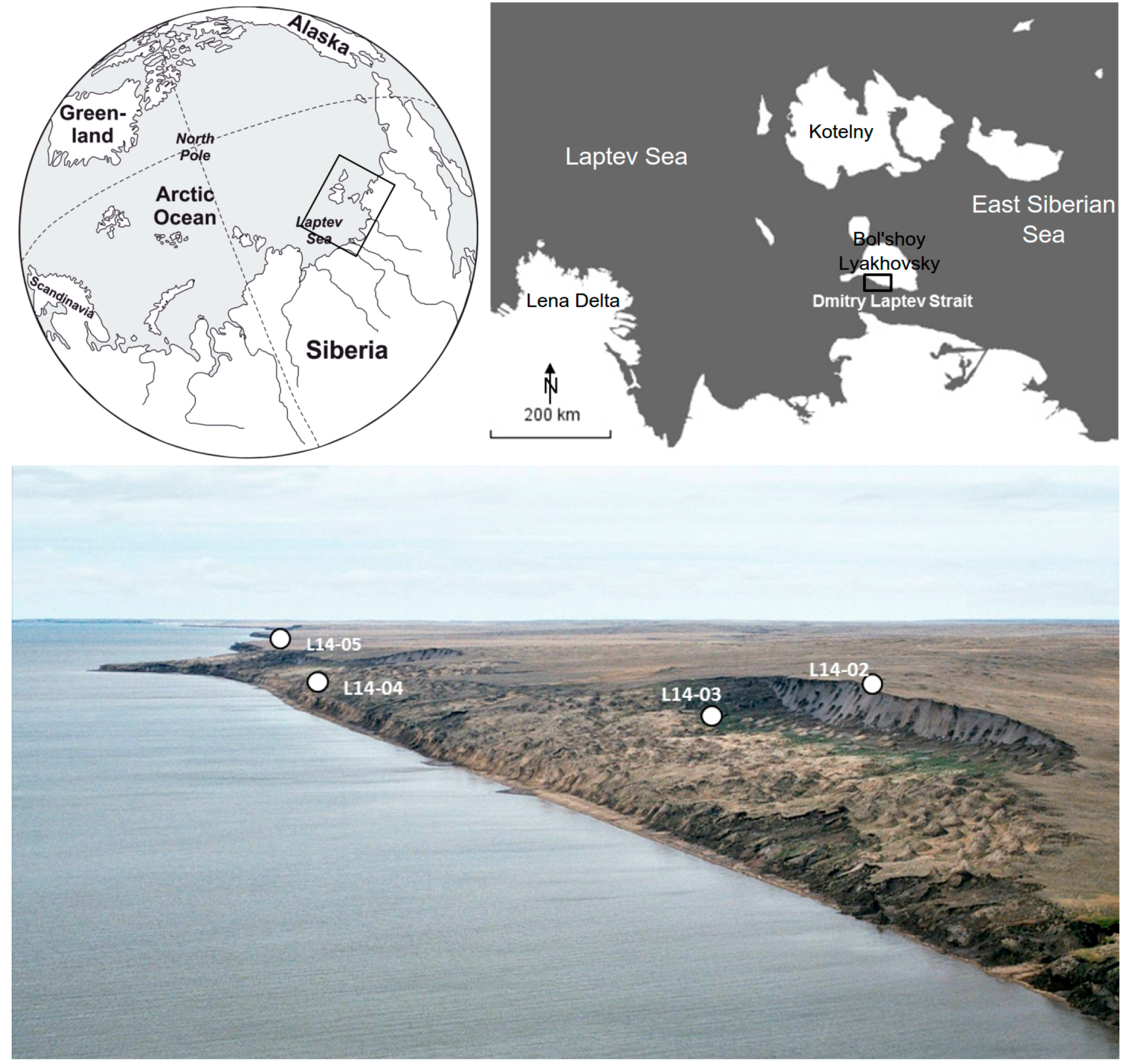
2.1. Geographic Setting
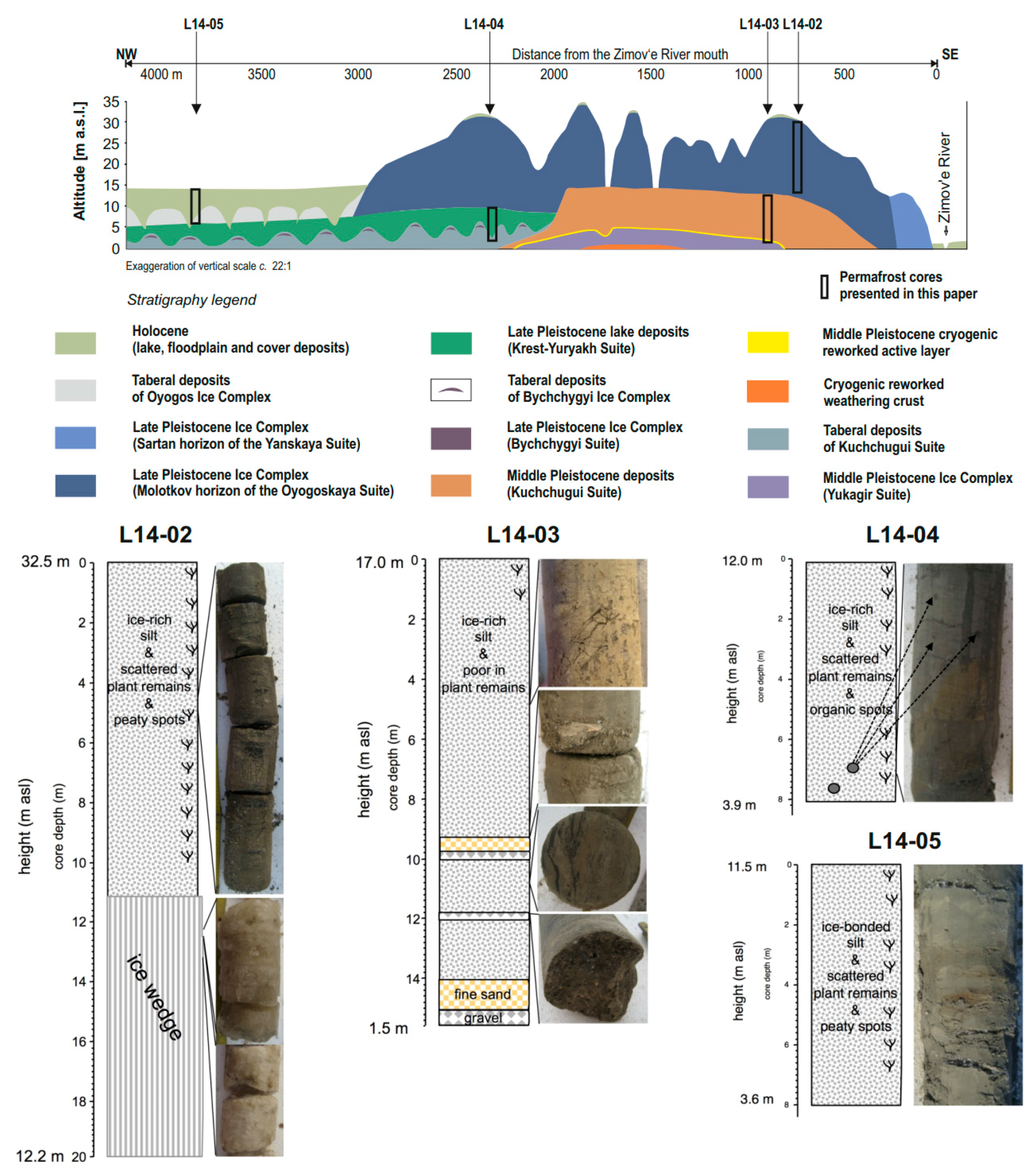
2.2. Core Material
2.2.1. Core L14-04 and Hand-Pieces L14-04B and L14-04C
2.2.2. Core L14-03
2.2.3. Core L14-02
2.2.4. Core L14-05
2.3. Radiocarbon Dating
2.4. Core Sub-Sampling
2.5. Molecular Genetic Laboratory Work
2.5.1. Sedimentary Ancient DNA Metabarcoding Approach
2.5.2. Specific Amplification of Larix from sedaDNA
2.5.3. Filtering of Illumina Sequencing Data and Taxonomic Assignments
2.6. Pollen Sample Treatment and Analysis
2.7. Statistical Analyses and Visualisation
3. Results
3.1. Radiocarbon Ages
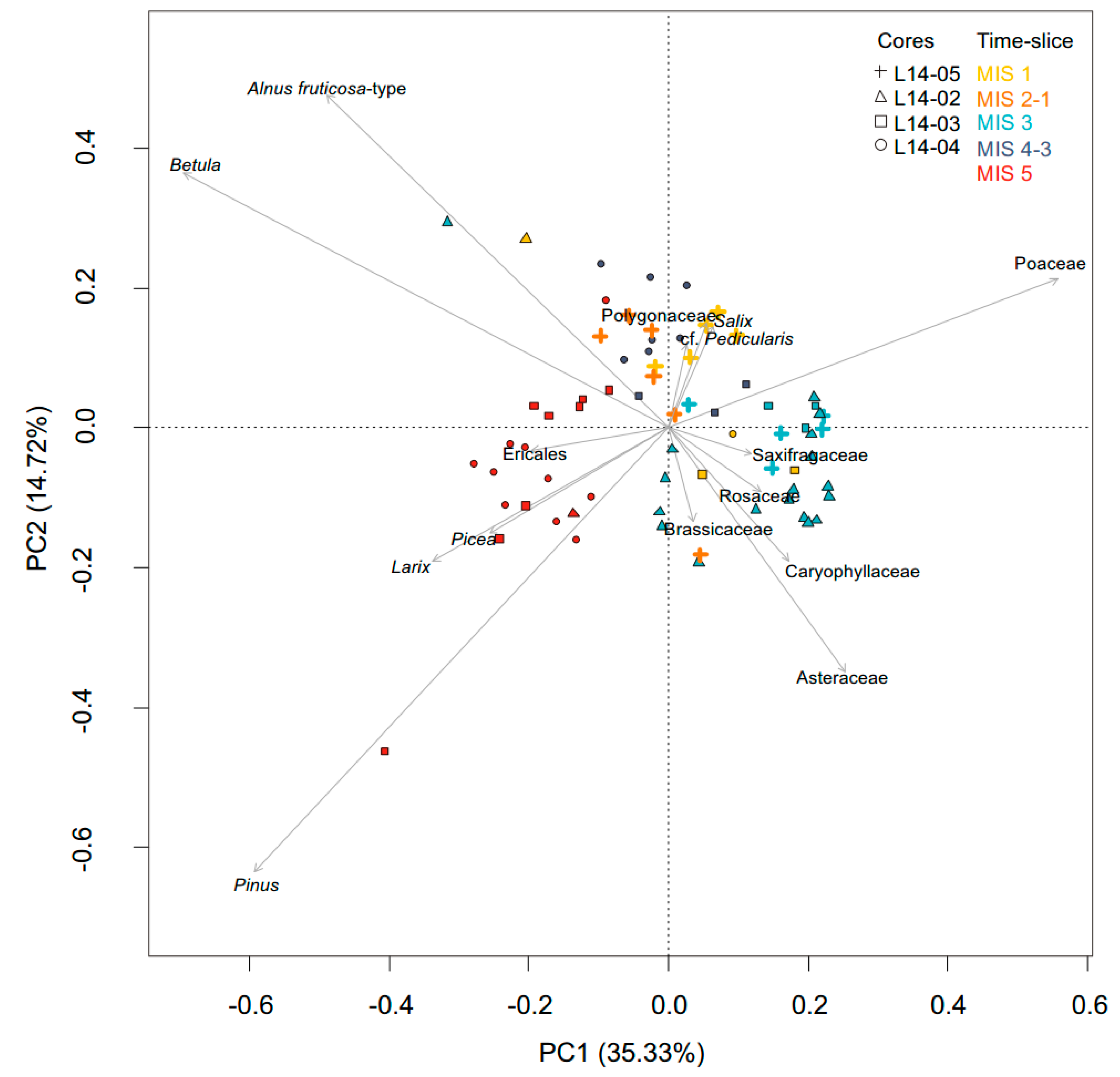
3.2. Overall Composition of the sedaDNA Metabarcoding and Pollen Data
3.3. Temporal Changes of Trees and Shrubs
3.3.1. SedaDNA Metabarcoding
3.3.2. Pollen
3.4. Validation of Sediment-Derived Larix Sequences and First Insights into Past Genetic Variation
4. Discussion
4.1. Geochronology
4.2. Assessment of Tree and Shrub Taxa in the sedaDNA and Pollen Records
4.3. Terrestrial Plant Community Changes of Warm Phases since the Last Interglacial
4.4. Past Genetic Diversity of Larch Populations on Bol’shoy Lyakhovsky Island
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shackleton, N. Oxygen isotope analyses and pleistocene temperatures re-assessed. Nature 1967, 215, 15–17. [Google Scholar] [CrossRef]
- Mix, A.C.; Ruddiman, W.F. Oxygen-isotope analyses and Pleistocene ice volumes. Quat. Res. 1984, 21, 1–20. [Google Scholar] [CrossRef]
- Lambeck, K.; Esat, T.M.; Potter, E.-K. Links between climate and sea levels for the past three million years. Nature 2002, 419, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Andreev, A.A.; Grosse, G.; Schirrmeister, L.; Kuznetsova, T.V.; Kuzmina, S.A.; Bobrov, A.A.; Tarasov, P.E.; Novenko, E.Y.; Meyer, H.; Derevyagin, A.Y.; et al. Weichselian and Holocene palaeoenvironmental history of the Bol’shoy Lyakhovsky Island, New Siberian Archipelago, Arctic Siberia. Boreas 2009, 38, 72–110. [Google Scholar] [CrossRef] [Green Version]
- Andreev, A.A.; Schirrmeister, L.; Tarasov, P.E.; Ganopolski, A.; Brovkin, V.; Siegert, C.; Wetterich, S.; Hubberten, H.-W. Vegetation and climate history in the Laptev Sea region (Arctic Siberia) during Late Quaternary inferred from pollen records. Quat. Sci. Rev. 2011, 30, 2182–2199. [Google Scholar] [CrossRef] [Green Version]
- Kienast, F.; Wetterich, S.; Kuzmina, S.; Schirrmeister, L.; Andreev, A.A.; Tarasov, P.; Nazarova, L.; Kossler, A.; Frolova, L.; Kunitsky, V.V. Paleontological records indicate the occurrence of open woodlands in a dry inland climate at the present-day Arctic coast in western Beringia during the Last Interglacial. Quat. Sci. Rev. 2011, 30, 2134–2159. [Google Scholar] [CrossRef] [Green Version]
- Wetterich, S.; Schirrmeister, L.; Andreev, A.A.; Pudenz, M.; Plessen, B.; Meyer, H.; Kunitsky, V.V. Eemian and Late Glacial/Holocene palaeoenvironmental records from permafrost sequences at the Dmitry Laptev Strait (NE Siberia, Russia). Palaeogeogr. Palaeoclimatol. Palaeoecol. 2009, 279, 73–95. [Google Scholar] [CrossRef] [Green Version]
- Wetterich, S.; Tumskoy, V.; Rudaya, N.; Andreev, A.A.; Opel, T.; Meyer, H.; Schirrmeister, L.; Hüls, M. Ice Complex formation in arctic East Siberia during the MIS3 Interstadial. Quat. Sci. Rev. 2014, 84, 39–55. [Google Scholar] [CrossRef] [Green Version]
- Emiliani, C. Pleistocene Temperatures. J. Geol. 1955, 63, 538–578. [Google Scholar] [CrossRef]
- Lisiecki, L.E.; Raymo, M.E. A Pliocene-Pleistocene stack of 57 globally distributed benthic δ 18 O records: Pliocene-Pleistocene benthic stack. Paleoceanography 2005, 20. [Google Scholar] [CrossRef]
- Railsback, L.B.; Gibbard, P.L.; Head, M.J.; Voarintsoa, N.R.G.; Toucanne, S. An optimized scheme of lettered marine isotope substages for the last 1.0 million years, and the climatostratigraphic nature of isotope stages and substages. Quat. Sci. Rev. 2015, 111, 94–106. [Google Scholar] [CrossRef]
- Hewitt, G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Hewitt, G.M. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Payette, S.; Lavoie, C. The arctic tree line as a record of past and recent climatic changes. Environ. Rev. 1994, 2, 78–90. [Google Scholar] [CrossRef]
- Arctic Climate Impact Assessment. Arctic Climate Impact Assessment Impacts of a Warming Arctic; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Andersen, K.K.; Azuma, N.; Barnola, J.-M.; Bigler, M.; Biscaye, P.; Caillon, N.; Chappellaz, J.; Clausen, H.B.; Dahl-Jensen, D.; Fischer, H.; et al. High-resolution record of Northern Hemisphere climate extending into the last interglacial period. Nature 2004, 431, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Woodward, F.I.; Lomas, M.R.; Betts, R.A. Vegetation-climate feedbacks in a greenhouse world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1998, 353, 29–39. [Google Scholar] [CrossRef]
- MacDonald, G.; Kremenetski, K.; Beilman, D. Climate change and the northern Russian treeline zone. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 2283–2299. [Google Scholar] [CrossRef] [PubMed]
- Levis, S.; Foley, J.A.; Pollard, D. Large-Scale Vegetation Feedbacks on a Doubled CO2 Climate. J. Clim. 2000, 13, 1313–1325. [Google Scholar] [CrossRef]
- Levis, S.; Foley, J.A.; Pollard, D. Potential high-latitude vegetation feedbacks on CO2-induced climate change. Geophys. Res. Lett. 1999, 26, 747–750. [Google Scholar] [CrossRef]
- Foley, J.A.; Costa, M.H.; Delire, C.; Ramankutty, N.; Snyder, P. Green surprise? How terrestrial ecosystems could affect earth’s climate. Front. Ecol. Environ. 2003, 1, 38–44. [Google Scholar]
- Bonan, G.B. Forests and Climate Change: Forcings, Feedbacks, and the Climate Benefits of Forests. Science 2008, 320, 1444–1449. [Google Scholar] [CrossRef] [PubMed]
- Kruse, S.; Wieczorek, M.; Jeltsch, F.; Herzschuh, U. Treeline dynamics in Siberia under changing climates as inferred from an individual-based model for Larix. Ecol. Model. 2016, 338, 101–121. [Google Scholar] [CrossRef]
- Herzschuh, U.; Birks, H.J.B.; Laepple, T.; Andreev, A.; Melles, M.; Brigham-Grette, J. Glacial legacies on interglacial vegetation at the Pliocene-Pleistocene transition in NE Asia. Nat. Commun. 2016, 7, 11967. [Google Scholar] [CrossRef] [PubMed]
- Andreev, A.A.; Grosse, G.; Schirrmeister, L.; Kuzmina, S.A.; Novenko, E.Y.; Bobrov, A.A.; Tarasov, P.E.; Ilyashuk, B.P.; Kuznetsova, T.V.; Krbetschek, M.; et al. Late Saalian and Eemian palaeoenvironmental history of the Bol’shoy Lyakhovsky Island (Laptev Sea region, Arctic Siberia). Boreas 2004, 33, 319–348. [Google Scholar] [CrossRef] [Green Version]
- Kienast, F.; Tarasov, P.; Schirrmeister, L.; Grosse, G.; Andreev, A.A. Continental climate in the East Siberian Arctic during the last interglacial: Implications from palaeobotanical records. Glob. Planet. Chang. 2008, 60, 535–562. [Google Scholar] [CrossRef] [Green Version]
- Willerslev, E.; Hansen, A.J.; Binladen, J.; Brand, T.B.; Gilbert, M.T.P.; Shapiro, B.; Bunce, M.; Wiuf, C.; Gilichinsky, D.A.; Cooper, A. Diverse Plant and Animal Genetic Records from Holocene and Pleistocene Sediments. Science 2003, 300, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Willerslev, E.; Davison, J.; Moora, M.; Zobel, M.; Coissac, E.; Edwards, M.E.; Lorenzen, E.D.; Vestergard, M.; Gussarova, G.; Haile, J.; et al. Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 2014, 506, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, T.; Haile, J.; Möller, P.; Andreev, A.; Boessenkool, S.; Rasmussen, M.; Kienast, F.; Coissac, E.; Taberlet, P.; Brochmann, C.; et al. A comparative study of ancient sedimentary DNA, pollen and macrofossils from permafrost sediments of northern Siberia reveals long-term vegetational stability. Mol. Ecol. 2012, 21, 1989–2003. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, H.H.; Raschke, E.; Epp, L.S.; Stoof-Leichsenring, K.R.; Schwamborn, G.; Schirrmeister, L.; Overduin, P.P.; Herzschuh, U. Sedimentary ancient DNA and pollen reveal the composition of plant organic matter in Late Quaternary permafrost sediments of the Buor Khaya Peninsula (north-eastern Siberia). Biogeosciences 2017, 14, 575–596. [Google Scholar] [CrossRef]
- Willerslev, E.; Cappellini, E.; Boomsma, W.; Nielsen, R.; Hebsgaard, M.B.; Brand, T.B.; Hofreiter, M.; Bunce, M.; Poinar, H.N.; Dahl-Jensen, D.; et al. Ancient biomolecules from deep ice cores reveal a forested Southern Greenland. Science 2007, 317, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Parducci, L.; Matetovici, I.; Fontana, S.L.; Bennett, K.D.; Suyama, Y.; Haile, J.; Kjaer, K.H.; Larsen, N.K.; Drouzas, A.D.; Willerslev, E. Molecular- and pollen-based vegetation analysis in lake sediments from central Scandinavia. Mol. Ecol. 2013, 22, 3511–3524. [Google Scholar] [CrossRef] [PubMed]
- Parducci, L.; Väliranta, M.; Salonen, J.S.; Ronkainen, T.; Matetovici, I.; Fontana, S.L.; Eskola, T.; Sarala, P.; Suyama, Y. Proxy comparison in ancient peat sediments: Pollen, macrofossil and plant DNA. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015, 370. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.W.; Ginolhac, A.; Orlando, L.; Olsen, J.; Andersen, K.; Holm, J.; Funder, S.; Willerslev, E.; Kjær, K.H. A comparative study of ancient environmental DNA to pollen and macrofossils from lake sediments reveals taxonomic overlap and additional plant taxa. Quat. Sci. Rev. 2013, 75, 161–168. [Google Scholar] [CrossRef]
- Van der Knaap, W.O. Transported pollen and spores on Spitsbergen and Jan Mayen. Pollen Spores 1987, 24, 449–453. [Google Scholar]
- Birks, H.H. Plant macrofossils. In Tracking Environmental Change Using Lake Sediments; Smol, J.P., Birks, H.J.B., Last, W.M., Bradley, R.S., Alverson, K., Eds.; Springer: Dordrecht, The Netherlands, 2001; Volume 3: Terrestrial, Algal, and Siliceous Indicators; ISBN 978-1-4020-0681-4. [Google Scholar]
- Niemeyer, B.; Klemm, J.; Pestryakova, L.A.; Herzschuh, U. Relative pollen productivity estimates for common taxa of the northern Siberian Arctic. Rev. Palaeobot. Palynol. 2015, 221, 71–82. [Google Scholar] [CrossRef]
- Campbell, I.D.; McDonald, K.; Flannigan, M.D.; Kringayark, J. Long-distance transport of pollen into the Arctic. Nature 1999, 399, 29–30. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Q.; Yang, X.; Chen, H.; Lu, X. Pollen-vegetation relationship and pollen preservation on the Northeastern Qinghai-Tibetan Plateau. Grana 2005, 44, 160–171. [Google Scholar] [CrossRef]
- De Klerk, P.; Donner, N.; Joosten, H.; Karpov, N.S.; Minke, M.; Seifert, N.; Theuerkauf, M. Vegetation patterns, recent pollen deposition and distribution of non-pollen palynomorphs in a polygon mire near Chokurdakh (NE Yakutia, NE Siberia). Boreas 2009, 38, 39–58. [Google Scholar] [CrossRef]
- Sønstebø, J.H.; Gielly, L.; Brysting, A.K.; Elven, R.; Edwards, M.; Haile, J.; Willerslev, E.; Coissac, E.; Rioux, D.; Sannier, J.; et al. Using next-generation sequencing for molecular reconstruction of past Arctic vegetation and climate. Mol. Ecol. Resour. 2010, 10, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, B.; Epp, L.S.; Stoof-Leichsenring, K.R.; Pestryakova, L.A.; Herzschuh, U. A comparison of sedimentary DNA and pollen from lake sediments in recording vegetation composition at the Siberian treeline. Mol. Ecol. Resour. 2017, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Parducci, L.; Suyama, Y.; Lascoux, M.; Bennett, K.D. Ancient DNA from pollen: A genetic record of population history in Scots pine. Mol. Ecol. 2005, 14. [Google Scholar] [CrossRef] [PubMed]
- Coissac, E.; Riaz, T.; Puillandre, N. Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol. Ecol. 2012, 21, 1834–1847. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, M.N. Cryomorphogenesis and Lithodynamics of the Nearshore Shelf Zone of the East Siberian Seas; Melnikov Permafrost Institute: Yakutsk, Russia, 2008. [Google Scholar]
- Hubberten, H.W.; Andreev, A.; Astakhov, V.I.; Demidov, I.; Dowdeswell, J.A.; Henriksen, M.; Hjort, C.; Houmark-Nielsen, M.; Jakobsson, M.; Kuzmina, S.; et al. The periglacial climate and environment in northern Eurasia during the last glaciation. Quat. Sci. Rev. 2004, 23, 1333–1357. [Google Scholar] [CrossRef] [Green Version]
- Schwamborn, G.; Wetterich, S. Russian-German cooperation CARBOPERM: Field campaigns to Bol’shoy Lyakhovsky Island in 2004. Available online: http://epic.awi.de/37311/ (accessed on 12 October 2017).
- Rivas-Martínez, S.; Rivas-Sáenz, S. Worldwide Bioclimatic Classification System; Phytosociological Research Center: Madrid, Spain, 1996. [Google Scholar]
- CAVM Team. Circumpolar Arctic Vegetation Map (1:7,500,000 scale), Conservation of Arctic Flora and Fauna (CAFF) Map No. 1.; U.S. Fish and Wildlife Service: Anchorage, AK, USA, 2003.
- Opel, T.; Wetterich, S.; Meyer, H.; Dereviagin, A.Y.; Fuchs, M.C.; Schirrmeister, L. Ground-ice stable isotopes and cryostratigraphy reflect late Quaternary palaeoclimate in the Northeast Siberian Arctic (Oyogos Yar coast, Dmitry Laptev Strait). Clim. Past 2017, 13, 587–611. [Google Scholar] [CrossRef]
- Ilyashuk, B.P.; Andreev, A.A.; Bobrov, A.A.; Tumskoy, V.E.; Ilyashuk, E.A. Interglacial History of a Palaeo-lake and Regional Environment: A Multi-proxy Study of a Permafrost Deposit from Bol’shoy Lyakhovsky Island, Arctic Siberia. J. Paleolimnol. 2006, 35, 855–872. [Google Scholar] [CrossRef] [Green Version]
- Rethemeyer, J.; Fülöp, R.-H.; Höfle, S.; Wacker, L.; Heinze, S.; Hajdas, I.; Patt, U.; König, S.; Stapper, B.; Dewald, A. Status report on sample preparation facilities for 14C analysis at the new CologneAMS center. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2013, 294, 168–172. [Google Scholar] [CrossRef]
- Schirrmeister, L.; Schwamborn, G.; Overduin, P.P.; Strauss, J.; Fuchs, M.C.; Grigoriev, M.; Yakshina, I.; Rethemeyer, J.; Dietze, E.; Wetterich, S. Yedoma Ice Complex of the Buor Khaya Peninsula (southern Laptev Sea). Biogeosciences 2017, 14, 1261–1283. [Google Scholar] [CrossRef]
- Stuiver, M.; Polach, H.A. Discussion Reporting of 14C Data. Radiocarbon 1977, 19, 355–363. [Google Scholar] [CrossRef]
- Reimer, P.J.; Bard, E.; Bayliss, A.; Beck, J.W.; Blackwell, P.G.; Ramsey, C.B.; Buck, C.E.; Cheng, H.; Edwards, R.L.; Friedrich, M.; et al. IntCal13 and Marine13 radiocarbon age calibration curves 0–50,000 years cal BP. Radiocarbon 2013, 55, 1869–1887. [Google Scholar] [CrossRef] [Green Version]
- Stuiver, M.; Reimer, P.J.; Reimer, R.W. CALIB 7.1. Available online: http://calib.org (accessed on 26 February 2017).
- Zimmermann, H.H.; Harms, L.; Trense, D.; Epp, L.S.; Stoof-Leichsenring, K.R.; Frickenhaus, S.; Pestryakova, L.; Herzschuh, U. Genetic variation of larches at the Siberian tundra-taiga ecotone inferred from the assembly of chloroplast genomes and mitochondrial sequences. BMC Genom. under revision.
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Ficetola, G.F.; Coissac, E.; Zundel, S.; Riaz, T.; Shehzad, W.; Bessière, J.; Taberlet, P.; Pompanon, F. An In silico approach for the evaluation of DNA barcodes. BMC Genom. 2010, 11, 434. [Google Scholar] [CrossRef] [PubMed]
- Boyer, F.; Mercier, C.; Bonin, A.; Le Bras, Y.; Taberlet, P.; Coissac, E. OBITools: A Unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 2016, 16, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Informatics: Cambridg, UK, 2010. [Google Scholar]
- Epp, L.S.; Gussarova, G.; Boessenkool, S.; Olsen, J.; Haile, J.; Schrøder-Nielsen, A.; Ludikova, A.; Hassel, K.; Stenøien, H.K.; Funder, S.; et al. Lake sediment multi-taxon DNA from North Greenland records early post-glacial appearance of vascular plants and accurately tracks environmental changes. Quat. Sci. Rev. 2015, 117, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Soininen, E.M.; Gauthier, G.; Bilodeau, F.; Berteaux, D.; Gielly, L.; Taberlet, P.; GUssarova, G.; Bellemain, E.; Hassel, K.; Stenøien, H.K.; et al. Highly overlapping winter diet in two sympatric lemming species revealed by DNA metabarcoding. PLoS ONE 2015, 10, e0115335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanz, C.; Aldebert, P.; Althorpe, N.; Baker, W.; Baldwin, A.; Bates, K.; Browne, P.; van den Broek, A.; Castro, M.; Cochrane, G.; et al. The EMBL nucleotide sequence database. Nucleic Acids Res. 2005, 33, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009, 37, D5–D15. [Google Scholar] [CrossRef] [PubMed]
- Epp, L.S.; Boessenkool, S.; Bellemain, E.P.; Haile, J.; Esposito, A.; Riaz, T.; Erséus, C.; Gusarov, V.I.; Edwards, M.E.; Johnsen, A.; et al. New environmental metabarcodes for analysing soil DNA: Potential for studying past and present ecosystems. Mol. Ecol. 2012, 21, 1821–1833. [Google Scholar] [CrossRef] [PubMed]
- Schnell, I.B.; Bohmann, K.; Gilbert, M.T.P. Tag jumps illuminated—Reducing sequence-to-sample misidentifications in metabarcoding studies. Mol. Ecol. Resour. 2015, 15, 1289–1303. [Google Scholar] [CrossRef] [PubMed]
- Faegri, K.; Iversen, J. Textbook of Pollen Analysis, 4th ed.; Faegri, K., Kaland, P.E., Krzywinski, K., Eds.; Blackburn Press: Caldwell, NJ, USA, 1989; ISBN 978-1-930665-01-9. [Google Scholar]
- Stockmarr, J. Tablets with spores used in absolute pollen analysis. Pollen Spores 1971, 13, 614–621. [Google Scholar]
- Beug, H.-J. Leitfaden der Pollenbestimmung; Verlag Dr. Friedrich Pfeil: München, Germany, 2004. [Google Scholar]
- Kupriyanova, L.A.; Alyoshina, L.A. Pollen and Spores of Plants in the Flora of the European Part of the USSR (Vol. 1); Academy of Sciences USSR, Komarov Botanical Institute: Leningrad, The Soviet Union, 1972. [Google Scholar]
- Kupriyanova, L.A.; Alyoshina, L.A. Pollen and Spores of Plants from the Flora of European Part of USSR. Lamiaceae-Zygophyllaceae; Academy of Sciences USSR, Komarov Botanical Institute: Leningrad, The Soviet Union, 1978. [Google Scholar]
- Moore, P.D.; Webb, J.A.; Collison, M.E. Pollen Analysis, 2nd ed.; Blackwell Scientific Publications: Oxford, UK, 1991. [Google Scholar]
- Savelieva, L.A.; Raschke, E.A.; Titova, D.V. Photographic Atlas of Plants and Pollen of the Lena River Delta; St. Petersburg State University: Saint-Petersburg, Russia, 2013; ISBN 978-5-4391-0036-1. [Google Scholar]
- Sokolovskaya, A.P. Vegetation of Far North and its development. In Pollen of the Arctic Plants; Komarov Botanical Institute, Academy of Sciences USSR: Moscow-Leningrad, The Soviet Union, 1958; pp. 245–292. [Google Scholar]
- Van Geel, B. Non-pollen palynomorphs. Volume 3: Terrestrial, algal and silicaceous indicators. In Tracking Environmental Change Using Lake Sediments: Terrestrial, Algal, and Siliceous Indicators; Smol, J.P., Birks, H.J.B., Last, W.M., Bradley, R.S., Alverson, K., Eds.; Springer: Dordrecht, The Netherlands, 2001; pp. 99–119. ISBN 978-0-306-47668-6. [Google Scholar]
- Van Geel, B.; Hallewas, D.P.; Pals, J.P. A late Holocene deposit under the Westfriese Zeedijk near Enkhuizen (Prov. of Noord-Holland, The Netherlands): Palaeoecological and archaeological aspects. Rev. Palaeobot. Palynol. 1983, 38, 269–335. [Google Scholar] [CrossRef]
- Van Geel, B.; Aptroot, A. Fossil ascomycetes in Quaternary deposits. Nova Hedwig. 2006, 82, 313–329. [Google Scholar] [CrossRef]
- Jankovská, V.; Komárek, J. Indicative value of Pediastrum and other coccal green algae in palaeoecology. Folia Geobot. 2000, 35, 59–82. [Google Scholar] [CrossRef]
- Komárek, J.; Jankovská, V. Review of the Green Algal Genus Pediastrum; Implication for Pollenanalytical Research; Bibliotheca Phycologica: Berlin, Germany; Schweizerbart Science Publishers: Stuttgart, Germany, 2001; Volume 108, ISBN 978-3-443-60035-8. [Google Scholar]
- The Angiosperm Phylogeny Group An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 2003, 141, 399–436.
- Hurlbert, S.H. The nonconcept of species diversity: A critique and alternative parameters. Ecology 1971, 52, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Heck, K.L.; van Belle, G.; Simberloff, D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 1975, 56, 1459–1461. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.0–2.; R Foundation for Statistical Computing: Vienna, Austria, 2011. [Google Scholar]
- Juggins, S. Rioja: Analysis of Quaternary Science Data, R Package Version 0.7–3; R Foundation for Statistical Computing: Vienna, Austria, 2012. [Google Scholar]
- Simpson, G.L. Analogue methods in palaeoecology: Using the analogue package. J. Stat. Softw. 2007, 22, 1–29. [Google Scholar] [CrossRef]
- Simpson, G.L.; Oksanen, J. Analogue: Analogue and Weighted Averaging Methods for Palaeoecology; R Package Version 0.6–8; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Zimmermann, H.H.; Raschke, E.; Epp, L.S.; Stoof-Leichsenring, K.R.; Schirrmeister, L.; Schwamborn, G.; Herzschuh, U. Ancient DNA Dataset, Pollen and Non-Pollen Palynomorphs of Four Permafrost Cores Hand-Pieces, Bol’shoy Lyakhovsky Island. Available online: https://doi.pangaea.de/10.1594/PANGAEA.878888 (accessed on 12 October 2017).
- Wetterich, S.; Tumskoy, V.; Rudaya, N.; Kuznetsov, V.; Maksimov, F.; Opel, T.; Meyer, H.; Andreev, A.A.; Schirrmeister, L. Ice Complex permafrost of MIS5 age in the Dmitry Laptev Strait coastal region (East Siberian Arctic). Quat. Sci. Rev. 2016, 147, 298–311. [Google Scholar] [CrossRef]
- French, H.M. The Periglacial Environment; John Wiley and Sons: Chichester, England; Hoboken, NJ, USA, 2007; ISBN 978-0-470-86590-3. [Google Scholar]
- Sjögren, P.; van der Knaap, W.O.; Huusko, A.; van Leeuwen, J.F.N. Pollen productivity, dispersal, and correction factors for major tree taxa in the Swiss Alps based on pollen-trap results. Rev. Palaeobot. Palynol. 2008, 152, 200–210. [Google Scholar] [CrossRef]
- Edwards, M.E.; Brubaker, L.B.; Lozhkin, A.V.; Anderson, P.M. Structurally Novel Biomes: A Response to Past Warming in Beringia. Ecology 2005, 86, 1696–1703. [Google Scholar] [CrossRef]
- Van Geel, B.; Protopopov, A.; Protopopova, V.; Pavlov, I.; van der Plicht, J.; van Reenen, G.B.A. Larix during the Mid-Pleniglacial (Greenland Interstadial 8) on Kotelny Island, northern Siberia. Boreas 2016. [Google Scholar] [CrossRef]
- Brubaker, L.B.; Anderson, P.M.; Edwards, M.E.; Lozhkin, A.V. Beringia as a glacial refugium for boreal trees and shrubs: New perspectives from mapped pollen data. J. Biogeogr. 2005, 32, 833–848. [Google Scholar] [CrossRef]
- Campbell, I.D. Quaternary pollen taphonomy: Examples of differential redeposition and differential preservation. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1999, 149, 245–256. [Google Scholar] [CrossRef]
- Eisenhut, G. Untersuchungen über die Morphologie und Ökologie der Pollenkörner Heimischer und Fremdländischer Waldbäume; P. Parey: Elbe-Parey, Germany, 1961. [Google Scholar]
- Parducci, L.; Jørgensen, T.; Tollefsrud, M.M.; Elverland, E.; Alm, T.; Fontana, S.L.; Bennett, K.D.; Haile, J.; Matetovici, I.; Suyama, Y.; et al. Glacial survival of boreal trees in northern Scandinavia. Science 2012, 335, 1083–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parducci, L.; Edwards, M.E.; Bennett, K.D.; Alm, T.; Elverland, E.; Tollefsrud, M.M.; Jørgensen, T.; Houmark-Nielsen, M.; Larsen, N.K.; Kjær, K.H.; et al. Response to Comment on “Glacial Survival of Boreal Trees in Northern Scandinavia”. Science 2012, 338, 742. [Google Scholar] [CrossRef]
- Koltowski, Z.; Pluta, S.; Jablonski, B.; Szklanowska, K. Pollination requirements of eight cultivars of black currant (Ribes nigrum L.). J. Hortic. Sci. Biotechnol. 1999, 74, 472–474. [Google Scholar] [CrossRef]
- Faegri, K.; van der Pijl, L. The Principles of Pollination Ecology, Third revised edition; Pergamon Press: Oxford, UK, 1980. [Google Scholar]
- Iversen, C.M.; Sloan, V.L.; Sullivan, P.F.; Euskirchen, E.S.; McGuire, A.D.; Norby, R.J.; Walker, A.P.; Warren, J.M.; Wullschleger, S.D. The unseen iceberg: Plant roots in arctic tundra. New Phytol. 2015, 205, 34–58. [Google Scholar] [CrossRef] [PubMed]
- Polezhaeva, M.A.; Lascoux, M.; Semerikov, V.L. Cytoplasmic DNA variation and biogeography of Larix Mill. in Northeast Asia. Mol. Ecol. 2010, 19, 1239–1252. [Google Scholar] [CrossRef] [PubMed]
- Semerikov, V.L.; Semerikova, S.A.; Polezhaeva, M.A.; Kosintsev, P.A.; Lascoux, M. Southern montane populations did not contribute to the recolonization of West Siberian Plain by Siberian larch (Larix sibirica): A range-wide analysis of cytoplasmic markers. Mol. Ecol. 2013, 22, 4958–4971. [Google Scholar] [CrossRef] [PubMed]
- Kharuk, V.I.; Ranson, K.J.; Im, S.T.; Naurzbaev, M.M. Forest-tundra larch forests and climatic trends. Russ. J. Ecol. 2006, 37, 291–298. [Google Scholar] [CrossRef]
- Köppen, W. Das geografische System der Klimate. In Handbuch der Klimatologie; Köppen, W., Geiger, R., Eds.; Gebrüder Borntraeger: Berlin, Germany, 1936; Volume I, C, pp. 1–44. [Google Scholar]
- Crawford, R.M.M.; Jeffree, C.E.; Rees, W.G. Paludification and forest retreat in northern oceanic environments. Ann. Bot. 2003, 91, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, H.; Saito, H.; Kuwao, K.; Maximov, T.C.; Hasegawa, S. Forest decline caused by high soil water conditions in a permafrost region. Hydrol. Earth Syst. Sci. 2010, 14, 301–307. [Google Scholar] [CrossRef]
- Abaimov, A.P. Geographical distribution and genetics of Siberian larch species. In Permafrost Ecosystems: Siberian Larch Forests; Ecological Studies: Dordrecht, The Netherlands; Springer: New York, NY, USA, 2010; pp. 41–58. ISBN 978-1-4020-9692-1. [Google Scholar]




| Core | Depth (m) | Lab Code | Plant Remains | Radiocarbon Age (Yr BP) | ± (Yr BP) | Calibrated Age (Cal Yr BP) | ± (Yr BP) |
|---|---|---|---|---|---|---|---|
| L14-05 | 0.65–0.85 | COL3744.1.1 | Poales, Amblystegiaceae | 2220 | 36 | 2239.5 | 91 |
| L14-05 | 0.85–1.04 | COL3745.1.1 | Poales, Amblystegiaceae (Drepanocladus, Campylium) | 6271 | 46 | 7221 | 70 |
| L14-05 | 1.39–1.69 | COL3746.1.1 | Poales, Carex, Amblystegiaceae | 10,079 | 50 | 11,611.5 | 222 |
| L14-05 | 5.00–5.17 | COL3747.1.1 | fine organic bulk material | 1259 | 34 | 1226.5 | 55 |
| L14-05 | 5.83–6.05 | COL3748.1.1 | Poales | 54,619 | 2314 | n. c. | n. c. |
| L14-05 | 7.75–7.89 | COL3623.1.1 | mosses (Drepanocladus revolvens) | 51,136 | 1743 | n. c. | n. c. |
| L14-02 | 0.31–0.44 | COL3614.1.1 | Carex stem | >modern | n.c. | n. c. | n.c. |
| L14-02 | 1.70–1.86 | COL3616.1.1 | fine root remains | 33,082 | 281 | 37,307.5 | 893.5 |
| L14-02 | 2.06–2.23 | COL3733.1.1 | Poales, roots, lignified remains | 33,711 | 247 | 37,977.5 | 754 |
| L14-02 | 3.47–3.77 | COL3617.1.1 | wood | 43,354 | 727 | 46,719.5 | 1508.5 |
| L14-02 | 4.03–4.16 | COL3618.1.1 | wood | 44,245 | 807 | 47,619.5 | 1753.5 |
| L14-02 | 4.27–4.48 | COL3619.1.1 | fine root remains | 43,802 | 811 | 47,194.5 | 1727.5 |
| L14-02 | 4.62–4.78 | COL3620.1.1 | fine root remains | 42,939 | 715 | 46,310.0 | 1429.0 |
| L14-02 | 5.33–5.56 | COL3621.1.1 | fine root remains | 62,773 | 7099 | n. c. | n. c. |
| L14-02 | 6.17–6.32 | COL3734.1.1 | roots | 51,479 | 1594 | n. c. | n. c. |
| L14-02 | 7.28–7.48 | COL3732.1.1 | fine organic bulk material, rootlets, Poales | 51,509 | 1597 | n. c. | n. c. |
| L14-02 | 7.48–7.68 | COL3735.1.1 | Carex, lignified remains, rootlets | 54,594 | 2354 | n. c. | n. c. |
| L14-02 | 8.12–8.29 | COL3730.1.1 | Carex, lignified remains, rootlets | 61,056 | 5081 | n. c. | n. c. |
| L14-02 | 8.51–8.68 | COL3731.1.1 | Carex, lignified remains, rootlets | 56,379 | 2866 | n. c. | n. c. |
| L14-02 | 9.02–9.28 | COL3736.1.1 | lignified remains, roots, remains of higher plants | 54,430 | 2275 | n. c. | n. c. |
| L14-02 | 9.84–10.12 | COL3737.1.1 | roots, fine organic bulk material | 53,591 | 2049 | n. c. | n. c. |
| L14-02 | 10.39–10.64 | COL3738.1.1 | roots, fine organic bulk material | 51,995 | 1702 | n. c. | n. c. |
| L14-02 | 10.92–11.04 | COL3739.1.1 | Poales | 49,309 | 1235 | n. c. | n. c. |
| L14-03 | 0.24–0.50 | COL3740.1.1 | lignified remains, fine organic bulk material | 3975 | 36 | 4454.5 | 73 |
| L14-03 | 0.8–0.95 | COL3741.1.1 | fine organic bulk material, roots | 29,259 | 174 | 33,433 | 411 |
| L14-03 | 1.86–1.98 | COL3742.1.1 | lignified remains, fine organic bulk material, Amblystegiaceae | 52,072 | 1704 | n. c. | n. c. |
| L14-03 | 2.30–2.42 | COL3743.1.1 | wood | 58,311 | 3628 | n. c. | n. c. |
| Primer Name | Sequence (5’–3’) | Ta (°C) | Product Size (nt) | SNP Variants | ecoPCR Specificity |
|---|---|---|---|---|---|
| cp77444F cp77444R | GGAAGGAAGGCGGAATGAATAG TCCATTACAGTACTTCCCTTCAG | 58 | 37 | A/C | Larix decidua |
| (MF = 0, MR = 0, L = 37) | |||||
| Larix occidentalis | |||||
| (MF = 0, MR = 0, L = 38) | |||||
| Pseudotsuga sinensis | |||||
| (MF = 2, MR = 1, L = 37) | |||||
| cp103595F cp103595R | GATGGATCACTTTCTGTCAAGGT GTATAGACAATGTTTCTCTGGCG | 58 | 54 | T/C | Larix decidua |
| (MF = 0, MR = 0, L = 54) | |||||
| Pseudotsuga menziesii | |||||
| (MF = 0, MR = 2, L = 54) | |||||
| Pseudotsuga sinensis | |||||
| (MF = 0, MR = 2, L = 54) | |||||
| Picea morrisonicola | |||||
| (MF = 0, MR = 3, L = 54) | |||||
| Picea abies | |||||
| (MF = 0, MR = 3, L = 54) | |||||
| Picea jezoensis | |||||
| (MF = 0, MR = 3, L = 54) | |||||
| Keteleeria | |||||
| (MF = 0, MR = 3, L = 54) | |||||
| Cedrus deodara | |||||
| (MF = 0, MR = 3, L = 54) | |||||
| Keteleeria davidiana | |||||
| (MF = 0, MR = 3, L =54) | |||||
| Abies koreana | |||||
| (MF = 0, MR = 3, L = 54) | |||||
| Abies nephrolepis | |||||
| (MF = 0, MR = 3, L = 54) | |||||
| Picea glauca | |||||
| (MF = 0, MR = 3, L = 54) | |||||
| Tsuga chinensis | |||||
| (MF = 0, MR = 3, L = 54) | |||||
| Pseudolarix amabilis | |||||
| (MF = 0, MR = 3, L = 54) |
| Core/HP | Sample Depth | PCR-Product cp77444 | PCR-Product cp103595 | Cloning Success cp77444 | Cloning Success cp103595 | No. of Retrieved Sequences cp77444 | Detected Variants cp77444 | No. of Retrieved Sequences cp103595 | Detected Variants cp103595 |
|---|---|---|---|---|---|---|---|---|---|
| L14-02 | 7.53 m | + | + | + | - | 0 | - | - | - |
| - | EN2 | - | - | n. c. | n.c. | - | - | - | - |
| L14-03 | 6.72 m | + | + | + | + | 0 | - | 13 | T |
| - | EN3 | - | - | n.c. | n.c. | - | - | - | - |
| L14-03 | 7.59 m | + | - | + | n.c. | 11 | A | - | - |
| L14-03 | 8.56 m | + | + | + | + | 8 | A | 7 | 3× C, 4× T |
| L14-03 | 9.61 m | - | - | n.c. | n.c. | - | - | - | - |
| L14-03 | 10.11 m | - | - | n.c. | n.c. | - | - | - | - |
| L14-03 | 11.77 m | + | + | + | + | 9 | A | 3 | T |
| - | EN5 | - | - | n.c. | n.c. | - | - | - | - |
| L14-05 | 3.3 m | - | + | + | + | 1 | A | 1 | T |
| L14-05 | 3.7 m | - | - | n.c. | n.c. | - | - | - | - |
| - | EN6 | - | - | n.c. | n.c. | - | - | - | - |
| L14-04 | 6.15 m | - | + | n.c. | + | - | - | 2 | C |
| L14-04 | 7.27 m | + | + | + | + | 7 | A | 13 | T |
| - | EN7 | - | - | n.c. | n.c. | - | - | - | - |
| L14-04 | 8.03 m | + | + | + | + | 7 | 3× C, 4× A | 7 | 1× C, 6× T |
| L14-04C | 2.05 m | + | + | + | + | 9 | A | 11 | C |
| L14-04C | 3.15 m | + | + | + | + | 10 | A | 15 | 8× C, 7× T |
| L14-04C | 3.55 m | + | + | + | + | 6 | A | 13 | 2× C, 11× T |
| - | EN8 | - | - | n.c. | n.c. | - | - | - | - |
| - | NTCs | - | - | n.c. | n.c. | - | - | - | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zimmermann, H.H.; Raschke, E.; Epp, L.S.; Stoof-Leichsenring, K.R.; Schirrmeister, L.; Schwamborn, G.; Herzschuh, U. The History of Tree and Shrub Taxa on Bol'shoy Lyakhovsky Island (New Siberian Archipelago) since the Last Interglacial Uncovered by Sedimentary Ancient DNA and Pollen Data. Genes 2017, 8, 273. https://doi.org/10.3390/genes8100273
Zimmermann HH, Raschke E, Epp LS, Stoof-Leichsenring KR, Schirrmeister L, Schwamborn G, Herzschuh U. The History of Tree and Shrub Taxa on Bol'shoy Lyakhovsky Island (New Siberian Archipelago) since the Last Interglacial Uncovered by Sedimentary Ancient DNA and Pollen Data. Genes. 2017; 8(10):273. https://doi.org/10.3390/genes8100273
Chicago/Turabian StyleZimmermann, Heike H., Elena Raschke, Laura S. Epp, Kathleen R. Stoof-Leichsenring, Lutz Schirrmeister, Georg Schwamborn, and Ulrike Herzschuh. 2017. "The History of Tree and Shrub Taxa on Bol'shoy Lyakhovsky Island (New Siberian Archipelago) since the Last Interglacial Uncovered by Sedimentary Ancient DNA and Pollen Data" Genes 8, no. 10: 273. https://doi.org/10.3390/genes8100273






