Maintenance of Genome Integrity: How Mammalian Cells Orchestrate Genome Duplication by Coordinating Replicative and Specialized DNA Polymerases
Abstract
:1. Introduction
2. Overview of Polymerase Functions
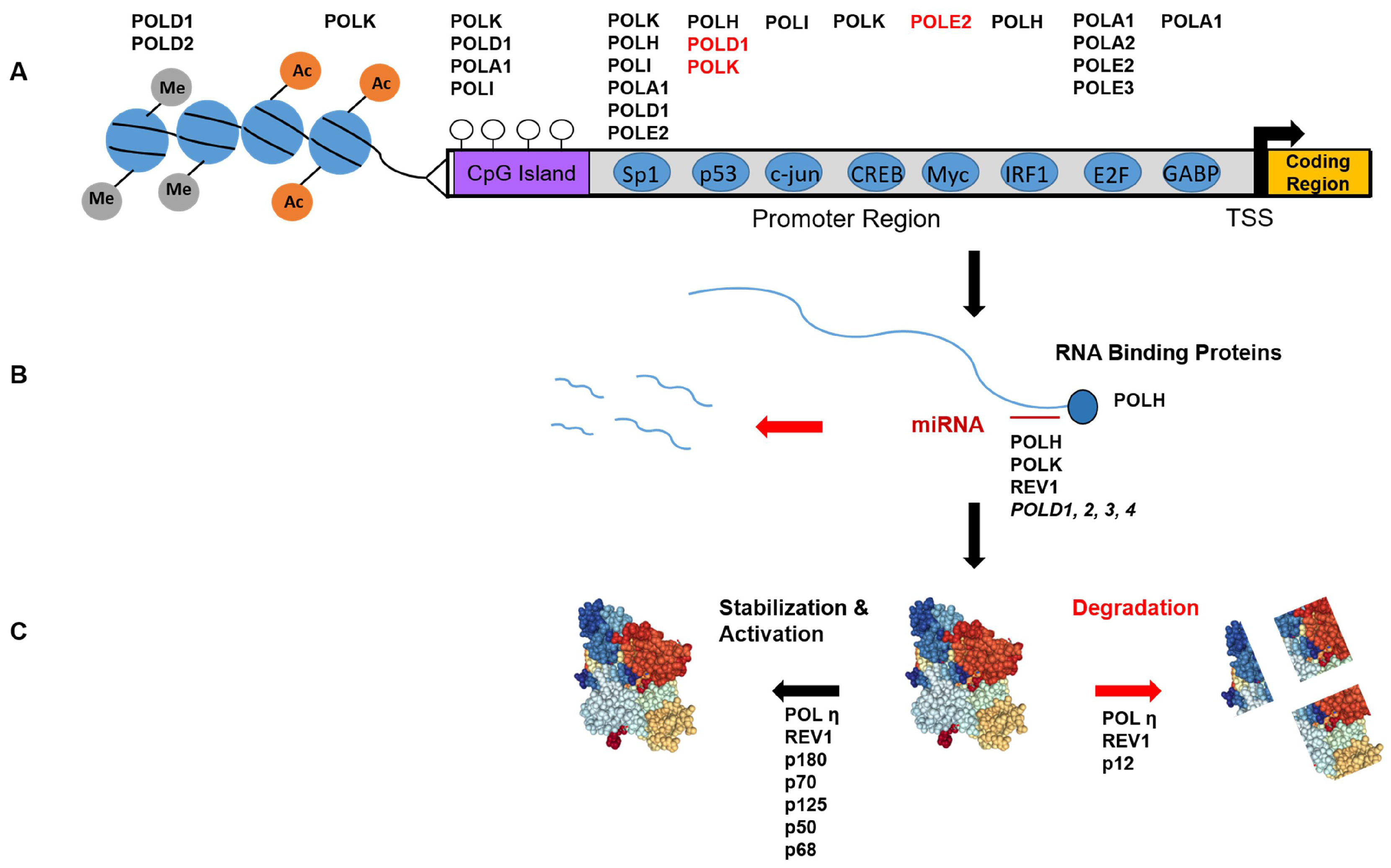
3. DNA Polymerase Expression during an Unperturbed Mitotic Cell Cycle
3.1. Transcriptional Regulation
3.1.1. Sp1
3.1.2. p53
3.1.3. E2F
3.1.4. Epigenetic Regulation
3.2. Post-Transcriptional Regulation
3.3. Post-Translational Modifications-Functional
3.4. Post-Translational Modifications-Degradation
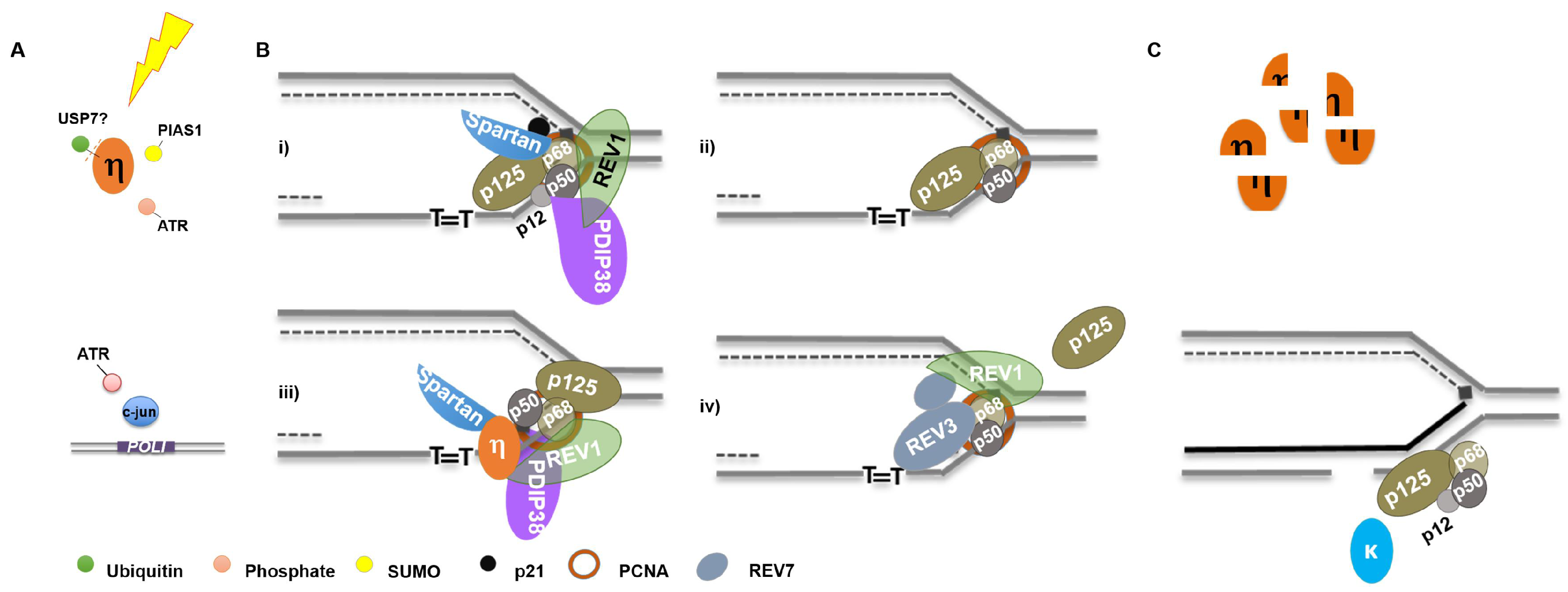
4. DNA Polymerase Expression under Stress
4.1. Transcriptional Regulation
4.2. Post-Translational Modifications—Functional
4.2.1. Phosphorylation
4.2.2. Ubiquitination
4.3. Post-Translational Modifications—Degradation and Stability
5. Orchestration of DNA Polymerases
6. Summary and Model for Orchestration
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Waters, L.S.; Minesinger, B.K.; Wiltrout, M.E.; D'Souza, S.; Woodruff, R.V.; Walker, G.C. Eukaryotic translesion polymerases and their roles and regulation in dna damage tolerance. Microbiol. Mol. Biol. Rev. 2009, 73, 134–154. [Google Scholar] [CrossRef] [PubMed]
- Boyer, A.S.; Grgurevic, S.; Cazaux, C.; Hoffmann, J.S. The human specialized dna polymerases and non-b dna: Vital relationships to preserve genome integrity. J. Mol. Biol. 2013, 425, 4767–4781. [Google Scholar] [CrossRef] [PubMed]
- Sale, J.E.; Lehmann, A.R.; Woodgate, R. Y-family dna polymerases and their role in tolerance of cellular dna damage. Nat. Rev. Mol. Cell Biol. 2012, 13, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Lujan, S.A.; Williams, J.S.; Kunkel, T.A. Dna polymerases divide the labor of genome replication. Trends Cell Biol. 2016, 26, 640–654. [Google Scholar] [CrossRef] [PubMed]
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, A.R.; Fuchs, R.P. Gaps and forks in dna replication: Rediscovering old models. DNA Repair (Amst.) 2006, 5, 1495–1498. [Google Scholar] [CrossRef] [PubMed]
- de Koning, A.P.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef] [PubMed]
- Durkin, S.G.; Glover, T.W. Chromosome fragile sites. Annu. Rev. Genet. 2007, 41, 169–192. [Google Scholar] [CrossRef] [PubMed]
- Hile, S.E.; Wang, X.; Lee, M.Y.; Eckert, K.A. Beyond translesion synthesis: Polymerase κ fidelity as a potential determinant of microsatellite stability. Nucleic Acids Res. 2012, 40, 1636–1647. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.N.; Opresko, P.L.; Meng, X.; Lee, M.Y.; Eckert, K.A. Dna structure and the werner protein modulate human dna polymerase delta-dependent replication dynamics within the common fragile site fra16d. Nucleic Acids Res. 2010, 38, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.; Wang, X.; Lee, M.Y.; Eckert, K.A. Mechanism of replicative dna polymerase delta pausing and a potential role for dna polymerase kappa in common fragile site replication. J. Mol. Biol. 2013, 425, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Bergoglio, V.; Boyer, A.S.; Walsh, E.; Naim, V.; Legube, G.; Lee, M.Y.; Rey, L.; Rosselli, F.; Cazaux, C.; Eckert, K.A.; et al. Dna synthesis by pol η promotes fragile site stability by preventing under-replicated dna in mitosis. J. Cell Biol. 2013, 201, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.; Andersen, P.L.; Qin, Z.; Xiao, W. Rev3, the catalytic subunit of polζ, is required for maintaining fragile site stability in human cells. Nucleic Acids Res. 2013, 41, 2328–2339. [Google Scholar] [CrossRef] [PubMed]
- Rey, L.; Sidorova, J.M.; Puget, N.; Boudsocq, F.; Biard, D.S.; Monnat, R.J.; Cazaux, C.; Hoffmann, J.S. Human dna polymerase eta is required for common fragile site stability during unperturbed dna replication. Mol. Cell. Biol. 2009, 29, 3344–3354. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.L.; Simpson, L.J.; Sale, J.E. Vertebrate dna damage tolerance requires the c-terminus but not brct or transferase domains of rev1. Nucleic Acids Res. 2005, 33, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, B.A.; Suo, Z. Recent insight into the kinetic mechanisms and conformational dynamics of y-family dna polymerases. Biochemistry 2014, 53, 2804–2814. [Google Scholar] [CrossRef] [PubMed]
- Yang, W. An overview of y-family dna polymerases and a case study of human dna polymerase η. Biochemistry 2014, 53, 2793–2803. [Google Scholar] [CrossRef] [PubMed]
- Biertümpfel, C.; Zhao, Y.; Kondo, Y.; Ramón-Maiques, S.; Gregory, M.; Lee, J.Y.; Masutani, C.; Lehmann, A.R.; Hanaoka, F.; Yang, W. Structure and mechanism of human dna polymerase eta. Nature 2010, 465, 1044–1048. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Biertümpfel, C.; Gregory, M.T.; Hua, Y.J.; Hanaoka, F.; Yang, W. Structural basis of human dna polymerase η-mediated chemoresistance to cisplatin. Proc. Natl. Acad. Sci. USA 2012, 109, 7269–7274. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.; Townson, S.A.; Uljon, S.N.; Johnson, R.E.; Brahma, A.; Nair, D.T.; Prakash, S.; Prakash, L.; Aggarwal, A.K. Human dna polymerase kappa encircles dna: Implications for mismatch extension and lesion bypass. Mol. Cell 2007, 25, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Y.; Tang, T.S.; Zhang, H.; Wang, Z.; Friedberg, E.; Yang, W.; Guo, C. Variants of mouse dna polymerase κ reveal a mechanism of efficient and accurate translesion synthesis past a benzo[a]pyrene dg adduct. Proc. Natl. Acad. Sci. USA 2014, 111, 1789–1794. [Google Scholar] [CrossRef] [PubMed]
- Jha, V.; Bian, C.; Xing, G.; Ling, H. Structure and mechanism of error-free replication past the major benzo[a]pyrene adduct by human dna polymerase κ. Nucleic Acids Res. 2016, 44, 4957–4967. [Google Scholar] [CrossRef] [PubMed]
- Thompson, H.C.; Sheaff, R.J.; Kuchta, R.D. Interactions of calf thymus dna polymerase alpha with primer/templates. Nucleic Acids Res. 1995, 23, 4109–4115. [Google Scholar] [CrossRef] [PubMed]
- Van, C.; Yan, S.; Michael, W.M.; Waga, S.; Cimprich, K.A. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J. Cell Biol. 2010, 189, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Michael, W.M. Topbp1 and dna polymerase-alpha directly recruit the 9-1-1 complex to stalled dna replication forks. J. Cell Biol. 2009, 184, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M.; Itkonen, H.; Hillukkala, T.; Liu, Z.; Nasheuer, H.P.; Schaarschmidt, D.; Pospiech, H.; Syväoja, J.E. Segregation of replicative dna polymerases during s phase: Dna polymerase ε, but not dna polymerases α/δ, are associated with lamins throughout s phase in human cells. J. Biol. Chem. 2012, 287, 33327–33338. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.L.; Preston, B.D.; O'Connor, T.R.; Dianov, G.L. Dna polymerase delta-dependent repair of dna single strand breaks containing 3'-end proximal lesions. Nucleic Acids Res. 2007, 35, 1054–1063. [Google Scholar] [CrossRef] [PubMed]
- Longley, M.J.; Pierce, A.J.; Modrich, P. Dna polymerase delta is required for human mismatch repair in vitro. J. Biol. Chem. 1997, 272, 10917–10921. [Google Scholar] [PubMed]
- Sneeden, J.L.; Grossi, S.M.; Tappin, I.; Hurwitz, J.; Heyer, W.D. Reconstitution of recombination-associated dna synthesis with human proteins. Nucleic Acids Res. 2013, 41, 4913–4925. [Google Scholar] [CrossRef] [PubMed]
- Ogi, T.; Limsirichaikul, S.; Overmeer, R.M.; Volker, M.; Takenaka, K.; Cloney, R.; Nakazawa, Y.; Niimi, A.; Miki, Y.; Jaspers, N.G.; et al. Three dna polymerases, recruited by different mechanisms, carry out ner repair synthesis in human cells. Mol. Cell 2010, 37, 714–727. [Google Scholar] [CrossRef] [PubMed]
- McIlwraith, M.J.; Mcllwraith, M.J.; Vaisman, A.; Liu, Y.; Fanning, E.; Woodgate, R.; West, S.C. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell 2005, 20, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Peña-Diaz, J.; Bregenhorn, S.; Ghodgaonkar, M.; Follonier, C.; Artola-Borán, M.; Castor, D.; Lopes, M.; Sartori, A.A.; Jiricny, J. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell 2012, 47, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Winter, D.B.; Kasmer, C.; Kraemer, K.H.; Lehmann, A.R.; Gearhart, P.J. Dna polymerase eta is an a-t mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2001, 2, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Bétous, R.; Rey, L.; Wang, G.; Pillaire, M.J.; Puget, N.; Selves, J.; Biard, D.S.; Shin-ya, K.; Vasquez, K.M.; Cazaux, C.; et al. Role of tls dna polymerases eta and kappa in processing naturally occurring structured dna in human cells. Mol. Carcinog. 2009, 48, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Ogi, T.; Lehmann, A.R. The y-family dna polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 2006, 8, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lv, L.; Chen, Q.; Yuan, F.; Zhang, T.; Yang, Y.; Zhang, H.; Wang, Y.; Jia, Y.; Qian, L.; et al. Mouse dna polymerase kappa has a functional role in the repair of dna strand breaks. DNA Repair (Amst.) 2013, 12, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Bétous, R.; Pillaire, M.J.; Pierini, L.; van der Laan, S.; Recolin, B.; Ohl-Séguy, E.; Guo, C.; Niimi, N.; Grúz, P.; Nohmi, T.; et al. Dna polymerase κ-dependent dna synthesis at stalled replication forks is important for chk1 activation. EMBO J. 2013, 32, 2172–2185. [Google Scholar] [CrossRef] [PubMed]
- Faili, A.; Aoufouchi, S.; Flatter, E.; Guéranger, Q.; Reynaud, C.A.; Weill, J.C. Induction of somatic hypermutation in immunoglobulin genes is dependent on dna polymerase iota. Nature 2002, 419, 944–947. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Miguel, S.; Richardson, J.A.; Gerlach, V.L.; Lai, W.C.; Gao, T.; Russell, L.D.; Hladik, C.L.; White, C.L.; Friedberg, E.C. Constitutive and regulated expression of the mouse dinb (polkappa) gene encoding dna polymerase kappa. DNA Repair (Amst.) 2003, 2, 91–106. [Google Scholar] [CrossRef]
- Gerlach, V.L.; Aravind, L.; Gotway, G.; Schultz, R.A.; Koonin, E.V.; Friedberg, E.C. Human and mouse homologs of escherichia coli dinb (dna polymerase iv), members of the umuc/dinb superfamily. Proc. Natl. Acad. Sci. USA 1999, 96, 11922–11927. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Wernick, M.; Collins, C.; Limoli, C.L.; Crowley, E.; Cleaver, J.E. Dna polymerase eta undergoes alternative splicing, protects against uv sensitivity and apoptosis, and suppresses mre11-dependent recombination. Genes Chromosomes Cancer 2001, 32, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Yamada, A.; Masutani, C.; Iwai, S.; Hanaoka, F. Complementation of defective translesion synthesis and uv light sensitivity in xeroderma pigmentosum variant cells by human and mouse dna polymerase eta. Nucleic Acids Res. 2000, 28, 2473–2480. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Xin, H.; Zhang, Y.; Wu, X.; Yuan, F.; Wang, Z. The human rev1 gene codes for a dna template-dependent dcmp transferase. Nucleic Acids Res. 1999, 27, 4468–4475. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.P.; Rapić-Otrin, V.; Epstein, J.A.; Broughton, B.C.; Wang, X.; Lehmann, A.R.; Wolgemuth, D.J.; Woodgate, R. Novel human and mouse homologs of saccharomyces cerevisiae dna polymerase eta. Genomics 1999, 60, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Frank, E.G.; Tissier, A.; McDonald, J.P.; Rapić-Otrin, V.; Zeng, X.; Gearhart, P.J.; Woodgate, R. Altered nucleotide misinsertion fidelity associated with poliota-dependent replication at the end of a dna template. EMBO J. 2001, 20, 2914–2922. [Google Scholar] [CrossRef] [PubMed]
- Akagi, J.; Masutani, C.; Kataoka, Y.; Kan, T.; Ohashi, E.; Mori, T.; Ohmori, H.; Hanaoka, F. Interaction with dna polymerase eta is required for nuclear accumulation of rev1 and suppression of spontaneous mutations in human cells. DNA Repair (Amst.) 2009, 8, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, V.P.; Farina, A.; Raghavan, V.; Tappin, I.; Hurwitz, J. Studies on human dna polymerase epsilon and gins complex and their role in dna replication. J. Biol. Chem. 2011, 286, 28963–28977. [Google Scholar] [CrossRef] [PubMed]
- Diamant, N.; Hendel, A.; Vered, I.; Carell, T.; Reissner, T.; de Wind, N.; Geacinov, N.; Livneh, Z. Dna damage bypass operates in the s and g2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 2012, 40, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Beishline, K.; Azizkhan-Clifford, J. Sp1 and the 'hallmarks of cancer'. FEBS J. 2015, 282, 224–258. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Yokoi, M.; Nishikawa, N.S.; Miyazawa, H.; Sugino, A.; Yamagishi, M.; Yamaguchi, M.; Matsukage, A.; Yatagai, F.; Hanaoka, F. Transcription of the catalytic 180-kda subunit gene of mouse dna polymerase alpha is controlled by e2f, an ets-related transcription factor, and sp1. Biochim. Biophys. Acta 2000, 1492, 341–352. [Google Scholar] [CrossRef]
- Zhao, L.; Chang, L.S. The human pold1 gene. Identification of an upstream activator sequence, activation by sp1 and sp3, and cell cycle regulation. J. Biol. Chem. 1997, 272, 4869–4882. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Jokela, M.; Tuusa, J.; Skog, S.; Poikonen, K.; Syväoja, J.E. E2f mediates induction of the sp1-controlled promoter of the human dna polymerase epsilon b-subunit gene pole2. Nucleic Acids Res. 2001, 29, 2810–2821. [Google Scholar] [CrossRef] [PubMed]
- Lemée, F.; Bavoux, C.; Pillaire, M.J.; Bieth, A.; Machado, C.R.; Pena, S.D.; Guimbaud, R.; Selves, J.; Hoffmann, J.S.; Cazaux, C. Characterization of promoter regulatory elements involved in downexpression of the dna polymerase kappa in colorectal cancer. Oncogene 2007, 26, 3387–3394. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Fan, Y.; Shen, J.; Qi, H.; Shao, J. Characterization of human dna polymerase κ promoter in response to benzo[a]pyrene diol epoxide. Environ. Toxicol. Pharmacol. 2012, 33, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, S.; Xie, L.; Liu, P.; Xie, F.; Wu, J.; Cao, J.; Ding, W.Q. Overexpression of dna polymerase iota (polι) in esophageal squamous cell carcinoma. Cancer Sci. 2012, 103, 1574–1579. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Fan, Y.; Jiang, H.; Shen, J.; Qi, H.; Mei, R.; Shao, J. Response of human dna polymerase ι promoter to n-methyl-n'-nitro-n-nitrosoguanidine. Environ. Toxicol. Pharmacol. 2010, 29, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Zhu, H.; Lou, M.; Fan, Y.; Liu, H.; Shen, J.; Li, Z.; Lv, X.; Shan, J.; Zhu, L.; et al. Interferon regulatory factor 1 transactivates expression of human dna polymerase η in response to carcinogen n-methyl-n'-nitro-n-nitrosoguanidine. J. Biol. Chem. 2012, 287, 12622–12633. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at ucsc. Genome Res. 2002, 12, 996–1006. [Google Scholar] [PubMed]
- Liu, G.; Chen, X. Dna polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the dna damage checkpoint and p53 activation. Mol. Cell. Biol. 2006, 26, 1398–1413. [Google Scholar] [CrossRef] [PubMed]
- Durando, M.; Tateishi, S.; Vaziri, C. A non-catalytic role of dna polymerase η in recruiting rad18 and promoting pcna monoubiquitination at stalled replication forks. Nucleic Acids Res. 2013, 41, 3079–3093. [Google Scholar] [CrossRef] [PubMed]
- Melanson, B.D.; Bose, R.; Hamill, J.D.; Marcellus, K.A.; Pan, E.F.; McKay, B.C. The role of mRNA decay in p53-induced gene expression. RNA 2011, 17, 2222–2234. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Seimiya, M.; Kawamura, K.; Yu, L.; Ogi, T.; Takenaga, K.; Shishikura, T.; Nakagawara, A.; Sakiyama, S.; Tagawa, M.; et al. Elevated expression of dna polymerase kappa in human lung cancer is associated with p53 inactivation: Negative regulation of polk promoter activity by p53. Int. J. Oncol. 2004, 25, 161–165. [Google Scholar] [PubMed]
- Antoniali, G.; Marcuzzi, F.; Casarano, E.; Tell, G. Cadmium treatment suppresses dna polymerase δ catalytic subunit gene expression by acting on the p53 and sp1 regulatory axis. DNA Repair (Amst.) 2015, 35, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lee, M.Y. Transcriptional regulation of the human dna polymerase delta catalytic subunit gene pold1 by p53 tumor suppressor and sp1. J. Biol. Chem. 2001, 276, 29729–29739. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, N.S.; Izumi, M.; Uchida, H.; Yokoi, M.; Miyazawa, H.; Hanaoka, F. Cloning and characterization of the 5'-upstream sequence governing the cell cycle-dependent transcription of mouse dna polymerase alpha 68 kda subunit gene. Nucleic Acids Res. 2000, 28, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Bolognese, F.; Forni, C.; Caretti, G.; Frontini, M.; Minuzzo, M.; Mantovani, R. The pole3 bidirectional unit is regulated by myc and e2fs. Gene 2006, 366, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Karkhanis, V.; Wang, L.; Tae, S.; Hu, Y.J.; Imbalzano, A.N.; Sif, S. Protein arginine methyltransferase 7 regulates cellular response to dna damage by methylating promoter histones h2a and h4 of the polymerase δ catalytic subunit gene, pold1. J. Biol. Chem. 2012, 287, 29801–29814. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Cho, S.J.; Jung, Y.S.; Chen, X. Dna polymerase η is regulated by poly(rc)-binding protein 1 via mrna stability. Biochem. J. 2014, 464, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Czochor, J.R.; Sulkowski, P.; Glazer, P.M. Mir-155 overexpression promotes genomic instability by reducing high-fidelity polymerase delta expression and activating error-prone dsb repair. Mol. Cancer Res. 2016, 14, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, J.W.; Calses, P.; Kemp, C.J.; Taniguchi, T. Mir-96 downregulates rev1 and rad51 to promote cellular sensitivity to cisplatin and parp inhibition. Cancer Res. 2012, 72, 4037–4046. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Jiang, Z.; Li, X.; Wang, X.I.; Xiao, Y. Mir-20b downregulates polymerases κ and θ in xp-v tumor cells. Oncol. Lett. 2016, 11, 3790–3794. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Han, C.; Zhao, R.; Cui, T.; Dai, Y.; Mao, C.; Zhao, W.; Zhang, X.; Yu, J.; Wang, Q.E. Enhanced expression of dna polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc. Natl. Acad. Sci. USA 2015, 112, 4411–4416. [Google Scholar] [CrossRef] [PubMed]
- Voitenleitner, C.; Rehfuess, C.; Hilmes, M.; O'Rear, L.; Liao, P.C.; Gage, D.A.; Ott, R.; Nasheuer, H.P.; Fanning, E. Cell cycle-dependent regulation of human dna polymerase alpha-primase activity by phosphorylation. Mol. Cell. Biol. 1999, 19, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Nasheuer, H.P.; Moore, A.; Wahl, A.F.; Wang, T.S. Cell cycle-dependent phosphorylation of human dna polymerase alpha. J. Biol. Chem. 1991, 266, 7893–7903. [Google Scholar] [PubMed]
- Zhou, Y.; Meng, X.; Zhang, S.; Lee, E.Y.; Lee, M.Y. Characterization of human dna polymerase delta and its subassemblies reconstituted by expression in the multibac system. PLoS ONE 2012, 7, e39156. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.W.T.; Zhang, S.; Lin, S.H.S.; Chea, J.; Wang, X.; LeRoy, C.; Wong, A.; Zhang, Z.; Lee, E.Y.C. Regulation of human DNA polymerase delta in the cellular responses to DNA damage. Environ. Mol. Mutagen. 2012, 53, 683–698. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.R.; Hao, H.; Jiang, Y.; Lee, M.Y. Regulation of human dna polymerase delta during the cell cycle. J. Biol. Chem. 1994, 269, 24027–24033. [Google Scholar] [PubMed]
- Li, H.; Xie, B.; Rahmeh, A.; Zhou, Y.; Lee, M.Y. Direct interaction of p21 with p50, the small subunit of human dna polymerase delta. Cell Cycle 2006, 5, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Ducoux, M.; Urbach, S.; Baldacci, G.; Hubscher, U.; Koundrioukoff, S.; Christensen, J.; Hughes, P. Mediation of proliferating cell nuclear antigen (pcna)-dependent dna replication through a conserved p21(cip1)-like pcna-binding motif present in the third subunit of human dna polymerase delta. J. Biol. Chem. 2001, 276, 49258–49266. [Google Scholar] [CrossRef] [PubMed]
- Lemmens, L.; Urbach, S.; Prudent, R.; Cochet, C.; Baldacci, G.; Hughes, P. Phosphorylation of the c subunit (p66) of human dna polymerase delta. Biochem. Biophys. Res. Commun. 2008, 367, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Rahmeh, A.A.; Zhou, Y.; Xie, B.; Li, H.; Lee, E.Y.C.; Lee, M.Y.W.T. Phosphorylation of the p68 subunit of poli´ acts as a molecular switch to regulate its interaction with pcna. Biochemistry 2011, 51, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhou, Y.; Xie, B.; Zhang, S.; Rahmeh, A.; Huang, H.-s.; Lee, M.Y.W.T.; Lee, E.Y.C. Protein phosphatase-1 is targeted to dna polymerase δ via an interaction with the p68 subunit†. Biochemistry 2008, 47, 11367–11376. [Google Scholar]
- Zhao, H.; Zhang, S.; Xu, D.; Lee, M.Y.; Zhang, Z.; Lee, E.Y.; Darzynkiewicz, Z. Expression of the p12 subunit of human dna polymerase δ (pol δ), cdk inhibitor p21(waf1), cdt1, cyclin a, pcna and ki-67 in relation to dna replication in individual cells. Cell Cycle 2014, 13, 3529–3540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, Y.; Sarkeshik, A.; Yates, J.R.; Thomson, T.M.; Zhang, Z.; Lee, E.Y.; Lee, M.Y. Identification of rnf8 as a ubiquitin ligase involved in targeting the p12 subunit of dna polymerase δ for degradation in response to dna damage. J. Biol. Chem. 2013, 288, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Zhang, S.; Lin, S.H.; Wang, X.; Darzynkiewicz, Z.; Zhang, Z.; Lee, E.Y. The tail that wags the dog: P12, the smallest subunit of DNA polymerase δ, is degraded by ubiquitin ligases in response to DNA damage and during cell cycle progression. Cell Cycle 2014, 13, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T.; Dutta, A. Crl4cdt2: Master coordinator of cell cycle progression and genome stability. Cell Cycle 2011, 10, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.H.; Wang, X.; Zhang, S.; Zhang, Z.; Lee, E.Y.; Lee, M.Y. Dynamics of enzymatic interactions during short flap human okazaki fragment processing by two forms of human dna polymerase δ. DNA Repair (Amst.) 2013, 12, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, Z.; Liu, Y.; Hickey, R.J.; Malkas, L.H. Altered dna polymerase iota expression in breast cancer cells leads to a reduction in dna replication fidelity and a higher rate of mutagenesis. Cancer Res. 2004, 64, 5597–5607. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Xu, Z.; Yang, M.; Wei, Q.; Zhang, Y.; Yu, J.; Zhi, Y.; Liu, Y.; Chen, Z.; Yang, J. Overexpressed DNA polymerase iota regulated by jnk/c-jun contributes to hypermutagenesis in bladder cancer. PLoS ONE 2013, 8, e69317. [Google Scholar] [CrossRef] [PubMed]
- Kannouche, P.; Fernández de Henestrosa, A.R.; Coull, B.; Vidal, A.E.; Gray, C.; Zicha, D.; Woodgate, R.; Lehmann, A.R. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2003, 22, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhou, G.; Zhang, W.; Song, Y.; Bian, Z. A novel polh mutation causes xp-v disease and xp-v tumor proneness may involve imbalance of numerous dna polymerases. Oncol. Lett. 2013, 6, 1583–1590. [Google Scholar] [PubMed]
- Jung, Y.S.; Liu, G.; Chen, X. Pirh2 e3 ubiquitin ligase targets DNA polymerase eta for 20s proteasomal degradation. Mol. Cell. Biol. 2010, 30, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Chen, Z.; Wang, S.; Wang, H.W.; Qiu, W.; Zhao, L.; Xu, R.; Luo, H.; Chen, Y.; Chen, D.; et al. The error-prone DNA polymerase κ promotes temozolomide resistance in glioblastoma through rad17-dependent activation of atr-chk1 signaling. Cancer Res. 2016, 76, 2340–2353. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Cleaver, J.E.; Hatahet, Z.; Honkanen, R.E.; Chang, J.Y.; Yen, Y.; Chou, K.M. Human DNA polymerase eta activity and translocation is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 16578–16583. [Google Scholar] [CrossRef] [PubMed]
- Göhler, T.; Sabbioneda, S.; Green, C.M.; Lehmann, A.R. ATR-mediated phosphorylation of DNA polymerase η is needed for efficient recovery from UV damage. J. Cell Biol. 2011, 192, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; You, C.; Wang, Y. The functions of serine 687 phosphorylation of human DNA polymerase η in UV damage tolerance. Mol. Cell. Proteom. 2016, 15, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Bienko, M.; Green, C.M.; Sabbioneda, S.; Crosetto, N.; Matic, I.; Hibbert, R.G.; Begovic, T.; Niimi, A.; Mann, M.; Lehmann, A.R.; et al. Regulation of translesion synthesis dna polymerase eta by monoubiquitination. Mol. Cell 2010, 37, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.S.; Hakem, A.; Hakem, R.; Chen, X. Pirh2 e3 ubiquitin ligase monoubiquitinates DNA polymerase eta to suppress translesion DNA synthesis. Mol. Cell. Biol. 2011, 31, 3997–4006. [Google Scholar] [CrossRef] [PubMed]
- Bienko, M.; Green, C.M.; Crosetto, N.; Rudolf, F.; Zapart, G.; Coull, B.; Kannouche, P.; Wider, G.; Peter, M.; Lehmann, A.R.; et al. Ubiquitin-binding domains in y-family polymerases regulate translesion synthesis. Science 2005, 310, 1821–1824. [Google Scholar] [CrossRef] [PubMed]
- Plosky, B.S.; Vidal, A.E.; Fernández de Henestrosa, A.R.; McLenigan, M.P.; McDonald, J.P.; Mead, S.; Woodgate, R. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 2006, 25, 2847–2855. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Tang, T.S.; Bienko, M.; Dikic, I.; Friedberg, E.C. Requirements for the interaction of mouse polkappa with ubiquitin and its biological significance. J. Biol. Chem. 2008, 283, 4658–4664. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Tang, T.S.; Bienko, M.; Parker, J.L.; Bielen, A.B.; Sonoda, E.; Takeda, S.; Ulrich, H.D.; Dikic, I.; Friedberg, E.C. Ubiquitin-binding motifs in rev1 protein are required for its role in the tolerance of DNA damage. Mol. Cell. Biol. 2006, 26, 8892–8900. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yang, K.; Dejsuphong, D.; D'Andrea, A.D. Regulation of rev1 by the fanconi anemia core complex. Nat. Struct. Mol. Biol. 2012, 19, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Wallace, H.A.; Merkle, J.A.; Yu, M.C.; Berg, T.G.; Lee, E.; Bosco, G.; Lee, L.A. Trip/nopo e3 ubiquitin ligase promotes ubiquitylation of dna polymerase η. Development 2014, 141, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Warbrick, E. The p66 and p12 subunits of dna polymerase delta are modified by ubiquitin and ubiquitin-like proteins. Biochem. Biophys. Res. Commun. 2006, 349, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Bursomanno, S.; Beli, P.; Khan, A.M.; Minocherhomji, S.; Wagner, S.A.; Bekker-Jensen, S.; Mailand, N.; Choudhary, C.; Hickson, I.D.; Liu, Y. Proteome-wide analysis of sumo2 targets in response to pathological dna replication stress in human cells. DNA Repair (Amst.) 2015, 25, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.S.; Qian, Y.; Chen, X. Dna polymerase eta is targeted by mdm2 for polyubiquitination and proteasomal degradation in response to ultraviolet irradiation. DNA Repair (Amst.) 2012, 11, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Pentz, K.; Zhu, Q.; Wang, Q.; He, J.; Srivastava, A.K.; Wani, A.A. Usp7 modulates uv-induced pcna monoubiquitination by regulating dna polymerase eta stability. Oncogene 2015, 34, 4791–4796. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Michael, W.M. Regulated proteolysis of dna polymerase eta during the DNA-damage response in c. Elegans. Mol. Cell 2008, 32, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Roerink, S.F.; Koole, W.; Stapel, L.C.; Romeijn, R.J.; Tijsterman, M. A broad requirement for tls polymerases η and κ, and interacting sumoylation and nuclear pore proteins, in lesion bypass during c. Elegans embryogenesis. PLoS Genet. 2012, 8, e1002800. [Google Scholar] [CrossRef] [PubMed]
- Despras, E.; Sittewelle, M.; Pouvelle, C.; Delrieu, N.; Cordonnier, A.M.; Kannouche, P.L. Rad18-dependent sumoylation of human specialized DNA polymerase eta is required to prevent under-replicated dna. Nat. Commun. 2016, 7, 13326. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, I.A.; D'Souza, R.C.; Yang, B.; Verlaan-de Vries, M.; Mann, M.; Vertegaal, A.C. Uncovering global sumoylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014, 21, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.S.; Wei, M.; Brandhorst, S.; Longo, V.D. Starvation promotes rev1 sumoylation and p53-dependent sensitization of melanoma and breast cancer cells. Cancer Res. 2015, 75, 1056–1067. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, H.; Darzynkiewicz, Z.; Zhou, P.; Zhang, Z.; Lee, E.Y.; Lee, M.Y. A novel function of crl4(cdt2): Regulation of the subunit structure of dna polymerase δ in response to dna damage and during the s phase. J. Biol. Chem. 2013, 288, 29550–29561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, Y.; Trusa, S.; Meng, X.; Lee, E.Y.; Lee, M.Y. A novel DNA damage response: Rapid degradation of the p12 subunit of dna polymerase delta. J. Biol. Chem. 2007, 282, 15330–15340. [Google Scholar] [CrossRef] [PubMed]
- Chea, J.; Zhang, S.; Zhao, H.; Zhang, Z.; Lee, E.Y.; Darzynkiewicz, Z.; Lee, M.Y. Spatiotemporal recruitment of human DNA polymerase delta to sites of uv damage. Cell Cycle 2012, 11, 2885–2895. [Google Scholar] [CrossRef] [PubMed]
- Pozo, F.M.; Oda, T.; Sekimoto, T.; Murakumo, Y.; Masutani, C.; Hanaoka, F.; Yamashita, T. Molecular chaperone hsp90 regulates rev1-mediated mutagenesis. Mol. Cell. Biol. 2011, 31, 3396–3409. [Google Scholar] [CrossRef] [PubMed]
- Sekimoto, T.; Oda, T.; Pozo, F.M.; Murakumo, Y.; Masutani, C.; Hanaoka, F.; Yamashita, T. The molecular chaperone hsp90 regulates accumulation of dna polymerase eta at replication stalling sites in uv-irradiated cells. Mol. Cell 2010, 37, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Baldeck, N.; Janel-Bintz, R.; Wagner, J.; Tissier, A.; Fuchs, R.P.; Burkovics, P.; Haracska, L.; Despras, E.; Bichara, M.; Chatton, B.; et al. Ff483-484 motif of human polη mediates its interaction with the pold2 subunit of polδ and contributes to dna damage tolerance. Nucleic Acids Res. 2015, 43, 2116–2125. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Kanao, R.; Kaji, K.; Ohmori, H.; Hanaoka, F.; Masutani, C. Different types of interaction between pcna and pip boxes contribute to distinct cellular functions of y-family dna polymerases. Nucleic Acids Res. 2015, 43, 7898–7910. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, E.; Murakumo, Y.; Kanjo, N.; Akagi, J.; Masutani, C.; Hanaoka, F.; Ohmori, H. Interaction of hrev1 with three human y-family dna polymerases. Genes Cells 2004, 9, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Park, J.; Conde, J.; Wakamiya, M.; Prakash, L.; Prakash, S. Rev1 promotes replication through uv lesions in conjunction with dna polymerases η, ι, and κ but not DNA polymerase ζ. Genes Dev. 2015, 29, 2588–2602. [Google Scholar] [PubMed]
- Pustovalova, Y.; Bezsonova, I.; Korzhnev, D.M. The c-terminal domain of human rev1 contains independent binding sites for DNA polymerase η and rev7 subunit of polymerase ζ. FEBS Lett. 2012, 586, 3051–3056. [Google Scholar] [CrossRef] [PubMed]
- Wojtaszek, J.; Lee, C.J.; D'Souza, S.; Minesinger, B.; Kim, H.; D'Andrea, A.D.; Walker, G.C.; Zhou, P. Structural basis of rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of rev1, heterodimeric polymerase (pol) ζ, and pol κ. J. Biol. Chem. 2012, 287, 33836–33846. [Google Scholar] [CrossRef] [PubMed]
- Pustovalova, Y.; Magalhães, M.T.; D'Souza, S.; Rizzo, A.A.; Korza, G.; Walker, G.C.; Korzhnev, D.M. Interaction between the rev1 c-terminal domain and the pold3 subunit of polζ suggests a mechanism of polymerase exchange upon rev1/polζ-dependent translesion synthesis. Biochemistry 2016, 55, 2043–2053. [Google Scholar] [CrossRef] [PubMed]
- Kosarek, J.N.; Woodruff, R.V.; Rivera-Begeman, A.; Guo, C.; D'Souza, S.; Koonin, E.V.; Walker, G.C.; Friedberg, E.C. Comparative analysis of in vivo interactions between rev1 protein and other y-family dna polymerases in animals and yeasts. DNA Repair (Amst.) 2008, 7, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Tissier, A.; Janel-Bintz, R.; Coulon, S.; Klaile, E.; Kannouche, P.; Fuchs, R.P.; Cordonnier, A.M. Crosstalk between replicative and translesional DNA polymerases: Pdip38 interacts directly with poleta. DNA Repair (Amst.) 2010, 9, 922–928. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Rodriguez-Belmonte, E.M.; Mazloum, N.; Xie, B.; Lee, M.Y. Identification of a novel protein, pdip38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen. J. Biol. Chem. 2003, 278, 10041–10047. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Machida, Y.; Vashisht, A.A.; Wohlschlegel, J.A.; Pang, Y.P.; Machida, Y.J. Regulation of error-prone translesion synthesis by spartan/c1orf124. Nucleic Acids Res. 2013, 41, 1661–1668. [Google Scholar] [CrossRef] [PubMed]
- Centore, R.C.; Yazinski, S.A.; Tse, A.; Zou, L. Spartan/c1orf124, a reader of pcna ubiquitylation and a regulator of uv-induced dna damage response. Mol. Cell 2012, 46, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, G.; Leung, J.W.; Nair, B.C.; Fong, K.W.; Chen, J. Proliferating cell nuclear antigen (pcna)-binding protein c1orf124 is a regulator of translesion synthesis. J. Biol. Chem. 2012, 287, 34225–34233. [Google Scholar] [CrossRef] [PubMed]
- Juhasz, S.; Balogh, D.; Hajdu, I.; Burkovics, P.; Villamil, M.A.; Zhuang, Z.; Haracska, L. Characterization of human spartan/c1orf124, an ubiquitin-pcna interacting regulator of DNA damage tolerance. Nucleic Acids Res. 2012, 40, 10795–10808. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.J.; Lachaud, C.; Appleton, P.; Macartney, T.J.; Näthke, I.; Rouse, J. Dvc1 (c1orf124) recruits the p97 protein segregase to sites of dna damage. Nat. Struct. Mol. Biol. 2012, 19, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Machida, Y.; Kim, M.S.; Machida, Y.J. Spartan/c1orf124 is important to prevent uv-induced mutagenesis. Cell Cycle 2012, 11, 3395–3402. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Dudimah, F.D.; Zhang, J.; Pickering, A.; Paneerselvam, J.; Palrasu, M.; Wang, H.; Fei, P. Recruitment of DNA polymerase eta by fancd2 in the early response to dna damage. Cell Cycle 2013, 12, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Bosques, L.; Sung, P.; Kupfer, G.M. A novel role for non-ubiquitinated fancd2 in response to hydroxyurea-induced dna damage. Oncogene 2016, 35, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, Z.; Wang, F.; Temviriyanukul, P.; Ma, X.; Tu, Y.; Lv, L.; Lin, Y.F.; Huang, M.; Zhang, T.; et al. Fancd2 and rev1 cooperate in the protection of nascent dna strands in response to replication stress. Nucleic Acids Res. 2015, 43, 8325–8339. [Google Scholar] [CrossRef] [PubMed]
- Mirchandani, K.D.; McCaffrey, R.M.; D'Andrea, A.D. The fanconi anemia core complex is required for efficient point mutagenesis and rev1 foci assembly. DNA Repair (Amst.) 2008, 7, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Renaud, E.; Rosselli, F. Fanc pathway promotes uv-induced stalled replication forks recovery by acting both upstream and downstream polη and rev1. PLoS ONE 2013, 8, e53693. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Sharma, S.; Zou, J.; Lin, S.Y.; Wang, B.; Rezvani, K.; Wang, H.; Parvin, J.D.; Ludwig, T.; Canman, C.E.; et al. Brca1 promotes the ubiquitination of pcna and recruitment of translesion polymerases in response to replication blockade. Proc. Natl. Acad. Sci. USA 2013, 110, 13558–13563. [Google Scholar] [CrossRef] [PubMed]
- Buisson, R.; Niraj, J.; Pauty, J.; Maity, R.; Zhao, W.; Coulombe, Y.; Sung, P.; Masson, J.Y. Breast cancer proteins palb2 and brca2 stimulate polymerase η in recombination-associated dna synthesis at blocked replication forks. Cell Rep. 2014, 6, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Mansilla, S.F.; Soria, G.; Vallerga, M.B.; Habif, M.; Martínez-López, W.; Prives, C.; Gottifredi, V. UV-triggered p21 degradation facilitates damaged-dna replication and preserves genomic stability. Nucleic Acids Res. 2013, 41, 6942–6951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baranovskiy, A.G.; Lada, A.G.; Siebler, H.M.; Zhang, Y.; Pavlov, Y.I.; Tahirov, T.H. Dna polymerase δ and ζ switch by sharing accessory subunits of dna polymerase δ. J. Biol. Chem. 2012, 287, 17281–17287. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Gregory, M.T.; Yang, W. Human pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements pol η in cisplatin bypass. Proc. Natl. Acad. Sci. USA 2014, 111, 2954–2959. [Google Scholar] [CrossRef] [PubMed]
- Janel-Bintz, R.; Wagner, J.; Haracska, L.; Mah-Becherel, M.C.; Bichara, M.; Fuchs, R.P.; Cordonnier, A.M. Evidence for a rad18-independent frameshift mutagenesis pathway in human cell-free extracts. PLoS ONE 2012, 7, e36004. [Google Scholar] [CrossRef] [PubMed]
- Hendel, A.; Krijger, P.H.; Diamant, N.; Goren, Z.; Langerak, P.; Kim, J.; Reissner, T.; Lee, K.Y.; Geacintov, N.E.; Carell, T.; et al. Pcna ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011, 7, e1002262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabbioneda, S.; Gourdin, A.M.; Green, C.M.; Zotter, A.; Giglia-Mari, G.; Houtsmuller, A.; Vermeulen, W.; Lehmann, A.R. Effect of proliferating cell nuclear antigen ubiquitination and chromatin structure on the dynamic properties of the y-family dna polymerases. Mol. Biol. Cell 2008, 19, 5193–5202. [Google Scholar] [CrossRef] [PubMed]
- Sebesta, M.; Burkovics, P.; Juhasz, S.; Zhang, S.; Szabo, J.E.; Lee, M.Y.; Haracska, L.; Krejci, L. Role of pcna and tls polymerases in d-loop extension during homologous recombination in humans. DNA Repair (Amst.) 2013, 12, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.M.; Vaisman, A.; Martomo, S.A.; Sullivan, P.; Lan, L.; Hanaoka, F.; Yasui, A.; Woodgate, R.; Gearhart, P.J. Msh2-msh6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J. Exp. Med. 2005, 201, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Wang, F.; Ma, X.; Yang, Y.; Wang, Z.; Liu, H.; Li, X.; Liu, Z.; Zhang, T.; Huang, M.; et al. Mismatch repair protein msh2 regulates translesion dna synthesis following exposure of cells to uv radiation. Nucleic Acids Res. 2013, 41, 10312–10322. [Google Scholar] [CrossRef] [PubMed]

| Polymerase | Gene/ Subunit | Cellular Functions | References |
|---|---|---|---|
| Replicative | (B-family) | ||
| Alpha (α) | POLA1-A2 p180, p70 | Replication: initiator DNA synthesis | [23] |
| Checkpoint signaling | [24,25] | ||
| Delta (δ) | POLD1-D4 p125, p50, p66, p12 | Replication: Lagging strand; late S/G2 | [4,26] |
| DNA repair synthesis (BER, NER, MMR) | [27,28,29] | ||
| Checkpoint Signaling | [24] | ||
| Epsilon (ε) | POLE1-E4 p261, p59, p17, p12 | Replication: Leading strand | [4,26] |
| DNA repair synthesis (NER) | [30] | ||
| Checkpoint signaling | [24] | ||
| Specialized | (B-family) | ||
| Zeta (ζ) | REV3L | Translesion synthesis Common Fragile Site stability | [1,13] |
| REV7 | |||
| Specialized | (Y-family) | ||
| Eta (η) | POLH | Translesion synthesis | [3] |
| Common Fragile Site stability | [14] | ||
| DNA repair synthesis (MMR; HR) | [31,32] | ||
| Somatic Hypermutation | [33] | ||
| Kappa (κ) | POLK | Translesion synthesis | [3] |
| G4 and Microsatellite DNA synthesis | [34] | ||
| DNA repair synthesis (NER, DSBR) | [30,35,36] | ||
| ATR signaling | [37] | ||
| Iota (ι) | POLI | Translesion synthesis | [3] |
| Somatic Hypermutation | [38] | ||
| Rev1 | REV1 | Translesion synthesis | [3] |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barnes, R.; Eckert, K. Maintenance of Genome Integrity: How Mammalian Cells Orchestrate Genome Duplication by Coordinating Replicative and Specialized DNA Polymerases. Genes 2017, 8, 19. https://doi.org/10.3390/genes8010019
Barnes R, Eckert K. Maintenance of Genome Integrity: How Mammalian Cells Orchestrate Genome Duplication by Coordinating Replicative and Specialized DNA Polymerases. Genes. 2017; 8(1):19. https://doi.org/10.3390/genes8010019
Chicago/Turabian StyleBarnes, Ryan, and Kristin Eckert. 2017. "Maintenance of Genome Integrity: How Mammalian Cells Orchestrate Genome Duplication by Coordinating Replicative and Specialized DNA Polymerases" Genes 8, no. 1: 19. https://doi.org/10.3390/genes8010019





