Remodeling and Control of Homologous Recombination by DNA Helicases and Translocases that Target Recombinases and Synapsis
Abstract
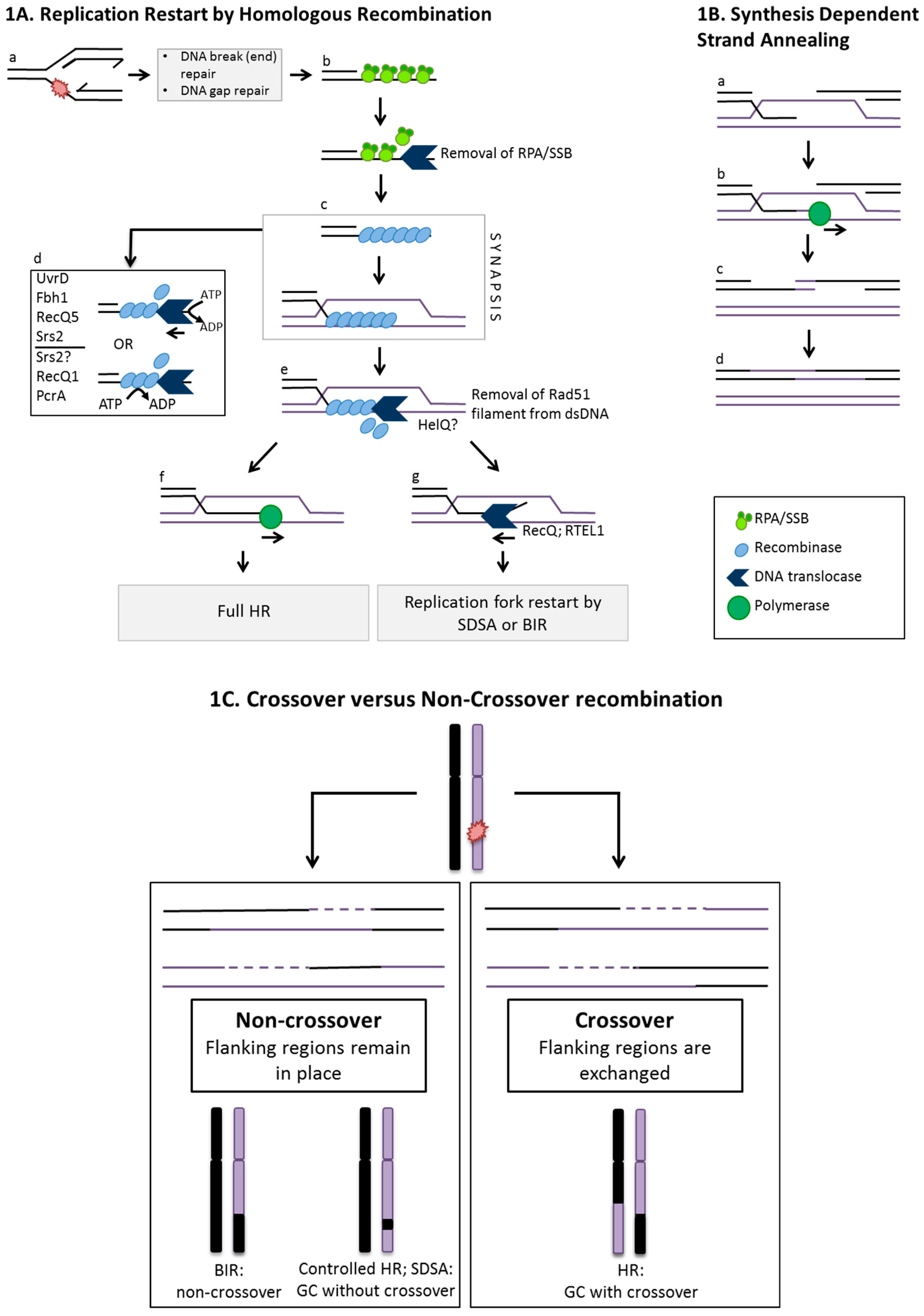
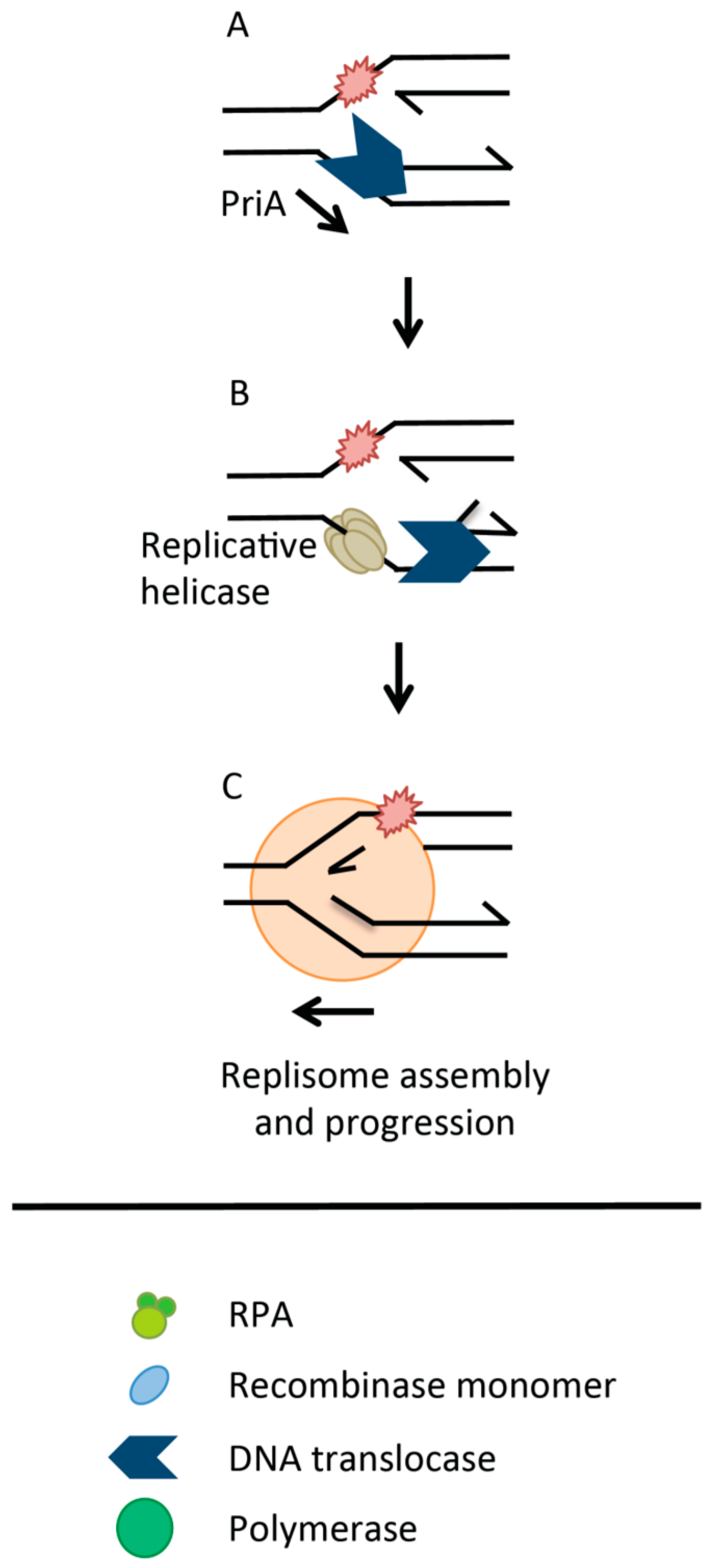
:1. Homologous Recombination Linked to DNA Replication
2. Controlling Homologous Recombination Choices at D-loops
3. Recombination Initiated from ssDNA: Nucleoprotein Filaments and D-loops
4. Disrupting Pre-synaptic Nucleoprotein Filaments: PcrA, UvrD and Fbh1
5. Disrupting Synapsis and D-loops: RecQ and RTEL1 Helicases
6. HelQ and Hel308 Helicases: Mediator and Negative Regulator of Synapsis
7. Homologous Recombination and the Efficiency of CRISPR Genome Editing
Acknowledgements
Conflicts of Interest
References
- Cox, M.M.; Goodman, M.F.; Kreuzer, K.N.; Sherratt, D.J.; Sandler, S.J.; Marians, K.J. The importance of repairing stalled replication forks. Nature 2000, 404, 37–41. [Google Scholar] [PubMed]
- West, S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003, 4, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Spies, A.M.; Kowalczykowski, S.C. Homologous recombination by the RecBCD and RecFOR pathways. In The Bacterial Chromosome; Patrick, H.N., Ed.; ASM Press: Washington, DC, USA, 2005; pp. 398–403. [Google Scholar]
- Cortez, D. Unwind and Slow down: Checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 2005, 19, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Bruning, J.G.; Howard, J.L.; McGlynn, P. Accessory replicative helicases and the replication of protein-bound DNA. J. Mol. Biol. 2014, 426, 3917–3928. [Google Scholar] [CrossRef] [PubMed]
- Heyer, W.D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Michel, B. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 2005, 3, e255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarbajna, S.; West, S.C. Holliday junction processing enzymes as guardians of genome stability. Trends Biochem. Sci. 2014, 39, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Jasin, M.; Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Greenberg, R.A. Noncanonical views of homology-directed DNA repair. Genes Dev. 2016, 30, 1138–1154. [Google Scholar] [CrossRef] [PubMed]
- Sale, J.E.; Lehmann, A.R.; Woodgate, R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012, 13, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.S.; Takata, K.; Wood, R.D. DNA polymerases and cancer. Nat. Rev. Cancer 2011, 11, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, S.; Bellani, M.A.; Thazhathveetil, A.K.; Ling, C.; de Winter, J.P.; Wang, Y.; Wang, W.; Seidman, M.M. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol. Cell 2013, 52, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Ose, T.; Okamoto, N.; Maenaka, K.; Tanaka, T.; Masai, H.; Saito, M.; Shirai, T.; Kohda, D. Structural basis of the 3′end recognition of a leading strand in stalled replication forks by PriA. EMBO J. 2007, 26, 2584–2593. [Google Scholar] [CrossRef] [PubMed]
- Heller, R.C.; Marians, K.J. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol. Cell 2005, 17, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Briggs, G.S.; Smits, W.K.; Soultanas, P. Chromosomal replication initiation machinery of low-G+C-content Firmicutes. J. Bacteriol. 2012, 194, 5162–5170. [Google Scholar] [CrossRef] [PubMed]
- Merrikh, H.; Machon, C.; Grainger, W.H.; Grossman, A.D.; Soultanas, P. Co-directional replication-transcription conflicts lead to replication restart. Nature 2011, 470, 554–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolt, E.L.; Lloyd, R.G. Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell 2002, 10, 187–198. [Google Scholar] [CrossRef]
- Jaktaji, R.P.; Lloyd, R.G. PriA supports two distinct pathways for replication restart in UV-irradiated Escherichia coli cells. Mol. Micro. 2003, 47, 1091–1100. [Google Scholar] [CrossRef]
- Rudolph, C.J.; Upton, A.L.; Briggs, G.S.; Lloyd, R.G. Is RecG a general guardian of the bacterial genome? DNA Repair 2010, 9, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.G.; Rudolph, C.J. 25 years on and no end in sight: A perspective on the role of RecG protein. Curr. Genet. 2016. [Google Scholar] [CrossRef] [PubMed]
- Courcelle, J.; Hanawalt, P.C. Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc. Natl. Acad. Sci. USA 2001, 98, 8196–8202. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Kong, H.; Nei, M.; Ma, H. Origins and evolution of the recA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl. Acad. Sci. USA 2006, 103, 10328–10333. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.C.; Kowalczykowski, S.C. RecA: Regulation and mechanism of a molecular search engine. Trends Biochem. Sci. 2016, 41, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Suwaki, N.; Klare, K.; Tarsounas, M. Rad51 Paralogs: Roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin.Cell Dev. Biol. 2011, 22, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Singleton, M.R.; Dillingham, M.S.; Gaudier, M.; Kowalczykowski, S.C.; Wigley, D.B. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 2004, 432, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Dillingham, M.S.; Kowalczykowski, S.C. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008, 72, 642–671. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.T.; Stahl, M.M.; McMilin, K.D.; Stahl, F.W. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics 1974, 77, 425–433. [Google Scholar] [PubMed]
- Krajewski, W.W.; Fu, X.; Wilkinson, M.; Cronin, N.B.; Dillingham, M.S.; Wigley, D.B. Structural basis for translocation by AddAB helicase-nuclease and its arrest at chi sites. Nature 2014, 508, 416–419. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.R. How RecBCD enzyme and Chi promote DNA break repair and recombination: A molecular biologist’s view. Microbiol. Mol. Biol. Rev. 2012, 76, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.A.; Kowalczykowski, S.C. The recombination hotspot C is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell 1993, 73, 87–96. [Google Scholar] [CrossRef]
- Anderson, D.G.; Kowalczykowski, S.C. The recombination hot spot Chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997, 11, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Churchill, J.J.; Anderson, D.G.; Kowalczykowski, S.C. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of chi, resulting in constitutive recombination activation. Genes Dev. 1999, 13, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, A.M.; Smith, G.R. Escherichia coli recBC deletion mutants. J. Bacteriol. 1984, 160, 788–791. [Google Scholar] [PubMed]
- Handa, N.; Morimatsu, K.; Lovett, S.T.; Kowalczykowski, S.C. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 2009, 23, 1234–1245. [Google Scholar] [CrossRef] [PubMed]
- Morimatsu, K.; Kowalczykowski, S.C. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc. Natl. Acad. Sci. USA 2014, 111, E5133–E5142. [Google Scholar] [CrossRef] [PubMed]
- Persky, N.S.; Lovett, S.T. Mechanisms of recombination: lessons from E. coli. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 347–370. [Google Scholar] [CrossRef] [PubMed]
- Komori, K.; Miyata, T.; DiRuggiero, J.; Holley-Shanks, R.; Hayashi, I.; Cann, I.K.; Mayanagi, K.; Shinagawa, H.; Ishino, Y. Both RadA and RadB are involved in homologous recombination in Pyrococcus furiosus. J. Biol. Chem. 2000, 275, 33782–33790. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.R.; Spirek, M.; Chaurasiya, K.R.; Ward, J.D.; Carzaniga, R.; Yu, X.; Egelman, E.H.; Collinson, L.M.; Rueda, D.; Krejci, L.; et al. Rad51 Paralogs remodel pre-synaptic Rad51 filaments to stimulate homologous recombination. Cell 2015, 162, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Amunugama, R.; Groden, J.; Fishel, R. The HsRAD51B-HsRAD51C stabilizes the HsRAD51 nucleoprotein filament. DNA Repair 2013, 12, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Myong, S.; Niedziela-Majka, A.; Lee, K.S.; Yu, J.; Lohman, T.M.; Ha, T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell 2010, 142, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Fagerburg, M.V.; Schauer, G.D.; Thickman, K.R.; Bianco, P.R.; Khan, S.A.; Leuba, S.H.; Anand, S.P. PcrA-mediated disruption of RecA nucleoprotein filaments—essential role of the ATPase activity of RecA. Nucleic Acids Res. 2012, 40, 8416–8424. [Google Scholar] [CrossRef] [PubMed]
- Veaute, X.; Delmas, S.; Selva, M.; Jeusset, J.; Le Cam, E.; Matic, I.; Fabre, F.; Petit, M.A. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. Embo. J. 2005, 24, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Centore, R.C.; Sandler, S.J. UvrD limits the number and intensities of RecA-green fluorescent protein structures in Escherichia coli K-12. J. Bacteriol. 2007, 189, 2915–2920. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.J.; Sanchez, N.; Michel, B. A fork-clearing role for UvrD. Mol. Microbiol. 2005, 57, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Sasanuma, H.; Furihata, Y.; Shinohara, M.; Shinohara, A. Remodeling of the Rad51 DNA strand-exchange protein by the Srs2 helicase. Genetics 2013, 194, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Veaute, X.; Jeusset, J.; Soustelle, C.; Kowalczykowski, S.C.; Le Cam, E.; Fabre, F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 2003, 423, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Krejci, L.; van Komen, S.; Li, Y.; Villemain, J.; Reddy, M.S.; Klein, H.; Ellenberger, T.; Sung, P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 2003, 423, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Seong, C.; Colavito, S.; Kwon, Y.; Sung, P.; Krejci, L. Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. J. Biol. Chem. 2009, 284, 24363–24371. [Google Scholar] [CrossRef] [PubMed]
- Antony, E.; Tomko, E.J.; Xiao, Q.; Krejci, L.; Lohman, T.M.; Ellenberger, T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell 2009, 35, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Goodwin, A.; Hickson, I.D.; Norbury, C.J. Involvement of Schizosaccharomyces pombe Srs2 in cellular responses to DNA damage. Nucleic Acids Res. 2001, 29, 2963–2972. [Google Scholar] [CrossRef]
- Tsutsui, Y.; Kurokawa, Y.; Ito, K.; Siddique, M.S.; Kawano, Y.; Yamao, F.; Iwasaki, H. Multiple regulation of Rad51-mediated homologous recombination by fission yeast Fbh1. PLoS Genet. 2014, 10, e1004542. [Google Scholar] [CrossRef] [PubMed]
- Osman, F.; Dixon, J.; Barr, A.R.; Whitby, M.C. The F-box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol. Cell. Biol. 2005, 25, 8084–8096. [Google Scholar] [PubMed]
- Lorenz, A.; Osman, F.; Folkyte, V.; Sofueva, S.; Whitby, M.C. Fbh1 limits Rad51-dependent recombination at blocked replication forks. Mol. Cell. Biol. 2009, 29, 4742–4756. [Google Scholar] [CrossRef] [PubMed]
- Simandlova, J.; Zagelbaum, J.; Payne, M.J.; Chu, W.K.; Shevelev, I.; Hanada, K.; Chatterjee, S.; Reid, D.A.; Liu, Y.; Janscak, P.; et al. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J. Biol. Chem. 2013, 288, 34168–34180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kim, J.H.; Lee, S.H.; Kim, D.H.; Kang, H.Y.; Bae, S.H.; Pan, Z.Q.; Seo, Y.S. The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 2002, 277, 24530–24537. [Google Scholar] [CrossRef] [PubMed]
- Fugger, K.; Mistrik, M.; Neelsen, K.J.; Yao, Q.; Zellweger, R.; Kousholt, A.N.; Haahr, P.; Chu, W.K.; Bartek, J.; Lopes, M.; et al. FBH1 catalyzes regression of stalled replication forks. Cell Rep. 2015, 10, 1749–1757. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Lloyd, R.G. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 2000, 101, 35–45. [Google Scholar] [CrossRef]
- Chu, W.K.; Payne, M.J.; Beli, P.; Hanada, K.; Choudhary, C.; Hickson, I.D. FBH1 influences DNA replication fork stability and homologous recombination through ubiquitylation of RAD51. Nat. Commun. 2015. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Irino, N.; Nakayama, H. The recQ gene of Escherichia coli K12: Molecular cloning and isolation of insertion mutants. Mol. Gen. Genet. 1985, 200, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Croteau, D.L.; Popuri, V.; Opresko, P.L.; Bohr, V.A. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014, 83, 519–552. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.B.; Hickson, I.D. RecQ helicases: Conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 2013, 767, 161–184. [Google Scholar] [PubMed]
- Wu, L.; Davies, S.L.; Hickson, I.D. Roles of RecQ family helicases in the maintenance of genome stability. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Brosh, R.M., Jr. Distinct roles of RECQ1 in the maintenance of genomic stability. DNA Repair 2010, 9, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Schwendener, S.; Raynard, S.; Paliwal, S.; Cheng, A.; Kanagaraj, R.; Shevelev, I.; Stark, J.M.; Sung, P.; Janscak, P. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 2010, 285, 15739–15745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barber, L.J.; Youds, J.L.; Ward, J.D.; McIlwraith, M.J.; O’Neil, N.J.; Petalcorin, M.I.; Martin, J.S.; Collis, S.J.; Cantor, S.B.; Auclair, M.; et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 2008, 135, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Schertzer, M.; Wu, X.; Gertsenstein, M.; Selig, S.; Kammori, M.; Pourvali, R.; Poon, S.; Vulto, I.; Chavez, E.; et al. Regulation of murine telomere length by Rtel: An essential gene encoding a helicase-like protein. Cell 2004, 117, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Marini, F.; Wood, R.D. A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2002, 277, 8716–8723. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.D.; Muzzini, D.M.; Petalcorin, M.I.; Martinez-Perez, E.; Martin, J.S.; Plevani, P.; Cassata, G.; Marini, F.; Boulton, S.J. Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol. Cell 2010, 37, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.P.; Bolt, E.L. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 2005, 33, 3678–3690. [Google Scholar] [CrossRef] [PubMed]
- Fujikane, R.; Komori, K.; Shinagawa, H.; Ishino, Y. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biol. Chem. 2005, 280, 12351–12358. [Google Scholar] [CrossRef] [PubMed]
- Takata, K.; Reh, S.; Tomida, J.; Person, M.D.; Wood, R.D. Human DNA helicase HELQ participates in DNA interstrand crosslink tolerance with ATR and RAD51 paralogs. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.D.; Doublie, S. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair. 2016, 44, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Adelman, C.A.; Lolo, R.L.; Birkbak, N.J.; Murina, O.; Matsuzaki, K.; Horejsi, Z.; Parmar, K.; Borel, V.; Skehel, J.M.; Stamp, G.; et al. HELQ promotes RAD51 paralogue-dependent repair to avert germ cell loss and tumorigenesis. Nature 2013, 502, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Luebben, S.W.; Kawabata, T.; Akre, M.K.; Lee, W.L.; Johnson, C.S.; O’Sullivan, M.G.; Shima, N. Helq acts in parallel to Fancc to suppress replication-associated genome instability. Nucleic Acids Res. 2013, 41, 10283–10297. [Google Scholar] [CrossRef] [PubMed]
- Woodman, I.L.; Brammer, K.; Bolt, E.L. Physical interaction between archaeal DNA repair helicase Hel308 and Replication Protein A (RPA). DNA Repair 2011, 10, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Buechelmaier, E.S.; Powell, S.N. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol. Cell. Biol. 2013, 33, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Maeder, M.L.; Gersbach, C.A. Genome-editing technologies for gene and cell therapy. Mol. Ther. 2016, 24, 430–446. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014. [Google Scholar] [CrossRef] [PubMed]
- Ivancic-Bace, I.; Al Howard, J.; Bolt, E.L. Tuning in to interference: R-Loops and cascade complexes in CRISPR Immunity. J. Mol. Biol. 2012, 422, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Kogoma, T. D-loops and R-loops: Alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J. Bacteriol. 1994, 176, 1807–1812. [Google Scholar] [PubMed]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Dougan, S.K.; Truttmann, M.C.; Bilate, A.M.; Ingram, J.R.; Ploegh, H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015, 33, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Kan, Y.; Ruis, B.; Lin, S.; Hendrickson, E.A. The mechanism of gene targeting in human somatic cells. PLoS Genet. 2014, 10, e1004251. [Google Scholar] [CrossRef] [PubMed]
- Vriend, L.E.; Prakash, R.; Chen, C.C.; Vanoli, F.; Cavallo, F.; Zhang, Y.; Jasin, M.; Krawczyk, P.M. Distinct genetic control of homologous recombination repair of Cas9-induced double-strand breaks, nicks and paired nicks. Nucleic Acids Res. 2016, 44, 5204–5217. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Northall, S.J.; Ivančić-Baće, I.; Soultanas, P.; Bolt, E.L. Remodeling and Control of Homologous Recombination by DNA Helicases and Translocases that Target Recombinases and Synapsis. Genes 2016, 7, 52. https://doi.org/10.3390/genes7080052
Northall SJ, Ivančić-Baće I, Soultanas P, Bolt EL. Remodeling and Control of Homologous Recombination by DNA Helicases and Translocases that Target Recombinases and Synapsis. Genes. 2016; 7(8):52. https://doi.org/10.3390/genes7080052
Chicago/Turabian StyleNorthall, Sarah J., Ivana Ivančić-Baće, Panos Soultanas, and Edward L. Bolt. 2016. "Remodeling and Control of Homologous Recombination by DNA Helicases and Translocases that Target Recombinases and Synapsis" Genes 7, no. 8: 52. https://doi.org/10.3390/genes7080052






