1. Introduction
The insulin-like growth factor (IGF) signaling pathway has an important role in cell growth, differentiation, apoptosis regulation and tumor development [
1]. The IGF axis comprises two growth factors (IGF-I, IGF-II), two IGF receptors (IGF-IR, IGF-IIR), and seven IGF-binding proteins (IGFBPs) which regulate the mitogenic activities of the IGFs, a group of IGFBP-related proteins that bind IGFs with low affinity and IGFBP proteases. IGFBP5 is the most conserved binding protein of the IGFBP family in all vertebrates and is frequently dysregulated in human cancers.
IGFBP5 has numerous functional roles in carcinogenesis and it acts both IGF-dependent and independent mechanisms [
2]. There are many findings and assumptions about the role of IGFBP5 and it is very crucial to identify the role of IGFBP5 in cancer progression, especially in breast cancer. Some studies concluded that IGFBP5 acts as an oncogene, since the high protein level is linked to proliferation, metastasis, poor prognosis, drug sensitivity and limited response to endocrine treatment, but on the other hand some studies revealed that IGFBP5 acts as a tumor suppressor gene due to being related with apoptotic, anti-metastatic, and anti-migratory effects and good outcomes [
1].
Some studies indicated that IGFBP5 expression is high in tumors and considered IGFBP-5 as having pro-metastatic capacity [
3,
4]. Exogenous IGFBP5 has been shown to have a protective effect for ceramid-induced apoptosis [
5]. IGFBP5 accumulates in the cytoplasm and is related with bad prognoses in the breast cancer tissue [
6]. Our group previously reported that apoptotic and migratory potential of IGFBP5 depends on cellular localization which is regulated by nuclear localization signal into C-terminal domain of the protein in breast cancer cells [
7]. Overexpression of IGFBP5 has been found to be associated with poor outcomes of breast cancer patients [
8]. A recently published study indicated the important role of IGFBP5 in tumor progression in urothelial carcinoma and associated IGFBP5 over-expression with advanced tumor stage, frequent mitosis and poorer clinical outcomes [
9].
A recently published genome-wide association study (GWAS) shows that the 2p35 locus is an important risk factor for breast cancer [
10]. Furthermore, an SNP in the locus rs4442975 (G/T) has been reported to contribute to changes in IGFBP5 expression. The data suggested that the G-allele of rs4442975 refers to increased breast cancer susceptibility through reduced IGFBP5 expression. IGFBP-5 expression was also shown to be correlated with increased survival rate and to help maintaining tumor sensitivity to tamoxifen in breast cancer patients [
11]. The same study showed over-expression of IGFBP-5 in MCF-7 xenografts inhibited tumor development in mice. In other recent studies the tumor suppressant role of IGFBP-5 has been shown in melanoma cells [
12], osteosarcoma [
13] and ovarian cancer cells [
14]. IGFBP5 has been also reported to suppress cell growth and cause G2/M arrest in PANC-1 pancreatic cancer cells [
15]. Sureshbabu
et al. [
16] verified that IGFBP-5 increases epithelial cell adhesion to the extracellular matrix (ECM) in MCF-7 human breast cancer cells and at the same time inhibits migration by maintaining E-cadherin expression. Sureshbabu
et al. assume that this is how IGFBP5 plays a key role in preventing metastasis. One of the latest reports on the subject showed that over-expression of IGFBP5 levels inhibited the epithelial-mesenchymal transition (EMT) and decreased E-cadherin expression and the key stem cell markers NANOG, SOX2, OCT4, KLF4, and CD133 in human melanoma cell line [
12].
Besides the anti-apoptotic effects, IGFBP5 was shown to have stimulatory effects on apoptosis in different cancer types. Overexpression of IGFBP5 is known to inhibit IGF-I activation of IRS-1 (IR substrate-1), FKHRL-1 (forkhead in rhabdomyosarcoma-like 1) and protein kinase B. This indicates that IGFBP5 acts as an apoptotic factor by inhibiting the action of local IGF-I [
17]. Butt [
18]
et al. found that IGFBP5 activates caspase 8 and 9 MDA-MB-231 breast cancer cell line, which leads to apoptosis through Bcl-2 in the intrinsic apoptotic pathway. The apoptosis-inducing role of IGFBP5 has also been shown in prostate cells [
19] and ovarian cells [
20]. Recently it was reported that IGFBP5 intercedes neuronal apoptosis through the regulation of mitochondrial cytochrome c release and caspase 3 activation [
21].
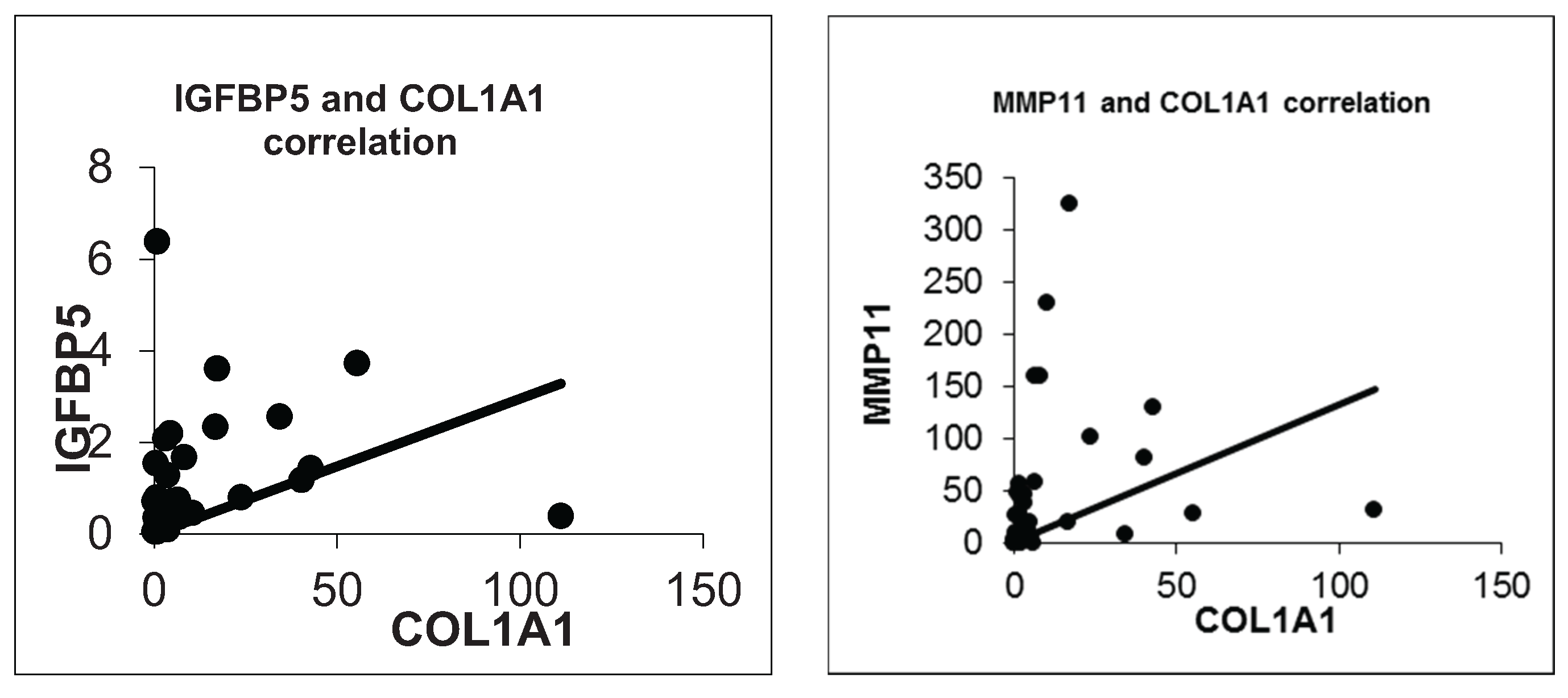
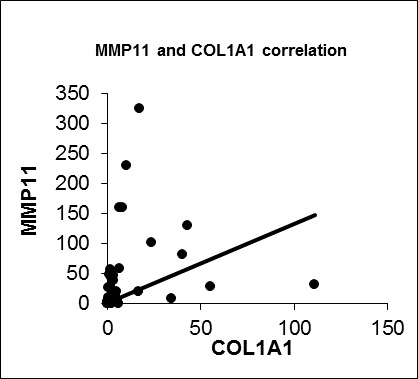
The functional and clinical meaning of expressional differences of IGFBP5 in tumor tissue is still controversial. To reveal differences in the gene expression profile between IGFBP5 overexpression in breast cancer tissues compared to matched normal breast tissue, a cDNA microarray experiment was conducted. Importantly, one of the top differentially expressed genes (DEGs), COL1A1, was validated by real time RT-qPCR and a positive correlation with IGFBP5 overexpression was found. In addition, there was one more positive correlation between the expression of COL1A1 and MMP11. Therefore, our results suggest that these two genes work together coordinately and contribute to breast cancer progression.
2. Experimental Section
2.1. Clinicopathological Parameters of Breast Cancer Patients
In total, 38 breast cancer patients, who were diagnosed at the Department of General Surgery, Marmara University School of Medicine from July 2010 to January 2012, were included in this study. Patients were between the ages of 38 and 73 and the median age was 59. Patients’ ages, histologic types, tumor grades, molecular subtypes, hormone statuses (Her2, ER, PR), proliferation marker Ki67 statuses, menopausal statuses, numbers of pregnancies, ages of menarche, tumor sizes and localizations have been recorded (
Table 1). Human epidermal growth factor receptor 2 (Her2), progesterone (PR), estrogen (ER) receptor and Ki67 statuses were evaluated by means of immunohistochemical staining with specific monoclonal antibody. Moreover, if the Her2 score was 2, fluorescence
in situ hybridization (FISH) was performed to clarify the Her2 status. Molecular subtypes were determined by using Goldhirsch classification [
22]. The study protocol was approved by the Local Ethics Committee of Marmara University (Istanbul, Turkey).
Table 1.
Differences of clinicopathological parameters depending on tumoral expression of IGFBP5 compared to adjacent normal tissue.
Table 1.
Differences of clinicopathological parameters depending on tumoral expression of IGFBP5 compared to adjacent normal tissue.
| Characteristics | Tumoral Expression of IGFBP5 Compared to Adjacent Normal Tissue | p-Value |
|---|
| | High (n = 21) | Low (n = 17) | |
| Age (years) | 55.66 | 55.52 | 0.9684 |
| Menarche age (years) | 12.95 | 14.18 | 0.0149 |
| Pregnancy (number) | 3.66 | 3.11 | 0.5876 |
| Menopausal state | | | |
| Pre | 7 | 3 | ns |
| Peri | 2 | 1 | |
| Post | 12 | 13 | |
| Tumor size (mm) | 26.71 | 23 | 0.3578 |
| Tumor localization | | | |
| Left | 10 | 13 | ns |
| Right | 10 | 4 | |
| Bilateral | 1 | 0 | |
| Estrogen receptor status | | | |
| Positive | 16 | 13 | ns |
| Negative | 5 | 4 | |
| Progesterone receptor status | | | |
| Positive | 14 | 13 | 0.7210 |
| Negative | 7 | 4 | |
| Her2 status | | | |
| Positive | 5 | 2 | 0.4267 |
| Negative | 16 | 15 | |
| Ki67 status | | | |
| Positive | 11 | 12 | 0.3264 |
| Negative | 10 | 5 | |
| Tumor grade | | | |
| 1 | 0 | 5 | 0.0123 |
| 2&3 | 21 | 12 | |
| Tumor histology | | | |
| IDC | 15 | 11 | ns |
| ILC | 1 | 0 | |
| IMC | 3 | 3 | |
| Others | 2 | 3 | |
| Molecular subtypes | | | |
| Luminal A | 6 | 5 | ns |
| Luminal B Her2(−) | 4 | 7 | |
| Luminal B Her2(+) | 6 | 1 | |
| Erb-B2 overexpression | 1 | 2 | |
| Basal-like | 4 | 2 | |
2.2. RNA Isolation and Quantification of IGFBP5 Expression in Breast Cancer Using qPCR
RNA was isolated from fresh frozen breast cancer and adjacent normal breast tissues from the 38 cases were isolated with High Pure RNA Tissue Kit according to instruction protocol (Roche, Mannheim, Germany). Transcriptor High Fidelity cDNA synthesis kit (Roche) was used for cDNA synthesis with 500 ng of total RNA in a reaction volume of 20 µL. Real time qRT-PCR was performed in 20 µL of final reaction volume with 5 µL of cDNA, 10 µL LightCycler 480 Probes Master mix, and 2 µL of primer-probe mix and 3 µL water to complete final volume. Reactions were carried out under the following cycling conditions: 5 min at 95 °C for pre-incubation, 10 s at 95 °C, 30 s 60 °C, 1 s 72 °C for amplification with 45 cycles, 10 s at 40 °C for cooling. All reactions were performed in duplicate for reference housekeeping gene, beta actin and IGFBP5 by using LightCycler 480 instrument. Relative quantification was calculated by delta-delta Ct method, subsequent to IGFBP5 expression normalization to beta actin expression.
2.3. Selection of Samples and Coding for Microarray
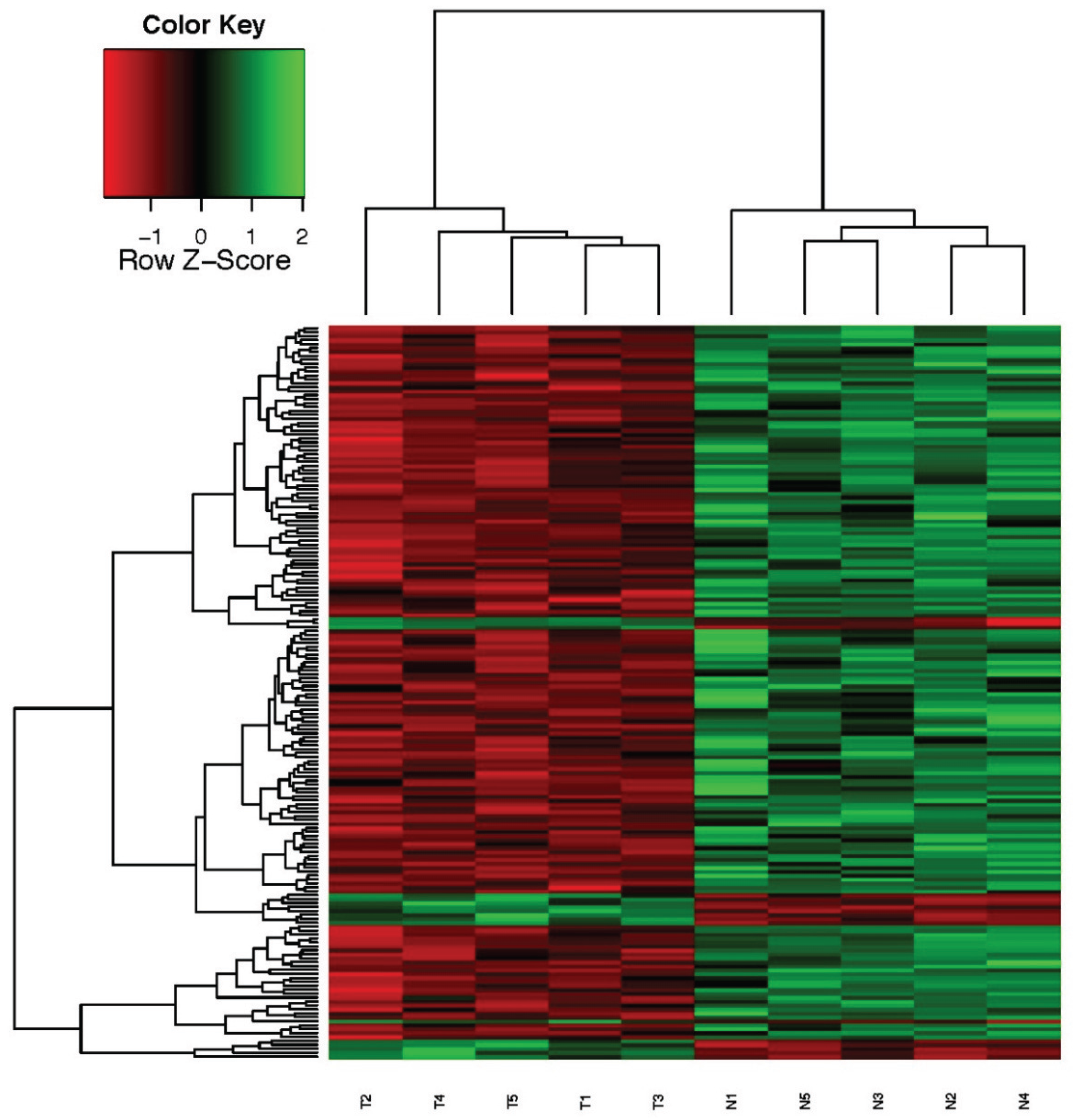
Of 38 samples for which we analyzed IGFBP5 expression level both in tumor and adjacent normal tissues, we selected those with the first 6 highest IGFBP5 expression in tumor samples compared to normal. T codes represent tumor tissues, N codes represent normal tissues. Microarray data analysis was excluded from one sample because it was found to be under the criteria for analysis. Finally, five tumor samples formed a group and five normal tissues formed another group. Examples belonging to the same group were evaluated as biological replicates. Tumor characteristics and fold changes of IGFBP5 expression in patients for use in microarray analysis are presented in
Table 2.
Table 2.
Tumor characteristics and fold changes of IGFBP5 expression in patients for use in microarray analysis.
Table 2.
Tumor characteristics and fold changes of IGFBP5 expression in patients for use in microarray analysis.
| Patients Code | Age | ER | PR | Her2 | Histology | Molecular Subtypes | Stage | Size (mm) | Fold Changes of IGFBP5 |
|---|
| T1 | 47 | 60% | 60% | (neg.) | ILC | lumA | T1N1 | 15 | 2.276 |
| T2 | 73 | 50% | 10% | (neg.) | IDC | lumA | T2N3 | 23 | 16.528 |
| T3 | 40 | 90% | 90% | (neg.) | IMC | lumB | T2N2 | 30 | 3.233 |
| T4 | 42 | 90% | 60% | (neg.) | IDC | lumA | T2N0 | 22 | 2.022 |
| T5 | 67 | 70% | 70% | (neg.) | IDC | lumA | T2N2 | 30 | 4.892 |
2.4. Gene Expression Profiling by Microarray and Validation with qPCR
RNA quality of samples was carried out with Agilent Bioanalyzer. Gene expression profiling of highly expressed IGFBP5 in tumor tissue compared to their matching adjacent normal tissue were performed by using a humanHT-12 v4 expression bead chip (Illumina, CA, USA). Each bead chip has more than 47,000 probes derived from the National Center for Biotechnology Information Reference Sequence (NCBI) RefSeq Release 38 (7 November 2009) and other sources. The microarray data was converted into electronic data by means of the GenomeStudio program. Signal intensity graphics of microarray data before normalization were generated for quality control purposes. The background noise was removed from each sample for comparative data analysis and data percentages of all samples (quantile) were subjected to normalization. Then, for use in the data analysis step, log transformation was conducted on the data that has been normalized. Hierarchical clustering analysis of the whole genome gene expression profiles of the samples was carried out employing a “Euclidean-average” approach.
Validation of microarray results was accomplished by performing qRT-PCR on COL1A1 and MMP11 genes in 37 patients. The expression levels of the target genes in each sample were calculated with a ∆∆Ct method. Beta actin was used as a housekeeping gene and the relative quantification was done by normalizing the cancer tissue data to its adjacent normal tissue data. All qRT-PCR experiments were performed in duplicate.
2.5. Pathways Analysis
Pathways analysis was performed using a hypergeometric test method; the p-value was been calculated and modified according to the Benjamin-Hochberg procedure. The adjusted p-value = 0.05 filter states the statistical significance that is used to determine the linked pathway, the condition that at least two genes must show different expressions.
2.6. Statistical Analysis
Student’s t tests (unpaired, two-tailed), Fisher’s exact probability test or the chi-square test were used to evaluate possible associations between the mRNA expression of IGFBP5 and various clinicopathological parameters. The data obtained after completion of the preliminary analysis used for the differential expression analysis was conducted to the data obtained after completion of the preliminary analysis. A SAM (Significance Analysis of Microarrays) method was used for determining the probe indicating expression of different genes. The analysis was conducted by comparing the probes’ expression levels in the normal tissue group and the tumor tissue group. FC-fold change and p-value for each probe were calculated statistically. After the statistical comparisons between the two groups, values of the probes that had FDR (False Discovery Rate) ≤ 0. 05 and FC (Fold Change) ≥ 1.5 were identified. Correlation between the mRNA expression levels were calculated using Spearman’s correlation. p-values ˂ 0.05 were considered statistically significant.
4. Discussion
IGFBP5 is one of the family members of IGF-binding proteins which play a critical role in tumor progression, especially cell survival, death and metastasis processes. The functional role of fluctuational IGFBP5 expression in origination and development of different types of cancer is still mysterious. IGFBP5 has diverse effects on growth of cancer cells depending on cell type and cell content. Because of the different functions of IGFBP5, a microarray system method, which provides a global picture in the cells, is the best method to clarify and discover new correlations with the genes. Using this method, we firstly determined potentially IGFBP5-related genes by using the microarray system in samples of breast cancer patients.
In this study, we identified that top differentially increased gene expressions with IGFBP5 are CST1, MMP11 and COL1A1. We focused on these three genes because of their associations and potential roles in extracellular matrix (ECM)-mediated pathways and metastasis process in cancer. Our previously published paper has shown the role of IGFBP5 on metastatic capacity of breast cancer [
7]. In the present study, KEGG pathway analysis reveals that focal adhesion and drug metabolism pathways are correlated with IGFBP5 expressional differentiation. These two pathways play a pivotal role in understanding cancer progression and development of treatment strategies considering IGFBP5.
Our microarray data shows that CST1 (cystatin SN) is one of the top DEGs in IGFBP5 overexpressed group. The general function of cystatins is the inhibition of the proteolytic activity of cysteine proteases which are involved in migration of cancer cells. Recent findings show that CST1 was highly expressed in colon, gastric and pancreatic cancers. CST1-overexpressing colon cancer cell lines exhibited increased tumor growth as well as metastasis in a xenograft nude mouse model [
23]. Overexpression of CST1 was also correlated with malignancy-associated proteins such as PCNA, cyclin D1, cyclin A2 and cyclin E in pancreatic cancer cell line [
24]. And finally, suppression of metastasis has been found to be correlated with reduced expression of CST1 in breast cancer cell line [
25]. We can suggest that metastatic potential of IGFBP5 might be correlated with CST1.
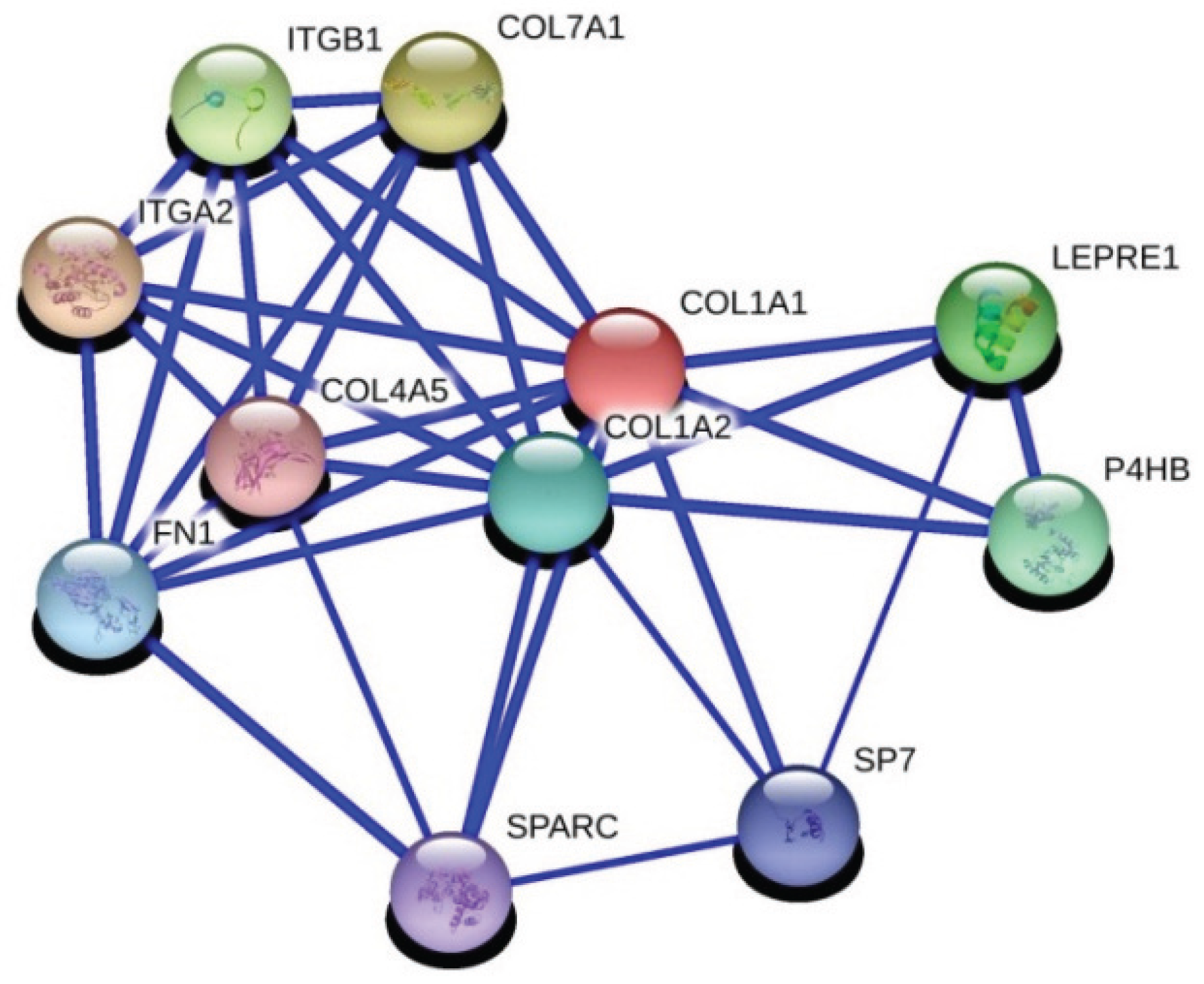
Extracellular matrix genes are critical players in tumor progression and metastasis. Here we identified an important correlation between IGFBP5 and two regulators of ECM-related pathways using gene co-expression analysis. The first is matrix metalloproteinase-11 (MMP11), which is related with distance metastasis [
26], resistance to anoikis [
27] and poor outcomes [
28] in breast cancer. The second is collagen 1a1 (COL1A1), which is an important regulator of pro-metastatic processes in cancer. Interactions with fibronectin and collagen 1a1 has been identified in breast cancer and related with adhesion of breast cancer in a previous study [
29]. We also know that IGFBP5 interacts with fibronectin and regulates cell migration [
30].
We also confirmed a positive correlation between IGFBP5 and COL1A1 expression (R = 0.476, p = 0.003). Besides that COL1A1 and MMP11 were significantly positively correlated (R = 0.602, p = 0.000). So we anticipate that IGFBP5 could be a possible modifier on metastatic capacity of breast cancer through regulating COL1A1 or MMP11. Further functional and protein-protein interactions studies are needed to clarify a likely correlation between IGFBP5, COL1A1 and MMP11 considering the metastasis process in breast cancer.
A recently published whole-genome analysis study identified a new breast cancer risk locus, 2q35. Fine scale mapping of the locus shows the strongest candidate for causality, SNP rs4442975, flanks a transcriptional enhancer that physically interacts with the promoter of IGFBP5. Furthermore, presence of this polymorphism reduced IGFBP5 expression [
10]. Whereas, we did not reach statistically significant difference between tumor and normal tissue samples from breast cancer patients considering IGFBP5 expression (
p = 0.2125), we found a positive correlation between higher expression of IGFBP5 and advanced stage of breast cancer. Further studies with a large sample size are needed to clarify tumoral expression of IGFBP5 and its clinical significance in breast cancer.