Re-Programming Photosynthetic Cells of Perennial Ryegrass (Lolium perenne L) for Fructan Biosynthesis through Transgenic Expression of Fructan Biosynthetic Genes under the Control of Photosynthetic Promoters
Abstract
:1. Introduction
2. Results
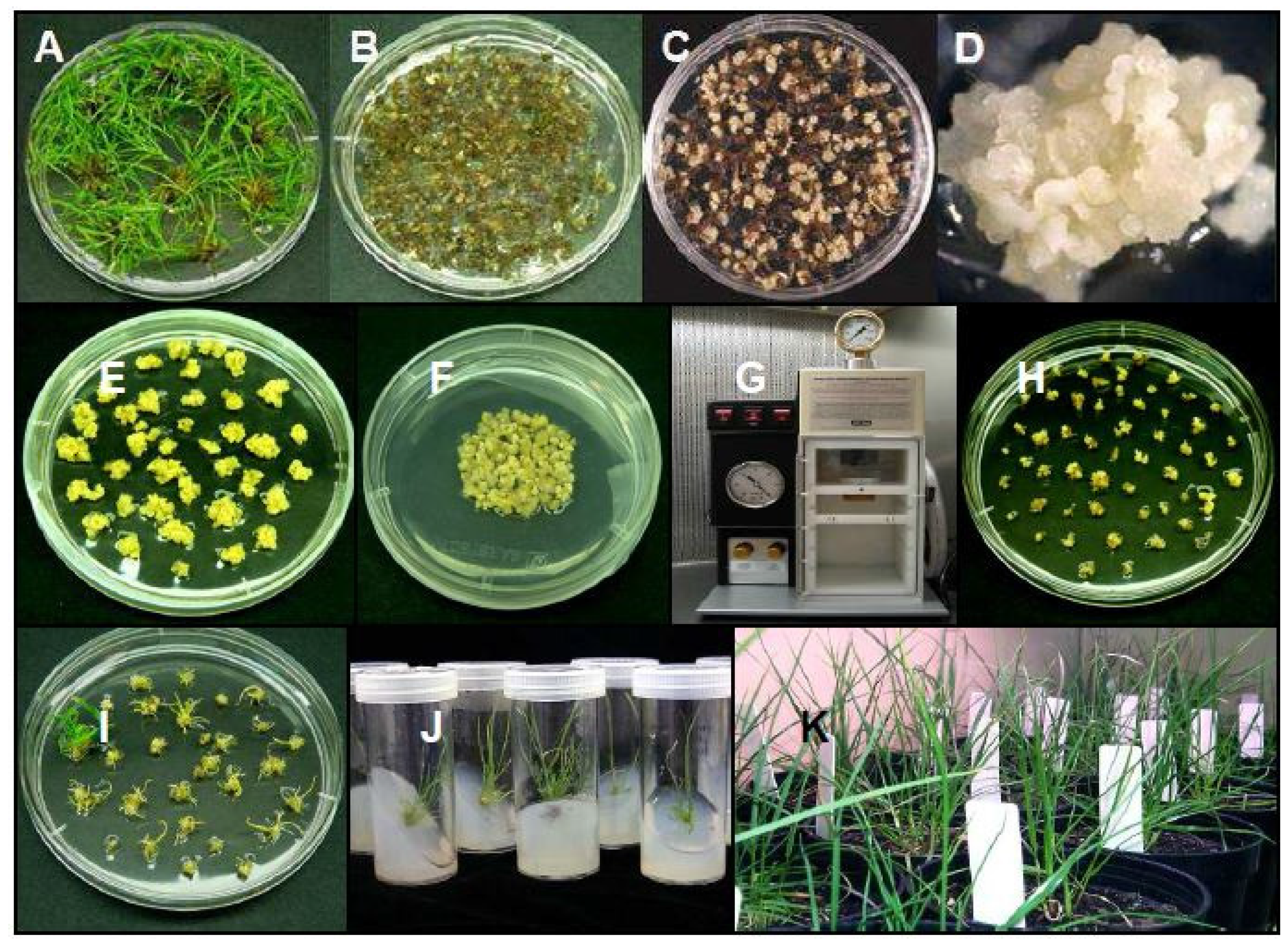
2.1. Production of Transgenic Plants
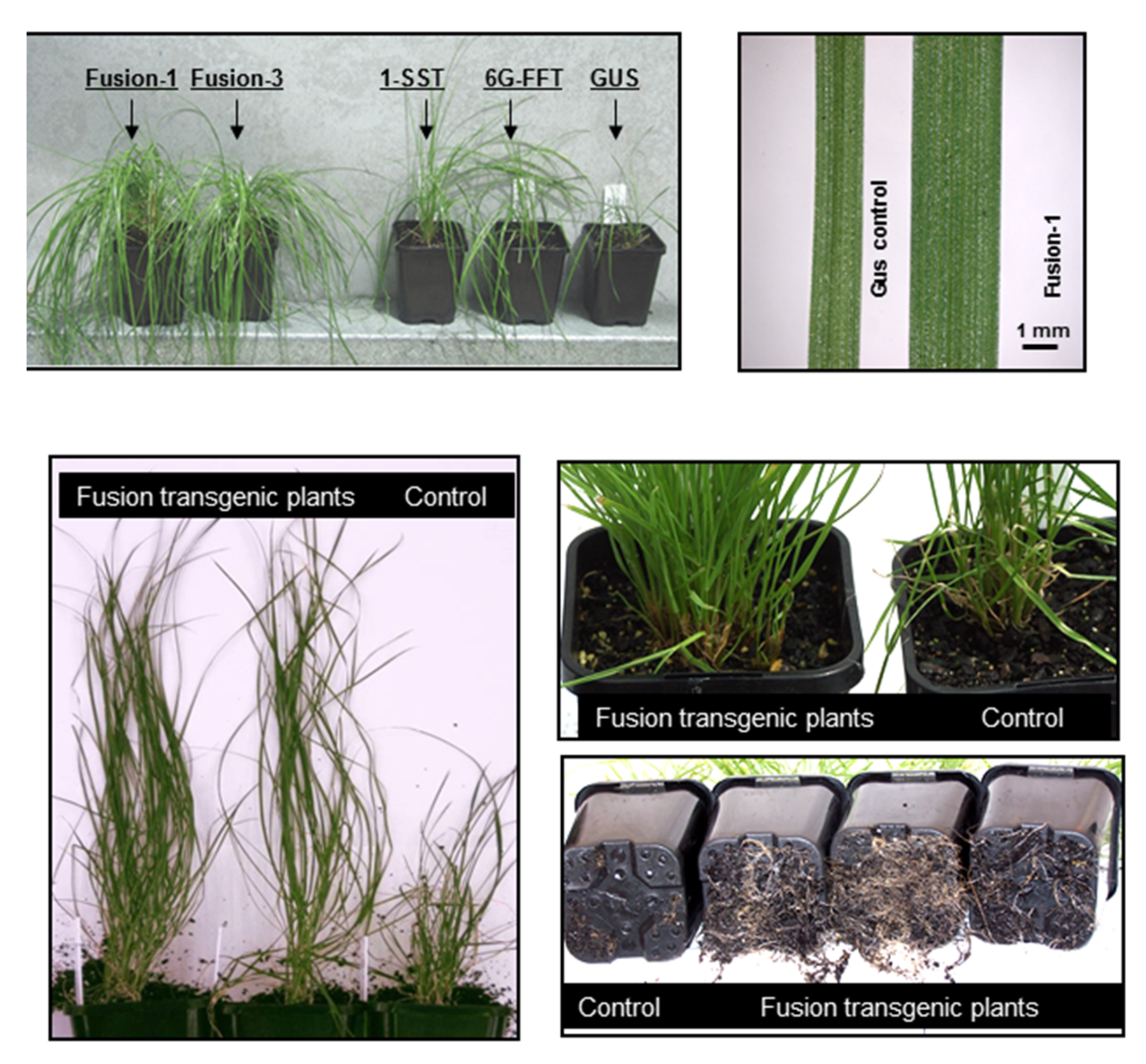
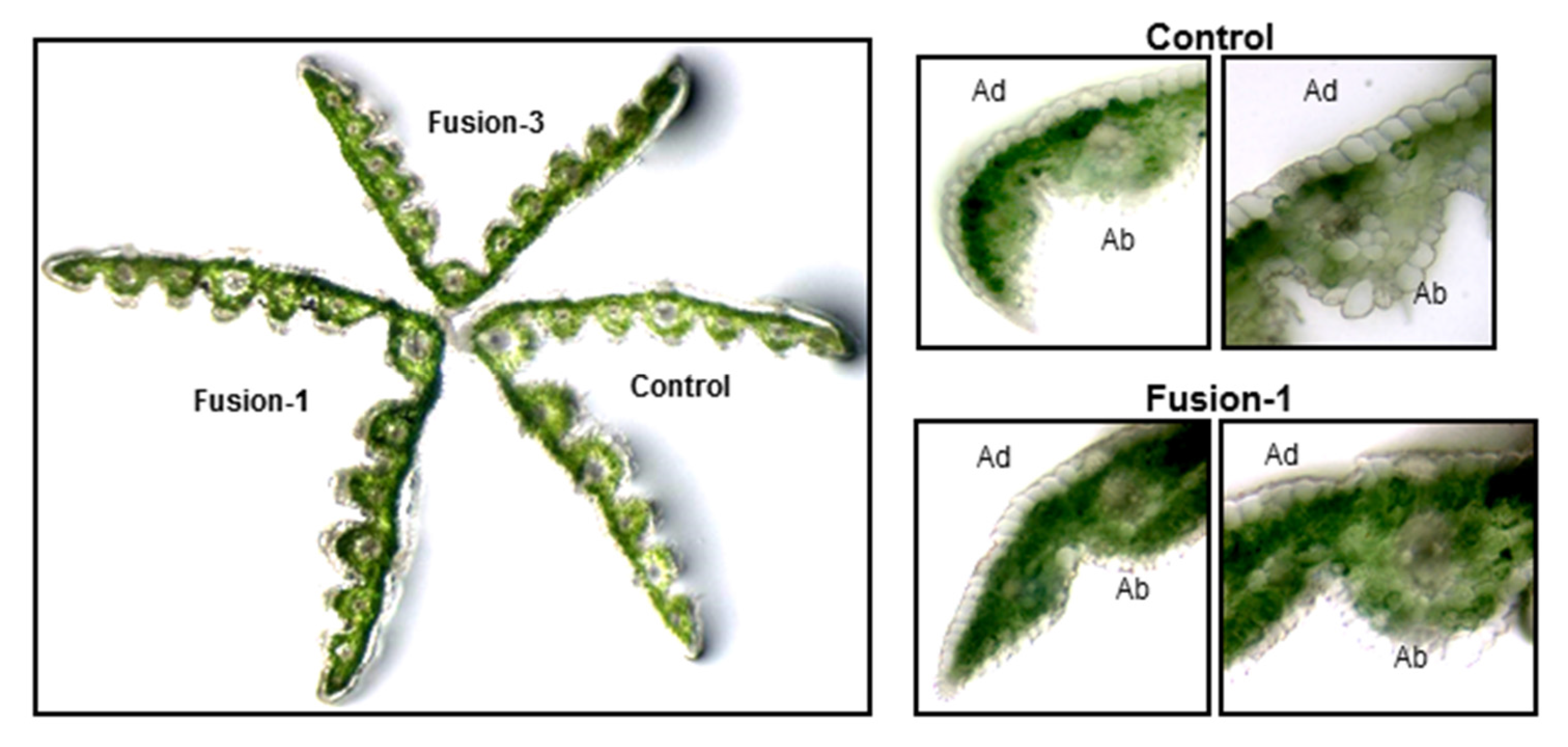
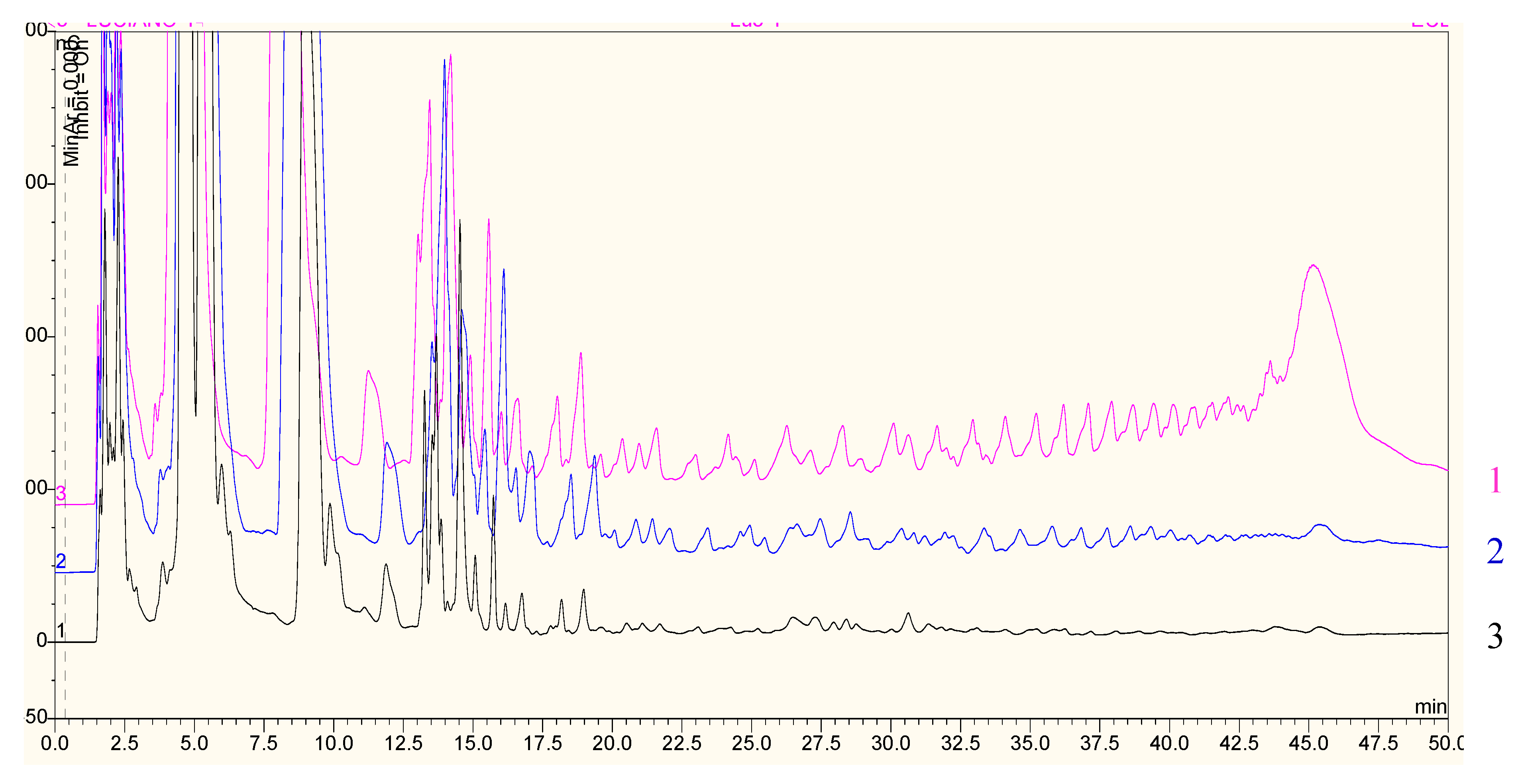
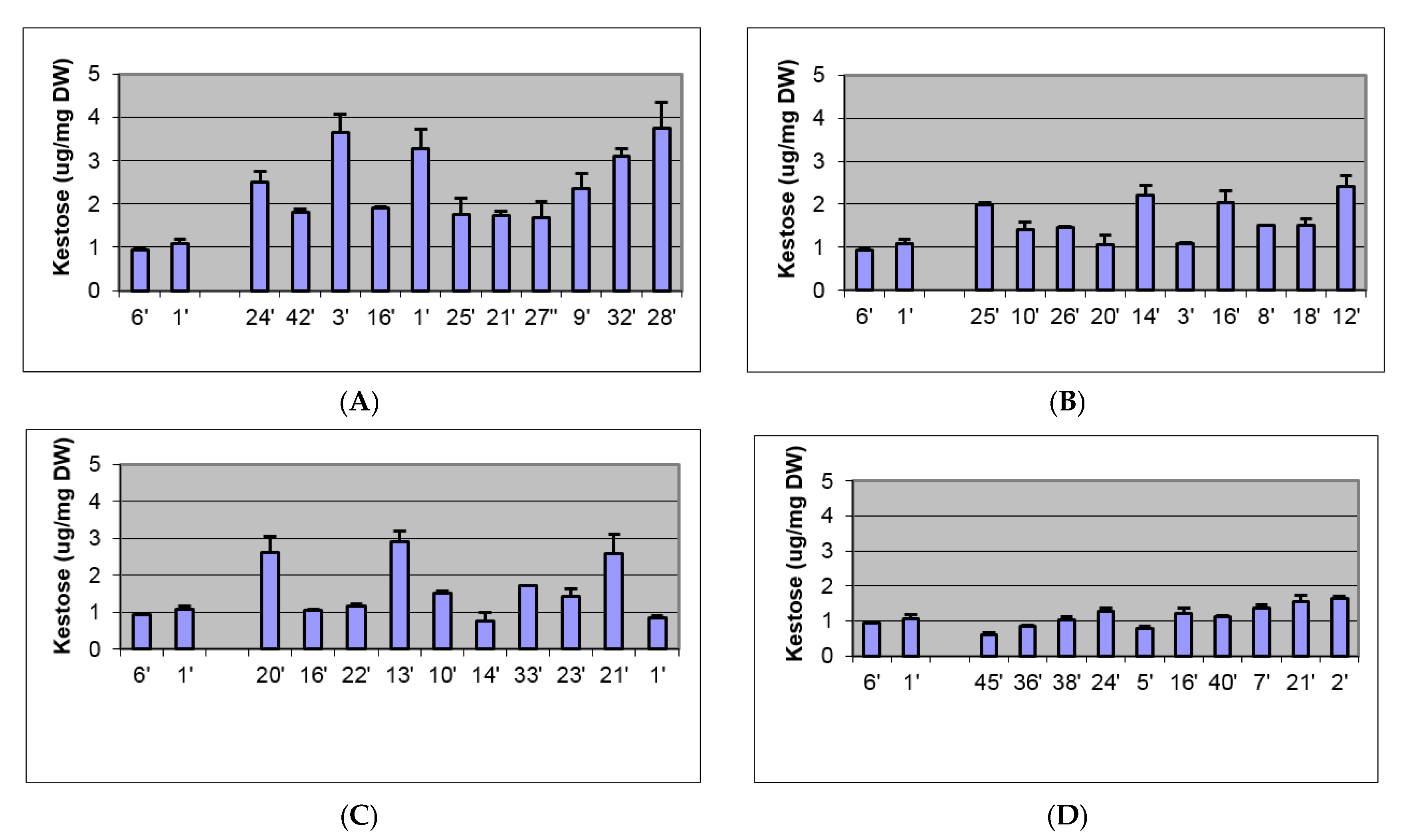
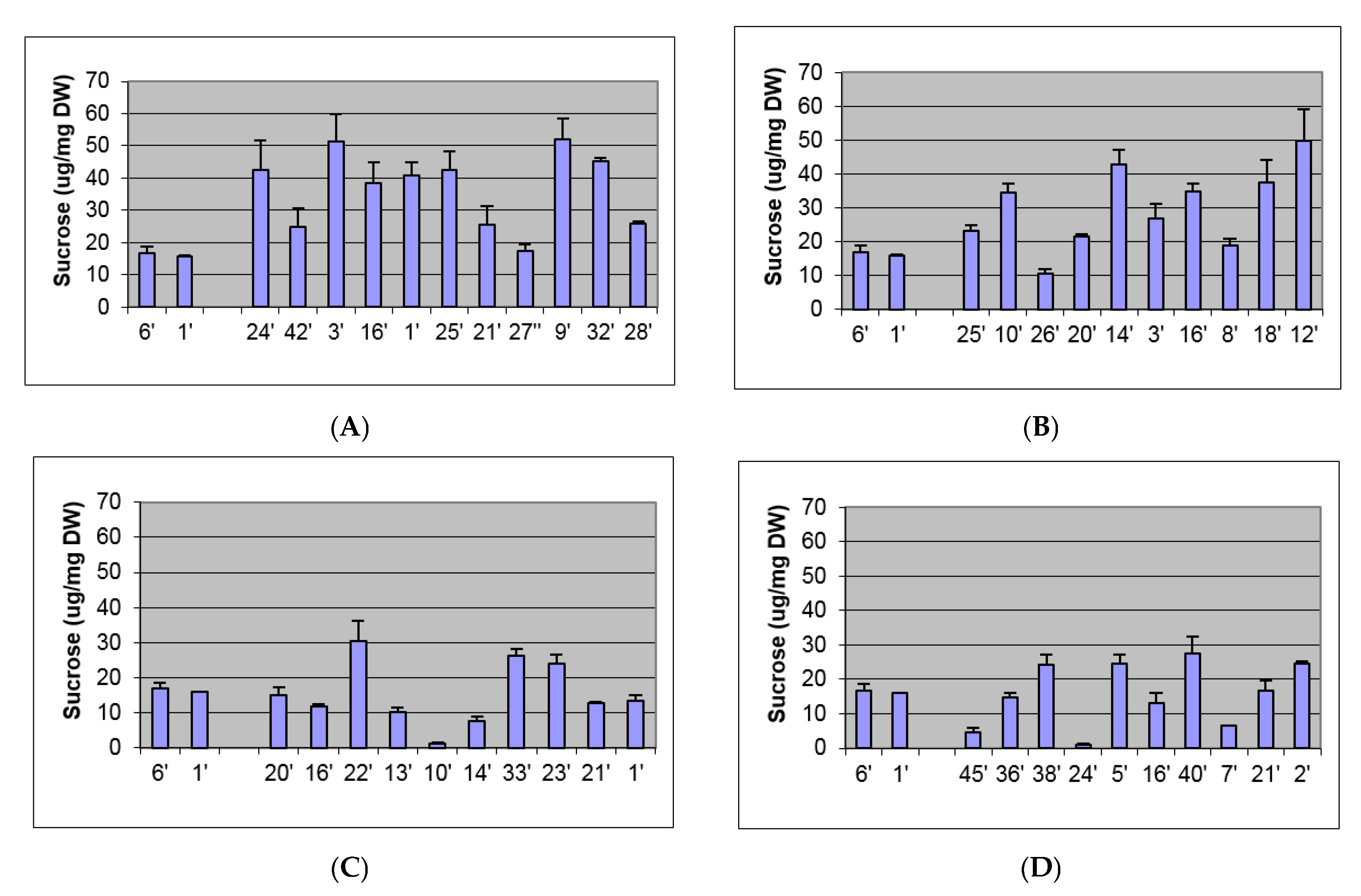
2.2. Biochemical and Morphological Characterisation of Transgenic Plants
3. Discussion
4. Materials and Methods
4.1. Identification and Cloning of Photosynthetic Promoters from Perennial Ryegrass
4.2. Isolation of Fructan Biosynthesis Genes
4.3. Cloning of FT Translational Fusion
4.4. Generation of Vectors for Transgenic Assays
- pPZP200: 35S2::Lp6G-FFT::TaRbcS
- pPZP200: 35S2::Lp1-SST::TaRbcS
- pPZP200: 35S2::Lp1-FFT::TaRbcS
- pPZP200: 35S2::Lp1-SST::6G-FFT::TaRbcS
- pPZP200: 35S2::GUS::TaRbcS
4.5. Function of Lp1-SST, Lp6G-FFT and FT-Fusion Protein in Transient Transgenic Assays
4.6. Generating Vectors for Stable Transformation
- pDEST-TaRbcS::Lp1-SST::TaRbcS
- pDEST-TaRbcS::Lp6G-FFT::TaRbcS
- pDEST-TaRbcS::Lp1-SST-Lp6G-FFT::TaRbcS (fusion-1)
- pDEST-TaRbcS::Lp1-SST-Lp6G-FFT::TaRbcS (fusion-3)
- pDEST-TaRbcS::GUS::TaRbcS
4.7. Production of Transgenic Ryegrass Plants
4.8. Transgene Detection
4.9. Quantification of Carbohydrates
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guerrand, D.; Prud’homme, M.; Boucaud, J. Fructan metabolism in expanding leaves, mature leaf sheaths and mature leaf blades of Lolium perenne. Fructan synthesis, fructosyltransferase and invertase activities. New Phytol. 1996, 134, 205–214. [Google Scholar] [CrossRef]
- Thomas, H.; James, A. Partitioning of sugars in Lolium perenne (perennial ryegrass) during drought and on rewatering. New Phytol. 1999, 142, 295–305. [Google Scholar] [CrossRef]
- Hendry, G.; Wallace, R. The origin, distribution and evolutionary significance of fructans. In Science and Technology of Fructans; Suzuki, M., Chatterton, N.J., Eds.; CRC Press: Baton Rouge, FL, USA, 1993; pp. 119–139. [Google Scholar]
- Nocek, J.; Russell, J. Protein and energy as in integrated system. Relationship of ruminal protein and carbohydrate availability to microbial synthesis and milk production. J. Dairy Sci. 1988, 70, 2070–2107. [Google Scholar] [CrossRef]
- Biggs, D.; Hancock, K. In vitro digestion of bacterial and plant fructans and effects on ammonia accumulation in cow and sheep rumen fluids. J. Gen. Appl. Microbiol. 1998, 44, 167–171. [Google Scholar] [CrossRef]
- Taweel, H.Z.; Tas, B.M.; Smit, H.J.; Elgersma, A.; Dijkstra, J.; Tamminga, S. Effects of feeding perennial ryegrass with an elevated concentration of water-soluble carbohydrates on intake, rumen function and performance of dairy cows. Anim. Feed Sci. Technol. 2005, 121, 243–256. [Google Scholar] [CrossRef]
- Ye, X.; Wu, X.; Zhao, H.; Frehner, M.; Nosberger, J.; Potrykus, I.; Spangenberg, G. Altered fructan accumulation in transgenic Lolium multiflorum plants expressing a Bacillus subtilis sacB gene. Plant Cell Rep. 2001, 20, 205–212. [Google Scholar] [CrossRef]
- Hisano, H.; Kanazawa, A.; Kawakami, A.; Yoshida, M.; Shimamoto, Y.; Yamada, T. Transgenic perennial ryegrass plants expressing wheat fructosyltransferase genes accumulate increased amounts of fructan and acquire increased tolerance on a cellular level to freezing. Plant Sci. 2004, 167, 861–868. [Google Scholar] [CrossRef]
- Gadegaard, G.; Didion, T.; Folling, M.; Storgaard, M.; Andersen, C.H.; Nielsen, K.K. Improved fructan accumulation in perennial ryegrass transformed with the onion fructosyltransferase genes 1-SST and 6G-FFT. J. Plant Physiol. 2008, 165, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Pollock, C.; Jones, T. Seasonal patterns of fructan metabolism in forage grasses. New Phytol. 1979, 83, 9–15. [Google Scholar] [CrossRef]
- Badenhorst, P.E. Phenomic Evaluation and Molecular Breeding of Field-Grown Transgenic Perennial Ryegrass (Lolium perenne) with Altered Fructan Biosynthesis. Ph.D. Thesis, La Trobe University Bundoora, Melbourne, Australia, 2014. [Google Scholar]
- Chalmers, J.; Lidgett, A.; Cummings, N.; Cao, Y.; Forster, J.; Spangenberg, G. Functional genomics of fructan metabolism in temperate grasses. Plant Biotechnol. J. 2005, 3, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Lasseur, B.; Lothier, J.; Wiemken, A.; van Laere, A.; Morvan-Bertrand, A.; Van den Ende, W.; Prudhomme, M.P. Towards a better understanding of the generation of fructan structure diversity in plants: Molecular and functional characterization of a sucrose:fructan 6-fructosyltransferase (6-SFT) cDNA from perennial ryegrass (Lolium perenne). J. Exp. Bot. 2011, 62, 1871–1885. [Google Scholar] [CrossRef] [PubMed]
- Morvan-Bertrand, A.; Boucaud, J.; Le Saos, J.; Prud’homme, M.P. Roles of fructans from leaf sheaths and from the elongating leaf bases in the regrowth following defoliation of Lolium perenne L. Planta 2001, 213, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Livingston, D.P.; Hincha, D.K.; Heyer, A.G. Fructan and its relationship to abiotic stress tolerance in plants. Cell. Mol. Life Sci. 2009, 66, 2007–2023. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.F.; Simpson, R.J.; Oram, R.N.; Lowe, K.F.; Kelly, K.B.; Evans, P.M.; Humphreys, M.O. Seasonal variation in the herbage yield and nutritive value of perennial ryegrass (Lolium perenne L.) cultivars with high or normal herbage water soluble carbohydrate concentrations grown in three contrasting Australian dairy environments. Aust. J. Exp. Agric. 1998, 38, 821–830. [Google Scholar] [CrossRef]
- Smith, K.F.; Reed, K.F.M.; Foot, J.Z. An assessment of the relative importance of specific traits for the genetic improvement of nutritive value in dairy pasture. Grass Forage Sci. 1997, 52, 167–175. [Google Scholar] [CrossRef]
- Ludemann, C.I.; Eckard, R.J.; Cullen, B.R.; Jacobs, J.L.; Malcolm, B.; Smith, K.F. Higher energy concentration traits in perennial ryegrass (Lolium perenne L.) may increase profitability and improve energy conversion on dairy farms. Agric. Syst. 2015, 137, 89–100. [Google Scholar] [CrossRef]
- Terzaghi, W.B.; Cashmore, A.R. Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 445–474. [Google Scholar] [CrossRef]
- Francis, S.A.; Chapman, D.F.; Doyle, P.T.; Leury, B.J. Dietary preferences of cows offered choices between white clover and “high sugar” and “typical” perennial ryegrass cultivars. Aust. J. Exp. Agric. 2006, 46, 1579–1587. [Google Scholar] [CrossRef]
- Schaffner, A.R.; Sheen, J. Maize rbcS Promoter Activity Depends on Sequence Elements Not Found in Dicot rbcS Promoters. Plant Cell 1991, 3, 997–1012. [Google Scholar] [CrossRef] [PubMed]
- Hudson, M.E.; Quail, P.H. Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 2003, 133, 1605–1616. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, J.; Johnson, X.; Lidgett, A. Isolation and characterisation of a sucrose:sucrose 1-fructosyltransferase gene from perennial ryegrass (Lolium perenne). J. Plant Physiol. 2003, 160, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Hernandez, A.; Lopez-Ochoa, L.; Arguello-Astorga, G.; Herrara-Estrella, L. Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol. 2002, 128, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Kay, R.; Chan, A.; Daly, M.; McPherson, J. Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 1987, 236, 1299–1302. [Google Scholar] [CrossRef] [PubMed]
- Hajdukiewicz, P.; Svab, Z.; Maliga, P. The small, versatile pPZP family of Agrobacterium binary vectors RT for plant transformation. Plant Mol. Biol. 1994, 25, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Kapila, J.; De Rycke, R.; van Montagu, M.; Angenon, G. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 1997, 124, 227. [Google Scholar] [CrossRef]
- Wydro, M.; Kozubek, E.; Lehmann, P. Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim. Pol. 2006, 53, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Sasanuma, T. Characterization of the rbcS multigene family in wheat: Subfamily classification, determination of chromosomal location and evolutionary analysis. Mol. Genet. Genom. 2001, 265, 161–171. [Google Scholar] [CrossRef]
- Spangenberg, G.; Wang, Z.Y.; Wu, X.; Nagel, J.; Potrykus, I. Transgenic perennial ryegrass (Lolium perenne) plants from microprojectile bombardment of embryogenic suspension cells. Plant Sci. 1995, 108, 209–217. [Google Scholar] [CrossRef]
- Bilang, R.; Shigeru, I.; Peterhans, A.; Potrykus, I.; Paszkowski, J. The 3′-terminal region of the hygromycin-B-resistance gene is important for its activity in Escherichia coli and Nicotiana tabacum. Gene 1991, 100, 247–250. [Google Scholar] [CrossRef]
- Spangenberg, G.; Wang, Z.Y.; Wu, X.L.; Nagel, J.; Iglesias, V.A.; Potrykus, I. Transgenic tall fescue and red fescue plants from microprojectile bombardment of embryogenic suspension cells. J. Plant Physiol. 1995, 145, 693–701. [Google Scholar] [CrossRef]
- Ye, X.; Wang, Z.Y.; Potrykus, I.; Spangenberg, G. Transgenic Italian ryegrass (Lolium multiflorum) plants from microprojectile bombardment of embryogenic suspension cells. Plant Cell Rep. 1997, 16, 379–384. [Google Scholar] [CrossRef]
- Bai, Y.; Qu, R. Genetic transformation of elite turf-type cultivars of Tall Fescue. Int. Turfgrass Soc. Res. J. 2001, 9, 129–136. [Google Scholar]
- Liu, Z.; Mouradov, A.; Smith, K.F.; Spangenberg, G. An improved method for quantitative analysis of total fructans in plant tissues. Anal. Biochem. 2011, 418, 253–259. [Google Scholar] [CrossRef] [PubMed]


| Species | Genotype | Transforming DNA | No Plates Bombarded | No calli Bombarded | No +ve Transgenics | No Plates Analysed | hph +ve Plants | GOI +ve Plants |
|---|---|---|---|---|---|---|---|---|
| L. perenne | FLP 418-20 | TaRbcS::Lp1-SST::Tarbcs +pACH1 | 10 | 500 | 46 | 46 | 37 | 32 |
| L. perenne | FLP 418-20 | TaRbcS::Lp6G-FFT::Tarbcs +pACH1 | 10 | 500 | 50 | 50 | 48 | 38 |
| L. perenne | FLP 418-20 | TaRbcS::Lp1-SST-Lp6G-FFT::Tarbcs (1) + pACH1 | 10 | 500 | 47 | 47 | 47 | 44 |
| L. perenne | FLP 418-20 | TaRbcS::Lp1-SST-Lp6G-FFT::Tarbcs (3) + pACH1 | 10 | 500 | 26 | 26 | 26 | 23 |
| L. perenne | FLP 418-20 | TaRbcS::GUS::Tarbcs +pACH1 | 10 | 500 | 13 | 13 | 11 | 9 |
| cis-Acting Regulatory seq. | Accession | Position LpRbcS | Position LpCAB |
|---|---|---|---|
| I-Box Core | S000199 | −184 | −137 |
| I-Box | S000124 | −311 | −137 |
| GT1 consensus | S000198 | −304 | n.p. |
| RbcS monocot seq | Schaffner et al., 1991 | −173 to −151 | n.p. |
| SORLIPs | S000482 | n.p. | −58, −217, −647, −695 |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Lp1-SST | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGAGTCCCCAAGCGCCGTC | TCTAAGCCTTTCCTCCTCCCAAGTCGTCGTTCGTG |
| Lp6G-FFT | ACTAAGCTTGGAGGAGGAGAGTCCAGCGCCG | GGGGACCACTTTGTACAAGAAAGCTGGGTCCTACATGTCGTCAGCCAAGAAGGCC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panter, S.; Mouradov, A.; Badenhorst, P.; Martelotto, L.; Griffith, M.; Smith, K.F.; Spangenberg, G. Re-Programming Photosynthetic Cells of Perennial Ryegrass (Lolium perenne L) for Fructan Biosynthesis through Transgenic Expression of Fructan Biosynthetic Genes under the Control of Photosynthetic Promoters. Agronomy 2017, 7, 36. https://doi.org/10.3390/agronomy7020036
Panter S, Mouradov A, Badenhorst P, Martelotto L, Griffith M, Smith KF, Spangenberg G. Re-Programming Photosynthetic Cells of Perennial Ryegrass (Lolium perenne L) for Fructan Biosynthesis through Transgenic Expression of Fructan Biosynthetic Genes under the Control of Photosynthetic Promoters. Agronomy. 2017; 7(2):36. https://doi.org/10.3390/agronomy7020036
Chicago/Turabian StylePanter, Stephen, Aidyn Mouradov, Pieter Badenhorst, Luciano Martelotto, Megan Griffith, Kevin F. Smith, and German Spangenberg. 2017. "Re-Programming Photosynthetic Cells of Perennial Ryegrass (Lolium perenne L) for Fructan Biosynthesis through Transgenic Expression of Fructan Biosynthetic Genes under the Control of Photosynthetic Promoters" Agronomy 7, no. 2: 36. https://doi.org/10.3390/agronomy7020036






