Start a Research on Biopolymer Polyhydroxyalkanoate (PHA): A Review
Abstract
:1. Introduction
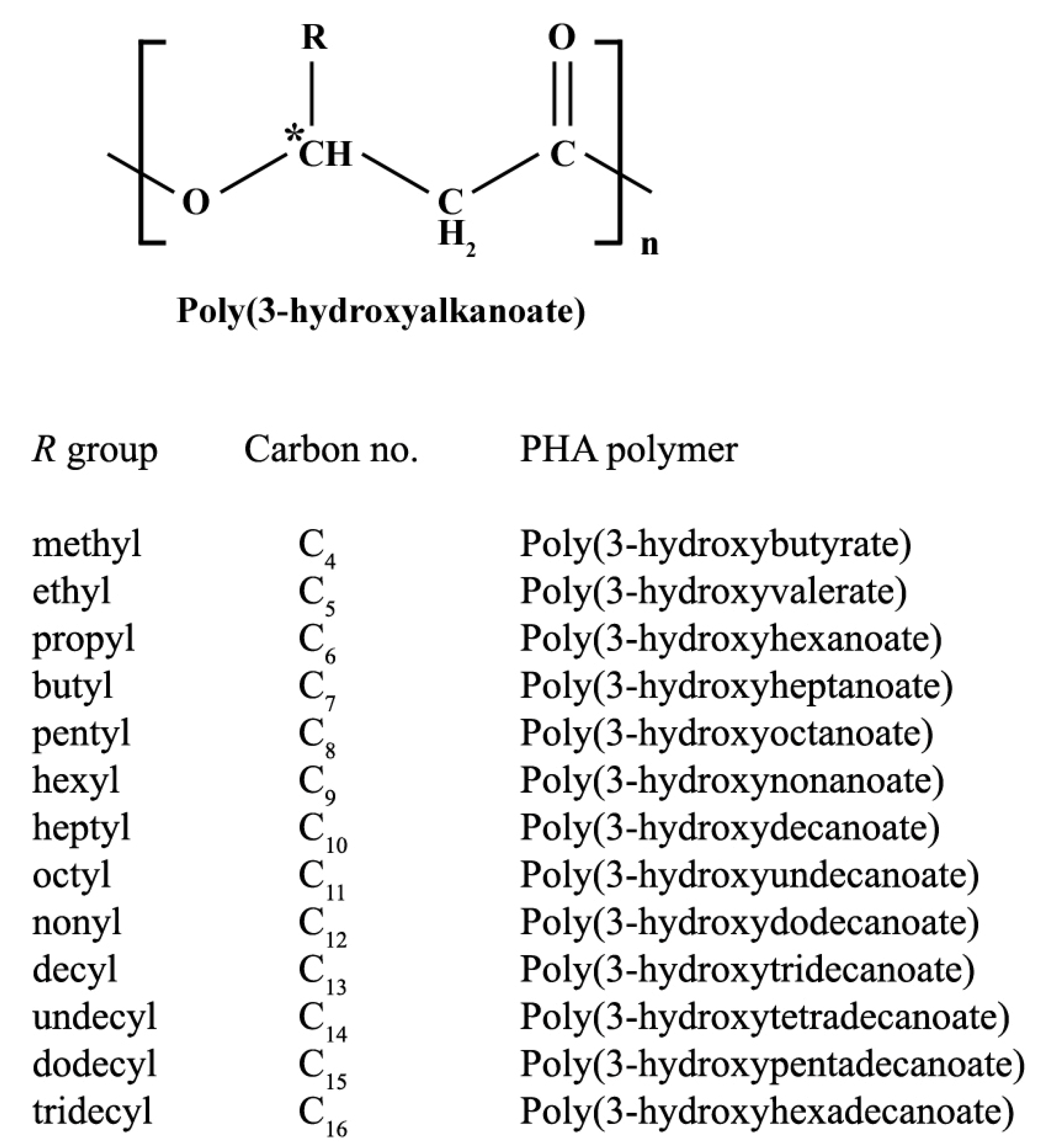
2. PHA Biosynthetic Pathways

| No. | Enzyme | Abbreviation | Species | Reference |
|---|---|---|---|---|
| 1 | Glyceraldehyde-3-phosphate dehydrogenase | - | Cupriavidus necator | [22] |
| 2 | Pyruvate dehydrogenase complex | - | Cupriavidus necator and Burkholderia cepacia | [22] |
| 3 | 3-Ketothiolase | PhaA | Cupriavidus necator | [23] |
| 4 | NADPH-dependent acetoacetyl-CoA reductase | PhaB | Cupriavidus necator | [23] |
| 5 | PHA synthase | PhaC | Cupriavidus necator and various | [12,23] |
| 6 | Acetyl-CoA carboxylase | ACC | Escherichia coli K-12 MG1655 | [24] |
| 7 | Malonyl-CoA:ACP transacylase | FabD | Escherichia coli K-12 MG1655 | [24] |
| 8 | 3-Ketoacyl carrier protein synthase | FabH | Escherichia coli K-12 MG1655 | [24,25] |
| 9 | NADPH-dependent 3-Ketoacyl reductase | FabG | Pseudomonas aeruginosa | [26] |
| 10 | Succinic semialdehyde dehydrogenase | SucD | Clostridium kluyveri | [27] |
| 11 | 4-Hydroxybutyrate dehydrogenase | 4HbD | Clostridium kluyveri | [27] |
| 12 | 4-Hydroxybutyrate-CoA:CoA transferase | OrfZ | Clostridium kluyveri | [27] |
| 13 | Alcohol dehydrogenase, putative | - | Aeromonas hydrophila 4AK4 | [28] |
| 14 | Hydroxyacyl-CoA synthase, putative | - | Mutants and recombinants of Cupriavidus necator | [29] |
| 15 | Methylmalonyl-CoA mutase | Sbm | Escherichia coli W3110 | [30] |
| 16 | Methylmalonyl-CoA racemase | - | Nocardia corallina | [31] |
| 17 | Methylmalonyl-CoA decarboxylase | YgfG | Escherichia coli W3110 | [30] |
| 18 | Ketothiolase, putative | - | - | [32] |
| 19 | 3-Ketothiolase | BktB | Cupriavidus necator | [33] |
| 20 | Ketothiolase, putative | - | - | [32] |
| 21 | NADPH-dependent acetoacetyl-CoA reductase | - | Rhizobium (Cicer) sp. CC 1192 | [34] |
| 22 | Acyl-CoA synthetase | FadD | Pseudomonas putida CA-3 and Escherichia coli MG1655 | [35,36] |
| 23 | Acyl-CoA oxidase, putative | - | - | [37] |
| 24 | Enoyl-CoA hydratase I, putative | - | - | [37] |
| 25 | (R)-Enoyl-CoA hydratase | PhaJ | Pseudomonas putida KT2440 | [38] |
| 26 | Epidermase | - | - | [37] |
| 27 | 3-Ketoacyl-CoA thiolase | FadA | Pseudomonas putida KT2442 | [39] |
| 28 | 3-Hydroxyacyl-ACP:CoA transacylase | PhaG | Pseudomonas mendocina | [40] |
| 29 | Cyclohexanol dehydrogenase | ChnA | Acinetobacter sp. SE19 and Brevibacterium epidermidis HCU | [41] |
| 30 | Cyclohexanone monooxygenases | ChnB | Acinetobacter sp. SE19 and Brevibacterium epidermidis HCU | [41] |
| 31 | Caprolactone hydrolase | ChnC | Acinetobacter sp. SE19 and Brevibacterium epidermidis HCU | [41] |
| 32 | 6-Hydroxyhexanoate dehydrogenase | ChnD | Acinetobacter sp. SE19 and Brevibacterium epidermidis HCU | [41] |
| 33 | 6-Oxohexanoate dehydrogenase | ChnE | Acinetobacter sp. SE19 and Brevibacterium epidermidis HCU | [41] |
| 34 | Semialdehyde dehydrogenase, putative | - | - | [11] |
| 35 | 6-hydroxyhexanoate dehydrogenase, putative | - | - | [11] |
| 36 | Hydroxyacyl-CoA synthase, putative | - | - | [11] |
| 37 | Lactonase, putative | - | Mutants and recombinants of Cupriavidus necator | [29] |
3. PHA-Producing Microbial Strains from Culture Collections
| Microorganism | Culture collection number b | Carbon source | PHA monomer or polymer c | PHA content
(%CDM) | Average PHA productivity (g L−1 h−1) | Reference |
|---|---|---|---|---|---|---|
| Gram-negative bacteria | ||||||
| Azohydromonas australica (formerly Alcaligenes latus) | ATCC 29713, DSM 1124, IAM 12664, LMG 3324 | Malt waste | P3HB | 70.1 | 0.445 | [46] |
| Azohydromonas lata (formerly Alcaligenes latus) | ATCC 29714, DSM 1123, IAM 12665, LMG 3325 | Sucrose | P3HB | 50.0–88.0 | 0.050–4.940 | [47,48,49] |
| Fructose, glucose | P3HB | 76.5–79.4 | 0.121–0.128 | [50] | ||
| Azotobacter beijerinckii | DSM 1041, NCIB 11292 | Glucose | P3HB | 24.8 | 0.090 | [51] |
| Burkholderia cepacia (formerly Pseudomonas multivorans and Pseudomonas cepacia) | ATCC 17759, DSM 50181, NCIB 9085 | Xylose | P3HB | 58.4 | NG | [52] |
| Glycerol | P3HB | 31.3 | 0.103 | [53] | ||
| Fructose, glucose, sucrose | P3HB | 50.4–59.0 | NG | [50] | ||
| Burkholderia sp. USM | JCM 15050 | Lauric acid, myristic acid, oleic acid, palmitic acid, stearic acid | P3HB | 1.0–69.0 | NG | [54] |
| Caulobacter vibrioides (formerly Caulobacter crescentus) | DSM 4727 | Glucose | P3HB | 18.3 | 0.008 | [55] |
| Cupriavidus necator H16 (formerly Hydrogenomonas eutropha H16, Alcaligenes eutrophus H16, Ralstonia eutropha H16 and Wautersia eutropha H16) | ATCC 17699, DSM 428, KCTC 22496, NCIB 10442 | Fructose, glucose | P3HB | 67.0–70.5 | 0.052–0.067 | [50] |
| 4-Hydroxyhexanoic acid | P3HB | 76.3–78.5 | NG | [56] | ||
| Corn oil, oleic acid, olive oil, palm oil | P3HB | 79.0–82.0 | 0.041–0.047 | [57] | ||
| Acetate, butyrate, lactic acid,
propionic acid | 3HB, 3HV | 3.9–40.7 | 0.001–0.037 | [58] | ||
| CO2 | P3HB | 88.9 | 0.230 | [59] | ||
| Cupriavidus necator (formerly Hydrogenomonas eutropha, Alcaligenes eutrophus N9A, Ralstonia eutropha N9A and Wautersia eutropha) | DSM 518 | 4-Hydroxyhexanoic acid | P3HB | 65.8–66.2 | NG | [56] |
| Cupriavidus necator (formerly Hydrogenomonas eutropha, Alcaligenes eutrophus TF93, Ralstonia eutropha TF93 and Wautersia eutropha) | ATCC 17697, DSM 531 | 4-Hydroxyhexanoic acid | P3HB | 67.2 | NG | [56] |
| CO2 | P3HB | 60.0 | 0.600 | [60] | ||
| Cupriavidus necator a (formerly Hydrogenomonas eutropha, Alcaligenes eutrophus, Ralstonia eutropha and Wautersia eutropha) | CECT 4623, KCTC 2649, NCIMB 11599 | Glucose | P3HB | 76.0 | 2.420 | [61] |
| Potato starch, saccharified waste | P3HB | 46.0 | 1.470 | [62] | ||
| Cupriavidus necator (formerly Hydrogenomonas eutropha, Alcaligenes eutrophus, Ralstonia eutropha and Wautersia eutropha) | DSM 545 | Molasses | P3HB | 31.0–44.0 | 0.080–0.120 | [63] |
| Glucose, propionic acid | P3HB3HV | 80.0 | 0.820 | [64] | ||
| Waste glycerol | P3HB | 14.8–36.1 | 0.330–4.200 | [65] | ||
| Halomonas boliviensis LC1 | ATCC BAA-759, DSM 15516 | Hydrolyzed starch | P3HB | 56.0 | NG | [66] |
| Hydrogenophaga pseudoflava | ATCC 33668, DSM 1034 | Lactose, sucrose | P3HB3HV | 20.2–62.5 | 0.018–0.117 | [67] |
| Hydrolyzed whey and valerate | P3HB3HV | 40.0 | 0.050 | [68] | ||
| Methylobacterium extorquens | ATCC 55366 | Methanol | P3HB | 40.0–46.0 | 0.250–0.600 | [69] |
| Methylobacterium extorquens | ATCC 8457, DSM 1340, NCIB 2879, NCTC 2879 | Methanol | P3HB | 35.0–62.3 | 0.183–0.980 | [70,71] |
| Methylocystis sp. GB25 a | DSM 7674 | Methane | P3HB | 51.0 | NG | [72] |
| Novosphingobium nitrogenifigens Y88 | DSM 19370, ICMP 16470 | Glucose | P3HB | 81.0 | 0.014–0.021 | [73] |
| Paracoccus denitrificans | ATCC 17741, DSM 413 | n-Pentanol | P3HV | 22.0–24.0 | NG | [74] |
| Pseudomonas aeruginosa | NCIM 2948 | Cane molasses, fructose, glucose, glycerol, sucrose | P3HB | 12.4–62.0 | 0.012–0.110 | [75] |
| Pseudomonas aeruginosa PAO1 | ATCC 47085 | Oil and wax products from polyethylene (PE) pyrolysis | mcl-PHA | 25.0 | NG | [76] |
| Pseudomonas frederiksbergensis GO23 a | NCIMB 41539 | Terephthalic acid from polyethylene terephthalate (PET) pyrolysis | mcl-PHA | 24.0 | 0.004 | [77] |
| Pseudomonas marginalis | DSM 50276 | 1,3-butanediol, octanoate | scl-mcl-PHA,
mcl-PHA | 11.9–31.4 | NG | [78] |
| Pseudomonas mendocina | ATCC 25411, DSM 50017 | 1,3-butanediol, octanoate | scl-mcl-PHA | 13.5–19.3 | NG | [78] |
| Pseudomonas oleovorans | ATCC 8062, DSM 1045 | 4-Hydroxyhexanoic acid | scl-mcl-PHA | 18.6 | NG | [56] |
| Pseudomonas putida CA-3 a | NCIMB 41162 | Styrene | mcl-PHA | 31.8 | 0.063 | [79] |
| Styrene from polystyrene (PS) pyrolysis | mcl-PHA | 36.4 | 0.033 | [80] | ||
| Pseudomonas putida GO16 a | NCIMB 41538 | Terephthalic acid from polyethylene terephthalate (PET) pyrolysis | mcl-PHA | 27.0 | ~0.005, 0.008 d | [77] |
| Pseudomonas putida GO19 a | NCIMB 41537 | Terephthalic acid from polyethylene terephthalate (PET) pyrolysis | mcl-PHA | 23.0 | ~0.005, 0.008 d | [77] |
| Pseudomonas putida GPo1 (formerly Pseudomonas oleovorans) | ATCC 29347 | Alkenes, n-alkanes | mcl-PHA | 2.0–28.0 | NG | [81] |
| n-alkanoates | scl-mcl-PHA,
mcl-PHA | 5.0–60.0 | NG | [82,83] | ||
| Pseudomonas putida KT2440 | ATCC 47054 | Nonanoic acid | mcl-PHA | 26.8–75.4 | 0.250–1.110 | [84] |
| 4-Hydroxyhexanoic acid | mcl-PHA | 25.3–29.8 | NG | [56] | ||
| Glucose | mcl-PHA | 32.1 | 0.006 | [85] | ||
| Pseudomonas putida F1 | ATCC 700007, DSM 6899 | Benzene, ethylbenzene, toluene | mcl-PHA | 1.0–22.0 | NG | [86] |
| Pseudomonas putida mt-2 | NCIMB 10432 | Toluene, p-xylene | mcl-PHA | 22.0–26.0 | NG | [86] |
| Acetic acid, citric acid, glucose, glycerol, octanoic acid, pentanoic acid, succinic acid | mcl-PHA | 4.0–77.0 | NG | [87] | ||
| Thermus thermophilus HB8 | ATCC 27634, DSM 579 | Whey | scl-mcl-PHA | 35.6 | 0.024 | [88] |
| Gram-Positive bacteria | ||||||
| Bacillus megaterium | DSM 90 | Citric acid, glucose, glycerol, succinic acid | P3HB | 9.0–50.0 | NG | [87] |
| Bacillus megaterium | CCM 1464, DSM 509, IFO 12109, NBRC 12109 | Citric acid, glucose, glycerol, succinic acid, octanoic acid | P3HB, scl-mcl-PHA, mcl-PHA | 3.0–48.0 | NG | [87] |
| Various Bacillus spp. type strains | Refer to [89] | Acetate, n-alkanoate, 3-Hydroxybutyrate, propionate, sucrose, valerate | 3HB, 3HV, 3HHx | 2.2–47.6 | NG | [89] |
| Corynebacterium glutamicum | ATCC 15990, DSM 20137, NCIB 10337 | Acetic acid, citric acid, glucose, glycerol, succinic acid | P3HB, mcl-PHA | 4.0–32.0 | NG | [87] |
| Corynebacterium hydrocarboxydans | ATCC 21767 | Acetate, glucose | 3HB, 3HV | 8.0–21.0 | NG | [90] |
| Microlunatus phosphovorus | DSM 10555, JCM 9379 | Glucose | 3HB, 3HV | 20.0–30.0 | NG | [91] |
| Nocardia lucida | NCIMB 10980 | Acetate, succinate | 3HB, 3HV | 7.0–20.0 | NG | [90] |
| Rhodococcus sp. a | NCIMB 40126 | Acetate, 2-alkenoate, 1,4-butanediol, 5-chlorovalerate, fructose, glucose, hexanoate, 4-Hydroxybuytrate, lactate, molasses, succinate, valerate | P3HB3HV | 4.0–53.0 | NG | [90] |
| Various Streptomyces spp. type culture | Refer to [89] | Glucose | P3HB | 1.2–82.0 | NG | [89] |
| Archaea | ||||||
| Haloferax mediterranei | ATCC 33500, CCM 3361, DSM 1411 | Vinasse | P3HB3HV | 50.0–73.0 | 0.050–0.210 | [92] |
| Hydrolyzed whey | P3HB3HV | 72.8 | 0.090 | [93] | ||
| Glycerol and crude glycerol from biodiesel production | P3HB3HV | 75.0–76.0 | 0.120 | [94] | ||
| Various archaeal strains | Refer to [95] | Fructose, glucose, glycerol | P3HB, P3HB3HV | 0.8–22.9 | <0.001–0.021 | [95] |
3.1. Gram-Negative Bacteria
3.2. Gram-Positive Bacteria
3.3. Archaea
3.4. Formulation of Defined Co-Cultures Using Deposited Microbial Strains
4. Techniques for Detecting PHA and PHA Production Potential in Microbes
| Method | Characteristic | Sample | Sample preparation | Typical conditions | Advantage | Limitation | Reference |
|---|---|---|---|---|---|---|---|
| Polymerase chain reaction (PCR) gene detection | phaC gene encoding enzyme PHA synthase | 50–500 ng of DNA material or a single bacterial colony | DNA extraction or freeze/thaw cells to release DNA material | PCR thermal cycler temperature program for specific primer sets | Requires small sample size, high sensitivity and specificity, high throughput | Primers are inadequate for detection of all phaC genes, and prone to detection errors | [120,121] |
| Nile red and Nile blue A staining | Intracellular PHA granule structures | Bacterial colonies on agar medium | Add 0.5 µg mL−1 of Nile red or Nile blue
A to sterilized agar growth medium | Expose the agar plates to ultraviolet light (312 nm) after appropriate cultivation periods | Enables direct observation of live and actively-growing cells, requires small sample size, rapid analysis, allows differentiation between scl- and mcl-PHA under flow cytometry analysis, high throughput | Method cannot discriminate between lipids and PHAs, and is also less effective at distinguishing between PHA-negative and PHA-positive strains of Gram-positive bacteria | [122,123,124,125] |
| Microscope slide containing heat-fixed bacterial cells smear | Stain slide with 1% Nile blue A at 55 °C for 10 min. Remove excess stain with tap water before staining with 8% acetic acid for 1 min. Rinse slide with tap water and blot dry with bibulous paper | Examine slide with an epifluorescence microscope with an excitation wavelength of 460 nm | |||||
| 1 mL of cell culture with optical density at 600 nm (OD600) of 1.0 or less | Add 2.0–10 µg mL−L of Nile red to 1 mL cell culture and incubate in the dark for 15 min | Epifluorescence microscopy imaging with FITC filter with an excitation wavelength of 470–490 nm and an emission wavelength of 505 nm or fluorescence spectroscopy analysis at excitation wavelength of 488 nm and an emission wavelength of 590 nm and 575 nm for scl-PHA and mcl-PHA, respectively | |||||
| Transmission electron microscopy (TEM) | Intracellular PHA granule structures | 1–3 mL of exponential or stationary phase cell culture | Cell fixation with glutaraldehyde in phosphate buffer, followed by post-fixation with osmium tetroxide. Dehydrate fixated cells through a graded acetone series before acetone-resin infiltration and resin polymerization. Cut resins into ultrathin sections (70–100 nm thickness) with an ultramicrotome | View with an accelerating voltage of 200 kV and perform imaging at magnifications of 25,000–40,000× | High magnification enables direct visualization and size measurements of PHA granules | Tedious sample preparation involving radioactive and hazardous chemicals, cells are killed during sample preparation | [43] |
| Crotonic acid assay | Quantitative determination of P3HB | 5–50 µg P3HB | Add 10 mL concentrated H2SO4, and heat at 100 °C for 10 min to form crotonic acid | Measure UV absorbance at 235 nm | Easy operation, inexpensive per analysis, specific to P3HB determination | Result can be interfered by other endogenous components and matrix interferences can result in overestimation of P3HB content. Method is limited to P3HB determination | [89,105,126] |
| Fourier transform infrared spectroscopy (FTIR) | Cellular PHA content | 0.4-10 mg biomass | Spread cells on thallium bromoiodide (KRS-5) window and air-dry | FTIR was used to record the PHA spectrum at ambient temperature (25 °C), at a spectra range of 400–4000 cm−1, for 10–64 scans and a resolution of 4 cm−1 | Requires small sample size, short analysis time, solvent usage is optional, can provide quantitative information, enables online and real-time PHA analysis, high throughput | Method cannot discriminate between different PHA monomeric units, unable to distinguish between homogenous PHA and PHA copolymer, low sensitivity, quantification limited to scl-PHA | [127,128] |
| Liquid chromatography (LC) | PHA monomeric units | 0.01–500 mg biomass or 0.01–14 µg P3HB | Hydrolytic digestion with concentrated sulfuric acid 90 °C for 30 min, cool on ice before adding 0.014 N of sulfuric acid with rapid mixing to yield crotonic acids | High performance liquid chromatography (HPLC) analysis with an ion-exclusion organic acid analysis column and a UV detector at 210 nm | Does not require cell lyophilization, requires small sample size, short sample preparation time, provides both quantitative and qualitative information. Coupling with mass spectrometer (MS) detector enables tentative identification of novel PHA monomers, applicable for quantitative and qualitative analysis of mcl-PHA monomers | Low separation power that is currently limited to analysis of scl-PHA monomers unless coupled to MS detector, unable to distinguish between homogenous PHA and PHA copolymer | [129,130] |
| 10–25 mg biomass or 2 mg PHA | Propanolic digestion with propanol and concentrated sulfuric acid at 90 °C for 1 h to yield a mixture of monomeric acids and propionyl esters | Ion chromatography (IC) analysis with an anion trap column and a conductivity detector | |||||
| 2 mg PHA | Reductive depolymerization by dissolution of PHA in toluene, followed by addition of lithium aluminum hydride in tetrahydrofuran (THF) with 15 min of gentle agitation at room temperature to yield 1,3-diols | HPLC-MS analysis with a C18 column | |||||
| Gas chromatography (GC) | PHA monomeric units | 5–15 mg biomass or 0.15–15 mg PHA | Methanolysis with either sulfuric acid/methanol or boron trifluoride/methanol at 100 °C for 2 h–4 h to yield methyl esters or propanolysis with hydrochloric acid/propanol at 80 °C for 20 h to yield propyl esters | Analysis with a Supelco SPB-35 or DB-5 column using a flamed ionization detector (FID), or with a HP-5MS column using a MS detector | High separation power, high sensitivity, provides both quantitative and qualitative information, and can be applied for tentative identification of novel PHA monomers when coupled to MS detector | Requires cell lyophilization, long sample preparation time requiring the use of hazardous and volatile solvents, unable to distinguish between homogenous PHA and PHA copolymer | [43,131,132,133,134] |
4.1. Detection of PHA in Biomass and PHA Production Capacity
4.2. Quantification and Characterization of PHA in Biomass
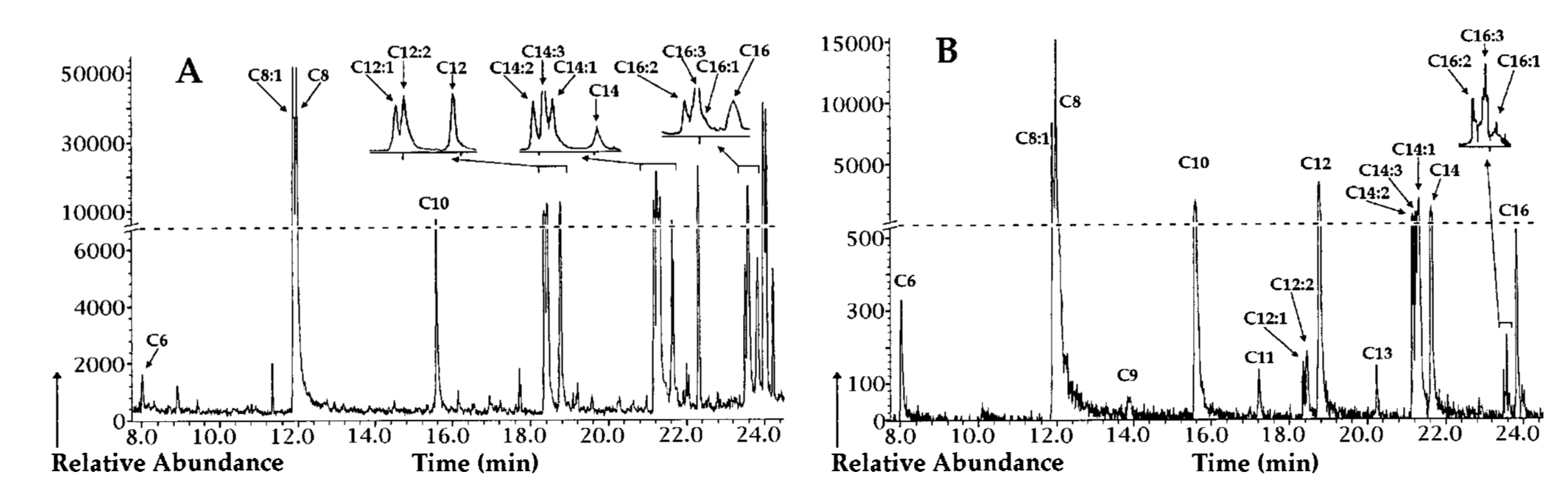
5. PHA Polymer Extraction Methods
| Method | Chemical | Species | Conditions | Purity and recovery | Reference |
|---|---|---|---|---|---|
| Solvent extraction | Chloroform | Cupriavidus necator (DSM 545) | Mixing continuously at 25 °C for 12 h | Purity: 94.0%-96.0%
Recovery: 65.0%-70.0% | [144] |
| Methylene chloride | Cupriavidus necator (DSM 545) | Mixing continuously at 25 °C for 12 h | Purity: 95.0%-98.0%
Recovery: 24.0%-25.0% | ||
| 1,2-Dichloroethane | Cupriavidus necator (DSM 545) | Mixing continuously at 25 °C for 12 h | Purity: 93.0%-98.0%
Recovery: 66.0%-70.0% | ||
| Acetone | Haloferax mediterranei (DSM 1411) | Mixing continuously at 120 °C, 7 bar for 20 min under anaerobic conditions followed by filtering hot solution and cooling it down at 4 °C to precipitate polymer | Purity: 98.4%
Recovery: 96.8% | [145] | |
| Medium-chain-length alcohols | Cupriavidus necator and Burkholderia sp. | Multi-stage extraction process in continuous-stirred tank reactors. Remove cell debris from the extract and cool extract to recover polymer | Purity: > 98.0%
Recovery: 95.0% | [146] | |
| Hypochlorite digestion | Sodium hypochlorite | Cupriavidus necator (DSM 545) | Biomass concentration: 10-40 g/L;
pH: 8-13.6; Temperature: 0-25 °C; Digestion time: 10 min-6 h; Hypochlorite concentration: 1%-10.5% weight/volume (w/v) | Purity: 90.0%-98.0%
Recovery: 90.0%-95.0% | [147] |
| Sodium hypochlorite and chloroform | Cupriavidus necator (NCIMB 11599) and recombinant Escherichia coli | Biomass concentration: 1% (w/v);
Temperature: 30 °C; Digestion time: 1 h; Hypochlorite concentration: 3%-20% volume/volume (v/v) | Purity: 86.0%
Recovery: NG Purity: 93.0% Recovery: NG | [153] | |
| Enzyme digestion | Trysin, bromelain, pancreatin | Cupriavidus necator (DSM 545) | Digestion with 2% trypsin (50 °C, pH 9.0, 1 h) or 2% bromelain (50 °C, pH 4.75, 10 h) or 2% pancreatin (50 °C, pH 8.0, 8 h), followed by centrifugation and washing with 0.85% saline solution | Purity: 87.7%-90.3%
Recovery: NG | [149] |
| Method | Chemical | Species | Conditions | Purity and recovery | Reference |
|---|---|---|---|---|---|
| Solvent | Chloroform | Pseudomonas oleovorans (strains NRRL B-14682, NRRL B-14683, and NRRL B-778) | 30 °C overnight at 250 rpm | NG | [150] |
| Chloroform | Pseudomonas oleovorans (NRRL B-14683), Pseudomonas resinovorans (NRRL B-2649), Pseudomonas citronellolis (NRRL B-2504), and Pseudomonas putida KT2442 | Soxhlet extraction for 24 h | NG | [154,155] | |
| Chloroform | Pseudomonas putida IPT 046 | Soxhlet extraction for 6 h | NG | [156] | |
| Chloroform | Pseudomonas aeruginosa 42A2 (NCIMB 40045) | 100 °C for 3 h in screw cap tubes for small quantities or in a soxhlet apparatus for large amounts of cell material | NG | [157] | |
| Dichlorome-thane | Pseudomonas oleovorans (ATCC 29347) | Soxhlet extraction at 60 °C for 5 h | Purity: > 98.0% Recovery: NG | [151] | |
| Acetone | Pseudomonas putida KT2440 (ATCC 47054) | 22 °C for 24 h at 170 rpm | Purity: 80.0%–90.0% Recovery: 60.0%–80.0% | [152] | |
| Enzyme digestion | Alcalase, SDS, EDTA, lysozyme | Pseudomonas putida | Digestion with alcalase and SDS at pH 8.5 and 55 °C followed by further treatments with EDTA and lysozyme at pH 7 and 30 °C | Purity: 92.6% Recovery: nearly 90.0% | [158] |
| Pseudomonas putida KT2442 | Digestion with excess alcalase, EDTA and SDS at pH 8.5 and 55 °C followed by diafiltration | Purity: > 95.0% Recovery: NG | [159,160] |
| Characteristic | Index | Method | Sample | Sample preparation | Typical conditions | Reference |
|---|---|---|---|---|---|---|
| PHA monomeric composition | Chemical derivative of PHA monomers | LC | Refer to Table 3 | |||
| GC | Refer to Table 3 | |||||
| PHA polymeric composition | Topology and functional groups of PHA molecule | 1D-Nuclear magnetic resonance (NMR) | 5–10 mg PHA for 1H-NMR and 20– 30 mg PHA for 13C-NMR | Dissolution of PHA polymer in 0.7 mL deuterated chloroform (CDCl3) containing 0.03% (v/v) tetramethylsilane (TMS) | 1H-NMR at 200 or 300 MHz and 13C-NMR measurements at 75.4 MHz at 20 °C with a sampling pulse of 3 s. Chemical shifts were referenced to the residual proton peak of CDCl3 at 7.26 ppm and to the carbon peak of CDCl3 at 77 ppm | [82] |
| 2D-NMR | 10 mg PHA for homonuclear 2D-NMR and 40–50 mg PHA for heteronuclear 2D-NMR | Refer to above “1D-NMR” | For homonuclear COSY and TOCSY, 16 scans were accumulated per increment over a spectral width of 7.8 ppm. For heteronuclear HSQC, 48 scans were accumulated per increment over a spectral width of 7.8 ppm for 1H and 75 ppm for 13C. For heteronuclear HMBC spectrum, 64 scans were acquired with the long-range coupling delay set for 8 Hz | [161] | ||
| PHA polymeric composition | Topology and functional groups of PHA molecule | Matrix assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF-MS) | 1 µg–1 mg PHA | The matrix used was either dithranol or dihydroxybenzoic acid (DHB) at a concentration of 10 mg mL−1 in THF. 1 mg mL−1 PHA solution (in chloroform) was mixed with equal volume THF. The matrix solution and the PHA solution were subsequently mixed in a 5:2 ratio (matrix/sample). 1 µL mixture was deposited onto the stainless steel sample holder. The solvent was allowed to air-dry before loading the sample plate into the MALDI ion source | MALDI-TOF-MS with 25 kV acceleration and detection in the positive-ion high-resolution reflection mode | [162] |
| Molecular distribution | Polydispersity, molecular mass and molecular mass distribution | Gel permeation chromatography (GPC) | 0.1–1 mg PHA | Dissolution of PHA polymer in 1 mL of THF | Analysis conducted with a refractive index detector (47 °C, 2.0 bar) and a solvent-compatible GPC column. THF, containing 250 ppm of 2,6-di-tert-butyl-4-methylphenol (BHT) as inhibitor, was used as an eluent at a flow rate of 0.5 mL min−1 and 40 °C | [43] |
| Dissolution of PHA polymer in chloroform | Analysis conducted with a differential refractive index detector (30 °C), a UV dual wavelength absorbance detector, and a combination of four GPC columns series. Chloroform was used as an eluent with a flow rate of 1.0 mL min−1 | [163] | ||||
| MALDI-TOF-MS | Refer to above “PHA polymeric composition” | |||||
| Thermal properties | Glass transition temperature and melting temperature | Differential scanning calorimetry (DSC) | 10 mg PHA | - | Heat sample from −100 °C– 400 °C at a heating rate of 10 °C min−1 under purified air or nitrogen gas with a flow rate of 80 mL min−1 | [43] |
| Differential thermal analysis (DTA) | 5 mg PHA | - | Crystallization was carried out isothermally by abruptly quenching the samples from melt to the crystallization temperature, at which the samples were annealed for 10 min. Melting of semicrystalline samples was performed by heating at a rate of 5 °C min−1 | [164] | ||
| Thermodegradation temperature | Thermogravimetric analysis (TGA) | 10 mg PHA | - | Heat sample from room temperature to 700 °C at a heating rate of 10 °C min−1 under purified air or nitrogen gas with a flow rate of 50 mL min−1 | [43] | |
| Crystallinity | Melting enthalpy | DSC | Refer to above “Thermal properties” | |||
| Infrared absorption bands correlated to crystallinity | FTIR | 5–10 mg PHA | Dissolve PHA in chloroform, apply onto KRS-5 window and blow dry to evaporate solvent. Alternatively, mix PHA with potassium bromide (KBr) powder and pelletize | Refer to Table 3 | [127,165] | |
| Place PHA sample between two pieces of barium fluoride slides | Melt sample at 100 °C for 2 min in FTIR hot stage under the protection of dry nitrogen gas. Quench the amorphous sample to 58 and 28 °C by a flow of liquid nitrogen and maintain at these temperatures for 30 min for isothermal melt-crystallization before re-heating at 1 °C min−1 | [166] | ||||
| Crystallinity | Diffraction intensity correlated to crystallinity | X-ray diffraction | Dry polymer powder | - | Diffractogram of the sample powder were measured at room temperature by an imaging plate diffractometer with Cu-Kα radiation (wavelength = 0.1542 nm) as an incident X-ray source emitted by a X-ray generator with a Ni filter. The scattering angle range of 2θ = 10°–40° at a scan speed of 3° min−1 | [156] |
| Mechanical properties | Tensile strength, tensile stress, percent elongation, modulus of elasticity | Mechanical testing machine of the constant-rate-of-crosshead-movement type with extensometer and micrometers | Polymer thickness 1–14 mm, width 19–29 mm, length 165–246 mm | Test samples were prepared using a hydraulic press at 150 °C and conditioned at a relative humidity of 50% ± 5% for 24 h prior to measurements | Perform stress-strain test at room temperature with a strain rate of 20 mm min−1 | [167] |
6. Techniques for PHA Polymer Characterization
6.1. Monomeric Composition and Distribution
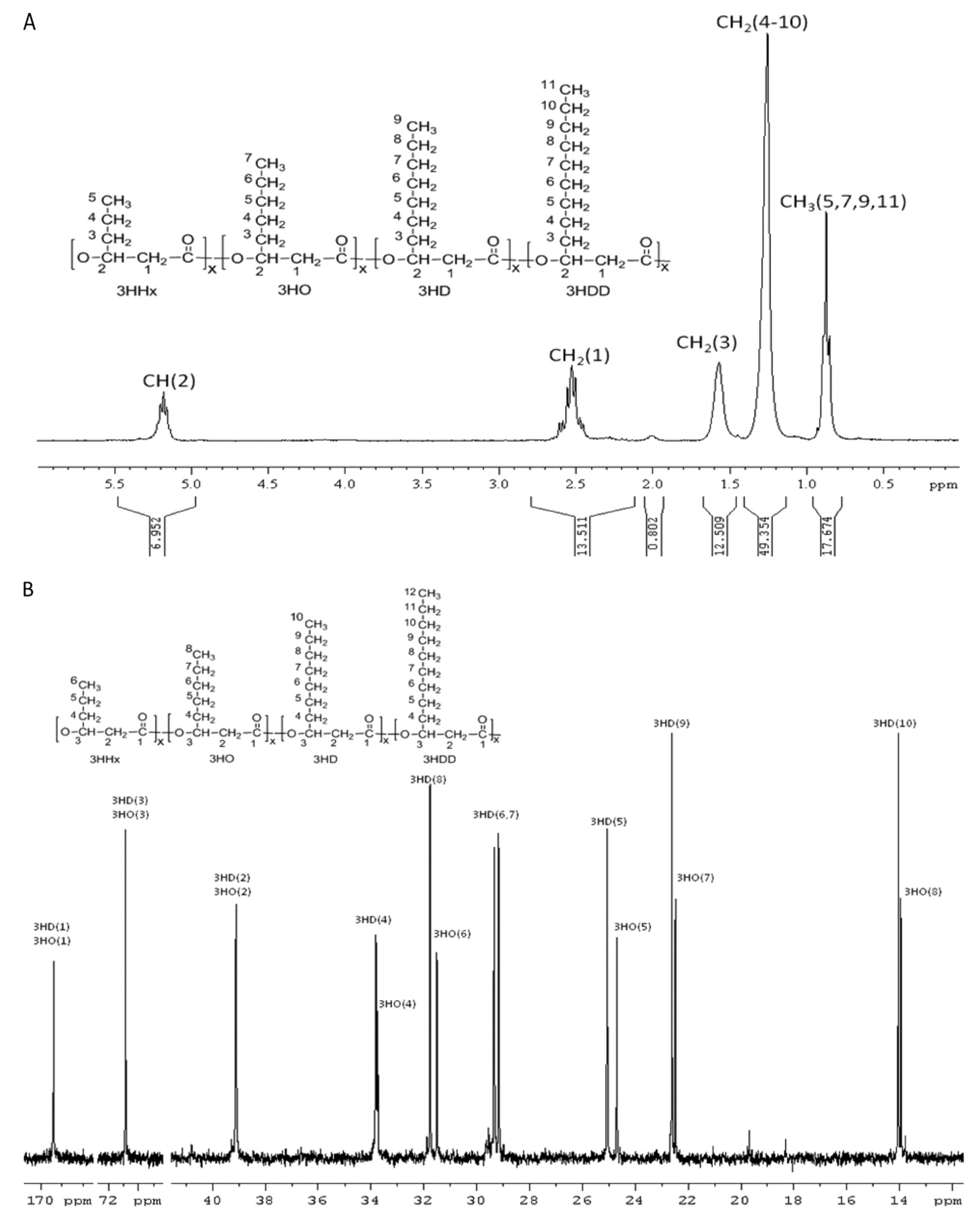
6.2. Molecular Mass (Mw), Molecular Mass Distribution (Mn), and Polydispersity Index (PDI)
6.3. Thermal Properties
6.4. Crystallinity
6.5. Mechanical Properties
7. Conclusions
Acknowledgment
Conflicts of Interest
References
- Anderson, A.J.; Dawes, E.A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 1990, 54, 450–472. [Google Scholar]
- Shah, A.A.; Hasan, F.; Hameed, A.; Ahmed, S. Biological degradation of plastics: A comprehensive review. Biotechnol. Adv. 2008, 26, 246–265. [Google Scholar] [CrossRef]
- Khanna, S.; Srivastava, A.K. Recent advances in microbial polyhydroxyalkanoates. Process Biochem. 2005, 40, 607–619. [Google Scholar] [CrossRef]
- Lu, J.; Tappel, R.C.; Nomura, C.T. Mini-review: Biosynthesis of poly(hydroxyalkanoates). Polym. Rev. 2009, 49, 226–248. [Google Scholar] [CrossRef]
- Zinn, M.; Hany, R. Tailored material properties of polyhydroxyalkanoates through biosynthesis and chemical modification. Adv. Eng. Mater. 2005, 7, 408–411. [Google Scholar] [CrossRef]
- Escapa, I.F.; Morales, V.; Martino, V.P.; Pollet, E.; Avérous, L.; García, J.L.; Prieto, M.A. Disruption of β-oxidation pathway in Pseudomonas putida KT2442 to produce new functionalized PHAs with thioester groups. Appl. Microbiol. Biotechnol. 2011, 89, 1583–1598. [Google Scholar] [CrossRef]
- Rai, R.; Keshavarz, T.; Roether, J.A.; Boccaccini, A.R.; Roy, I. Medium chain length polyhydroxyalkanoates, promising new biomedical materials for the future. Mater. Sci. Eng. R. Rep. 2011, 72, 29–47. [Google Scholar] [CrossRef]
- De Roo, G.; Kellerhals, M.B.; Ren, Q.; Witholt, B.; Kessler, B. Production of chiral R-3-hydroxyalkanoic acids and R-3-hydroxyalkanoic acid methylesters via hydrolytic degradation of polyhydroxyalkanoate synthesized by Pseudomonads. Biotechnol. Bioeng. 2002, 77, 717–722. [Google Scholar] [CrossRef]
- Philip, S.; Keshavarz, T.; Roy, I. Polyhydroxyalkanoates: Biodegradable polymers with a range of applications. J. Chem. Technol. Biotechnol. 2007, 82, 233–247. [Google Scholar] [CrossRef]
- Olivera, E.R.; Arcos, M.; Naharro, G.; Luengo, J.M. Unusual PHA biosynthesis. In Plastics from Bacteria: Natural Functions and Applications; Chen, G.-Q., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 14, pp. 133–186. [Google Scholar]
- Chen, G.-Q. Plastics completely synthesized by bacteria: Polyhydroxyalkanoates. In Plastics from Bacteria: Natural Functions and Applications; Chen, G.-Q., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 14, pp. 17–37. [Google Scholar]
- Kadouri, D.; Jurkevitch, E.; Okon, Y.; Castro-Sowinski, S. Ecological and agricultural significance of bacterial polyhydroxyalkanoates. Crit. Rev. Microbiol. 2005, 31, 55–67. [Google Scholar] [CrossRef]
- Madison, L.L.; Huisman, G.W. Metabolic engineering of poly(3-hydroxyalkanoates): From DNA to plastic. Microbiol. Mol. Biol. Rev. 1999, 63, 21–53. [Google Scholar]
- Rothermich, M.M.; Guerrero, R.; Lenz, R.W.; Goodwin, S. Characterization, seasonal occurrence, and diel fluctuation of poly(hydroxyalkanoate) in photosynthetic microbial mats. Appl. Environ. Microbiol. 2000, 66, 4279–4291. [Google Scholar] [CrossRef]
- Peplinski, K.; Ehrenreich, A.; Döring, C.; Bömeke, M.; Reinecke, F.; Hutmacher, C.; Steinbüchel, A. Genome-wide transcriptome analyses of the “Knallgas” bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology 2010, 156, 2136–2152. [Google Scholar] [CrossRef]
- Shimizu, R.; Chou, K.; Orita, I.; Suzuki, Y.; Nakamura, S.; Fukui, T. Detection of phase-dependent transcriptomic changes and Rubisco-mediated CO2 fixation into poly (3-hydroxybutyrate) under heterotrophic condition in Ralstonia eutropha H16 based on RNA-seq and gene deletion analyses. BMC Microbiol. 2013, 13, 169. [Google Scholar] [CrossRef]
- Yamane, T. Yield of poly-D(-)-3-hydroxybutyrate from various carbon sources: A theoretical study. Biotechnol. Bioeng. 1993, 41, 165–170. [Google Scholar] [CrossRef]
- Steinbüchel, A.; Hein, S. Biochemical and molecular basis of microbial synthesis of polyhydroxyalkanoates in microorganisms. In Biopolyesters; Babel, W., Steinbüchel, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; Volume 71, pp. 81–123. [Google Scholar]
- Ratledge, C.; Kristiansen, B. Basic Biotechnology, 2nd ed.; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]
- Jung, Y.M.; Lee, Y.H. Utilization of oxidative pressure for enhanced production of poly-β-hydroxybutyrate and poly(3-hydroxybutyrate-3-hydroxyvalerate) in Ralstonia eutropha. J. Biosci. Bioeng. 2000, 90, 266–270. [Google Scholar]
- Khosravi-Darani, K.; Mokhtari, Z.-B.; Amai, T.; Tanaka, K. Microbial production of poly(hydroxybutyrate) from C1 carbon sources. Appl. Microbiol. Biotechnol. 2013, 97, 1407–1424. [Google Scholar] [CrossRef]
- Raberg, M.; Bechmann, J.; Brandt, U.; Schlüter, J.; Uischner, B.; Voigt, B.; Hecker, M.; Steinbüchel, A. Versatile metabolic adaptations of Ralstonia eutropha H16 to a loss of PdhL, the E3 component of the pyruvate dehydrogenase complex. Appl. Environ. Microbiol. 2011, 77, 2254–2263. [Google Scholar] [CrossRef]
- Peoples, O.P.; Sinskey, A.J. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J. Biol. Chem. 1989, 264, 15298–15303. [Google Scholar]
- Lee, S.; Jeon, E.; Yun, H.S.; Lee, J. Improvement of fatty acid biosynthesis by engineered recombinant Escherichia coli. Biotechnol. Bioprocess Eng. 2011, 16, 706–713. [Google Scholar] [CrossRef]
- Nomura, C.T.; Taguchi, K.; Taguchi, S.; Doi, Y. Coexpression of genetically engineered 3-ketoacyl-ACP synthase III (fabH) and polyhydroxyalkanoate synthase (phaC) genes leads to short-chain-length-medium-chain-length polyhydroxyalkanoate copolymer production from glucose in Escherichia coli JM109. Appl. Environ. Microbiol. 2004, 70, 999–1007. [Google Scholar] [CrossRef]
- Ren, Q.; Sierro, N.; Witholt, B.; Kessler, B. FabG, an NADPH-dependent 3-ketoacyl reductase of Pseudomonas aeruginosa, provides precursors for medium-chain-length poly-3-hydroxyalkanoate biosynthesis in Escherichia coli. J. Bacteriol. 2000, 182, 2978–2981. [Google Scholar] [CrossRef]
- Valentin, H.E.; Dennis, D. Production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) in recombinant Escherichia coli grown on glucose. J. Biotechnol. 1997, 58, 33–38. [Google Scholar] [CrossRef]
- Xie, W.P.; Chen, G.-Q. Production and characterization of terpolyester poly(3-hydroxybutyrate-co-4-hydroxybutyrate-co-3-hydroxyhexanoate) by recombinant Aeromonas hydrophila 4AK4 harboring genes phaPCJ. Biochem. Eng. J. 2008, 38, 384–389. [Google Scholar] [CrossRef]
- Valentin, H.E.; Steinbüchel, A. Accumulation of poly(3-hydroxybutyric acid-co-3-hydroxyvaleric acid-co-4-hydroxyvaleric acid) by mutants and recombinant strains of Alcaligenes eutrophus. J. Polym. Environ. 1995, 3, 169–175. [Google Scholar] [CrossRef]
- Aldor, I.S.; Kim, S.W.; Jones Prather, K.L.; Keasling, J.D. Metabolic engineering of a novel propionate-independent pathway for the production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in recombinant Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 2002, 68, 3848–3854. [Google Scholar] [CrossRef]
- Valentin, H.E.; Dennis, D. Metabolic pathway for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) formation in Nocardia corallina: Inactivation of mutB by chromosomal integration of a kanamycin resistance gene. Appl. Environ. Microbiol. 1996, 62, 372–379. [Google Scholar]
- Satoh, H.; Mino, T.; Matsuo, T. PHA production by activated sludge. Int. J. Biol. Macromol. 1999, 25, 105–109. [Google Scholar] [CrossRef]
- Slater, S.; Houmiel, K.L.; Tran, M.; Mitsky, T.A.; Taylor, N.B.; Padgette, S.R.; Gruys, K.J. Multiple β-ketothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J. Bacteriol. 1998, 180, 1979–1987. [Google Scholar]
- Chohan, S.N.; Copeland, L. Acetoacetyl coenzyme a reductase and polyhydroxybutyrate synthesis in Rhizobium (Cicer) sp. strain CC 1192. Appl. Environ. Microbiol. 1998, 64, 2859–2863. [Google Scholar]
- Hume, A.R.; Nikodinovic-Runic, J.; O’Connor, K.E. FadD from Pseudomonas putida CA-3 is a true long-chain fatty acyl coenzyme A synthetase that activates phenylalkanoic and alkanoic acids. J. Bacteriol. 2009, 191, 7554–7565. [Google Scholar] [CrossRef]
- Yuan, M.-Q.; Shi, Z.-Y.; Wei, X.-X.; Wu, Q.; Chen, S.-F.; Chen, G.-Q. Microbial production of medium-chain-length 3-hydroxyalkanoic acids by recombinant Pseudomonas putida KT2442 harboring genes fadL, fadD and phaZ. FEMS Microbiol. Lett. 2008, 283, 167–175. [Google Scholar] [CrossRef]
- Mittendorf, V.; Robertson, E.J.; Leech, R.M.; Krüger, N.; Steinbüchel, A.; Poirier, Y. Synthesis of medium-chain-length polyhydroxyalkanoates in Arabidopsis thaliana using intermediates of peroxisomal fatty acid β-oxidation. Proc. Natl. Acad. Sci. USA 1998, 95, 13397–13402. [Google Scholar] [CrossRef]
- Sato, S.; Kanazawa, H.; Tsuge, T. Expression and characterization of (R)-specific enoyl coenzyme A hydratases making a channeling route to polyhydroxyalkanoate biosynthesis in Pseudomonas putida. Appl. Microbiol. Biotechnol. 2011, 90, 951–959. [Google Scholar] [CrossRef]
- Ouyang, S.-P.; Luo, R.C.; Chen, S.-S.; Liu, Q.; Chung, A.; Wu, Q.; Chen, G.-Q. Production of polyhydroxyalkanoates with high 3-hydroxydodecanoate monomer content by fadB and fadA knockout mutant of Pseudomonas putida KT2442. Biomacromolecules 2007, 8, 2504–2511. [Google Scholar] [CrossRef]
- Zheng, L.Z.; Li, Z.; Tian, H.-L.; Li, M.; Chen, G.-Q. Molecular cloning and functional analysis of (R)-3-hydroxyacyl-acyl carrier protein:coenzyme A transacylase from Pseudomonas mendocina LZ. FEMS Microbiol. Lett. 2005, 252, 299–307. [Google Scholar] [CrossRef]
- Brzostowicz, P.B.; Blasko, M.B.; Rouvière, P.R. Identification of two gene clusters involved in cyclohexanone oxidation in Brevibacterium epidermidis strain HCU. Appl. Microbiol. Biotechnol. 2002, 58, 781–789. [Google Scholar] [CrossRef]
- Poli, A.; Di Donato, P.; Abbamondi, G.R.; Nicolaus, B. Synthesis, production, and biotechnological applications of exopolysaccharides and polyhydroxyalkanoates by archaea. Archaea 2011, 2011, 1–13. [Google Scholar]
- Galia, M.B. Isolation and analysis of storage compounds. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3725–3741. [Google Scholar]
- Verlinden, R.A.J.; Hill, D.J.; Kenward, M.A.; Williams, C.D.; Radecka, I. Bacterial synthesis of biodegradable polyhydroxyalkanoates. J. Appl. Microbiol. 2007, 102, 1437–1449. [Google Scholar] [CrossRef]
- Koller, M.; Atlić, A.; Dias, M.; Reiterer, A.; Braunegg, G. Microbial PHA production from waste raw materials. In Plastics from Bacteria: Natural Functions and Applications; Chen, G.-Q., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 14, pp. 85–119. [Google Scholar]
- Yu, P.H.; Chua, H.; Huang, A.L.; Lo, W.; Chen, G.-Q. Conversion of food industrial wastes into bioplastics. Appl. Biochem. Biotechnol. 1998, 70, 603–614. [Google Scholar]
- Wang, F.; Lee, S.Y. Poly(3-hydroxybutyrate) production with high productivity and high polymer content by a fed-batch culture of Alcaligenes latus under nitrogen limitation. Appl. Environ. Microbiol. 1997, 63, 3703–3706. [Google Scholar]
- Grothe, E.; Moo-Young, M.; Chisti, Y. Fermentation optimization for the production of poly(beta-hydroxybutyric acid) microbial thermoplastic. Enzyme Microb. Technol. 1999, 25, 132–141. [Google Scholar] [CrossRef]
- Yamane, T.; Fukunaga, M.; Lee, Y.W. Increased PHB productivity by high-cell-density fed-batch culture of Alcaligenes latus, a growth-associated PHB producer. Biotechnol. Bioeng. 1996, 50, 197–202. [Google Scholar] [CrossRef]
- Gomez, J.; Rodrigues, M.; Alli, R.; Torres, B.; Netto, C.B.; Oliveira, M.; Da Silva, L. Evaluation of soil Gram-negative bacteria yielding polyhydroxyalkanoic acids from carbohydrates and propionic acid. Appl. Microbiol. Biotechnol. 1996, 45, 785–791. [Google Scholar] [CrossRef]
- Lasemi, Z.; Darzi, G.N.; Baei, M.S. Media optimization for poly(β-hydroxybutyrate) production using Azotobacter Beijerinckii. Int. J. Polym. Mater. 2012, 62, 265–269. [Google Scholar] [CrossRef]
- Pan, W.; Perrotta, J.A.; Stipanovic, A.J.; Nomura, C.T.; Nakas, J.P. Production of polyhydroxyalkanoates by Burkholderia cepacia ATCC 17759 using a detoxified sugar maple hemicellulosic hydrolysate. J. Ind. Microbiol. Biotechnol. 2012, 39, 459–469. [Google Scholar] [CrossRef]
- Zhu, C.; Nomura, C.T.; Perrotta, J.A.; Stipanovic, A.J.; Nakas, J.P. Production and characterization of poly-3-hydroxybutyrate from biodiesel-glycerol by Burkholderia cepacia ATCC 17759. Biotechnol. Prog. 2010, 26, 424–430. [Google Scholar]
- Chee, J.-Y.; Tan, Y.; Samian, M.-R.; Sudesh, K. Isolation and characterization of a Burkholderia sp. USM (JCM15050) capable of producing polyhydroxyalkanoate (PHA) from triglycerides, fatty acids and glycerols. J. Polym. Environ. 2010, 18, 584–592. [Google Scholar] [CrossRef]
- Qi, Q.S.; Rehm, B.H.A. Polyhydroxybutyrate biosynthesis in Caulobacter crescentus: Molecular characterization of the polyhydroxybutyrate synthase. Microbiology 2001, 147, 3353–3358. [Google Scholar]
- Valentin, H.E.; Lee, E.Y.; Choi, C.Y.; Steinbüchel, A. Identification of 4-hydroxyhexanoic acid as a new constituent of biosynthetic polyhydroxyalkanoic acids from bacteria. Appl. Microbiol. Biotechnol. 1994, 40, 710–716. [Google Scholar] [CrossRef]
- Fukui, T.; Doi, Y. Efficient production of polyhydroxyalkanoates from plant oils by Alcaligenes eutrophus and its recombinant strain. Appl. Microbiol. Biotechnol. 1998, 49, 333–336. [Google Scholar] [CrossRef]
- Chakraborty, P.; Gibbons, W.; Muthukumarappan, K. Conversion of volatile fatty acids into polyhydroxyalkanoate by Ralstonia eutropha. J. Appl. Microbiol. 2009, 106, 1996–2005. [Google Scholar]
- Sonnleitner, B.; Heinzle, E.; Braunegg, G.; Lafferty, R.M. Formal kinetics of poly-β-hydroxybutyric acid (PHB) production in Alcaligenes eutrophus H 16 and Mycoplana rubra R 14 with respect to the dissolved oxygen tension in ammonium-limited batch cultures. Eur. J. Appl. Microbiol. 1979, 7, 1–10. [Google Scholar] [CrossRef]
- Ishizaki, A.; Tanaka, K. Production of poly-β-hydroxybutyric acid from carbon dioxide by Alcaligenes eutrophus ATCC 17697T. J. Ferment. Bioeng. 1991, 71, 254–257. [Google Scholar] [CrossRef]
- Kim, B.S.; Lee, S.C.; Lee, S.Y.; Chang, H.N.; Chang, Y.K.; Woo, S.I. Production of poly(3-hydroxybutyric acid) by fed-batch culture of Alcaligenes eutrophus with glucose concentration control. Biotechnol. Bioeng. 1994, 43, 892–898. [Google Scholar] [CrossRef]
- Haas, R.; Jin, B.; Zepf, F.T. Production of poly(3-hydroxybutyrate) from waste potato starch. Biosci. Biotechnol. Biochem. 2008, 72, 253–256. [Google Scholar]
- Beaulieu, M.; Beaulieu, Y.; Melinard, J.; Pandian, S.; Goulet, J. Influence of ammonium salts and cane molasses on growth of Alcaligenes eutrophus and production of polyhydroxybutyrate. Appl. Environ. Microbiol. 1995, 61, 165–169. [Google Scholar]
- Du, G.C.C.; Chen, J.; Yu, J.; Lun, S.Y. Feeding strategy of propionic acid for production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) with Ralstonia eutropha. Biochem. Eng. J. 2001, 8, 103–110. [Google Scholar] [CrossRef]
- Cavalheiro, J.M.B.T.; de Almeida, M.C.M.D.; Grandfils, C.; da Fonseca, M.M.R. Poly(3-hydroxybutyrate) production by Cupriavidus necator using waste glycerol. Process Biochem. 2009, 44, 509–515. [Google Scholar] [CrossRef]
- Quillaguamán, J.; Hashim, S.; Bento, F.; Mattiasson, B.; Hatti-Kaul, R. Poly(β-hydroxybutyrate) production by a moderate halophile, Halomonas boliviensis LC1 using starch hydrolysate as substrate. J. Appl. Microbiol. 2005, 99, 151–157. [Google Scholar] [CrossRef]
- Povolo, S.; Romanelli, M.G.; Basaglia, M.; Ilieva, V.I.; Corti, A.; Morelli, A.; Chiellini, E.; Casella, S. Polyhydroxyalkanoate biosynthesis by Hydrogenophaga pseudoflava DSM1034 from structurally unrelated carbon sources. New Biotechnol. 2013, 30, 629–634. [Google Scholar] [CrossRef]
- Koller, M.; Hesse, P.; Bona, R.; Kutschera, C.; Atlic, A.; Braunegg, G. Potential of various archae- and eubacterial strains as industrial polyhydroxyalkanoate producers from whey. Macromol. Biosci. 2007, 7, 218–226. [Google Scholar] [CrossRef]
- Bourque, D.; Pomerleau, Y.; Groleau, D. High-cell-density production of poly-β-hydroxybutyrate (PHB) from methanol by Methylobacterium extorquens: Production of high-molecular-mass PHB. Appl. Microbiol. Biotechnol. 1995, 44, 367–376. [Google Scholar] [CrossRef]
- Mokhtari-Hosseini, Z.B.; Vasheghani-Farahani, E.; Heidarzadeh-Vazifekhoran, A.; Shojaosadati, S.A.; Karimzadeh, R.; Darani, K.K. Statistical media optimization for growth and PHB production from methanol by a methylotrophic bacterium. Bioresour. Technol. 2009, 100, 2436–2443. [Google Scholar] [CrossRef]
- Mokhtari-Hosseini, Z.B.; Vasheghani-Farahani, E.; Shojaosadati, S.A.; Karimzadeh, R.; Heidarzadeh-Vazifekhoran, A. Effect of feed composition on PHB production from methanol by HCDC of Methylobacterium extorquens (DSMZ 1340). J. Chem. Technol. Biotechnol. 2009, 84, 1136–1139. [Google Scholar] [CrossRef]
- Wendlandt, K.D.; Jechorek, M.; Helm, J.; Stottmeister, U. Production of PHB with a high molecular mass from methane. Polym. Degrad. Stabil. 1998, 59, 191–194. [Google Scholar] [CrossRef]
- Smit, A.M.; Strabala, T.J.; Peng, L.; Rawson, P.; Lloyd-Jones, G.; Jordan, T.W. Proteomic phenotyping of Novosphingobium nitrogenifigens reveals a robust capacity for simultaneous nitrogen fixation, polyhydroxyalkanoate production, and resistance to reactive oxygen species. Appl. Environ. Microbiol. 2012, 78, 4802–4815. [Google Scholar] [CrossRef]
- Yamane, T.; Chen, X.; Ueda, S. Growth-associated production of poly(3-hydroxyvalerate) from n-pentanol by a methylotrophic bacterium, Paracoccus denitrificans. Appl. Environ. Microbiol. 1996, 62, 380–384. [Google Scholar]
- Tripathi, A.D.; Yadav, A.; Jha, A.; Srivastava, S.K. Utilizing of sugar refinery waste (cane molasses) for production of bio-plastic under submerged fermentation process. J. Polym. Environ. 2012, 20, 446–453. [Google Scholar] [CrossRef]
- Guzik, M.W.; Kenny, S.T.; Duane, G.F.; Casey, E.; Woods, T.; Babu, R.P.; Nikodinovic-Runic, J.; Murray, M.; O’Connor, K.E. Conversion of post consumer polyethylene to the biodegradable polymer polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 2014. In Press. [Google Scholar]
- Kenny, S.T.; Nikodinovic-Runic, J.; Kaminsky, W.; Woods, T.; Babu, R.P.; Keely, C.M.; Blau, W.; O’Connor, K.E. Up-cycling of PET (polyethylene terephthalate) to the biodegradable plastic PHA (polyhydroxyalkanoate). Environ. Sci. Technol. 2008, 42, 7696–7701. [Google Scholar] [CrossRef]
- Lee, E.; Jendrossek, D.; Schirmer, A.; Choi, C.; Steinbüchel, A. Biosynthesis of copolyesters consisting of 3-hydroxybutyric acid and medium-chain-length 3-hydroxyalkanoic acids from 1,3-butanediol or from 3-hydroxybutyrate by Pseudomonas sp. A33. Appl. Microbiol. Biotechnol. 1995, 42, 901–909. [Google Scholar] [CrossRef]
- Nikodinovic-Runic, J.; Casey, E.; Duane, G.F.; Mitic, D.; Hume, A.R.; Kenny, S.T.; O’Connor, K.E. Process analysis of the conversion of styrene to biomass and medium chain length polyhydroxyalkanoate in a two-phase bioreactor. Biotechnol. Bioeng. 2011, 108, 2447–2455. [Google Scholar] [CrossRef]
- Ward, P.G.; Goff, M.; Donner, M.; Kaminsky, W.; O'Connor, K.E. A two step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ. Sci. Technol. 2006, 40, 2433–2437. [Google Scholar] [CrossRef]
- Lageveen, R.G.; Huisman, G.W.; Preusting, H.; Ketelaar, P.; Eggink, G.; Witholt, B. Formation of polyesters by Pseudomonas oleovorans: Effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl. Environ. Microbiol. 1988, 54, 2924–2932. [Google Scholar]
- Gross, R.A.; DeMello, C.; Lenz, R.W.; Brandl, H.; Fuller, R.C. The biosynthesis and characterization of poly(β-hydroxyalkanoates) produced by Pseudomonas oleovorans. Macromolecules 1989, 22, 1106–1115. [Google Scholar] [CrossRef]
- Elbahloul, Y.; Steinbuchel, A. Large-scale production of poly(3-hydroxyoctanoic acid) by Pseudomonas putida GPo1 and a simplified downstream process. Appl. Environ. Microbiol. 2009, 75, 643–651. [Google Scholar] [CrossRef]
- Sun, Z.; Ramsay, J.A.; Guay, M.; Ramsay, B.A. Carbon-limited fed-batch production of medium-chain-length polyhydroxyalkanoates from nonanoic acid by Pseudomonas putida KT2440. Appl. Microbiol. Biotechnol. 2007, 74, 69–77. [Google Scholar] [CrossRef]
- Davis, R.; Kataria, R.; Cerrone, F.; Woods, T.; Kenny, S.; O’Donovan, A.; Guzik, M.; Shaikh, H.; Duane, G.; Gupta, V.K.; et al. Conversion of grass biomass into fermentable sugars and its utilization for medium chain length polyhydroxyalkanoate (mcl-PHA) production by Pseudomonas strains. Bioresour. Technol. 2013, 150, 202–209. [Google Scholar] [CrossRef]
- Nikodinovic, J.; Kenny, S.T.; Babu, R.P.; Woods, T.; Blau, W.; O’Connor, K.E. The conversion of BTEX compounds by single and defined mixed cultures to medium-chain-length polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 2008, 80, 665–673. [Google Scholar] [CrossRef]
- Shahid, S.; Mosrati, R.; Ledauphin, J.; Amiel, C.; Fontaine, P.; Gaillard, J.-L.; Corroler, D. Impact of carbon source and variable nitrogen conditions on bacterial biosynthesis of polyhydroxyalkanoates: Evidence of an atypical metabolism in Bacillus megaterium DSM 509. J. Biosci. Bioeng. 2013, 116, 302–308. [Google Scholar] [CrossRef]
- Pantazaki, A.A.; Papaneophytou, C.P.; Pritsa, A.G.; Liakopoulou-Kyriakides, M.; Kyriakidis, D.A. Production of polyhydroxyalkanoates from whey by Thermus thermophilus HB8. Process Biochem. 2009, 44, 847–853. [Google Scholar] [CrossRef]
- Valappil, S.P.; Boccaccini, A.R.; Bucke, C.; Roy, I. Polyhydroxyalkanoates in Gram-positive bacteria: Insights from the genera Bacillus and Streptomyces. Antonie Van Leeuwenhoek 2007, 91, 1–17. [Google Scholar]
- Haywood, G.W.; Anderson, A.J.; Roger Williams, D.; Dawes, E.A.; Ewing, D.F. Accumulation of a poly(hydroxyalkanoate) copolymer containing primarily 3-hydroxyvalerate from simple carbohydrate substrates by Rhodococcus sp. NCIMB 40126. Int. J. Biol. Macromol. 1991, 13, 83–88. [Google Scholar] [CrossRef]
- Akar, A.; Akkaya, E.U.; Yesiladali, S.K.; Celikyilmaz, G.; Cokgor, E.U.; Tamerler, C.; Orhon, D.; Cakar, Z.P. Accumulation of polyhydroxyalkanoates by Microlunatus phosphovorus under various growth conditions. J. Ind. Microbiol. Biotechnol. 2006, 33, 215–220. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Pramanik, A.; Maji, S.K.; Haldar, S.; Mukhopadhyay, U.K.; Mukherjee, J. Utilization of vinasse for production of poly-3-(hydroxybutyrate-co-hydroxyvalerate) by Haloferax mediterranei. AMB Express 2012, 2, 1–10. [Google Scholar] [CrossRef]
- Koller, M.; Atlić, A.; Gonzalez‐Garcia, Y.; Kutschera, C.; Braunegg, G. Polyhydroxyalkanoate (PHA) biosynthesis from whey lactose. Macromol. Symp. 2008, 272, 87–92. [Google Scholar] [CrossRef]
- Hermann-Krauss, C.; Koller, M.; Muhr, A.; Fasl, H.; Stelzer, F.; Braunegg, G. Archaeal production of polyhydroxyalkanoate (PHA) co-and terpolyesters from biodiesel industry-derived by-products. Archaea 2013, 2013. [Google Scholar]
- Han, J.; Hou, J.; Liu, H.; Cai, S.; Feng, B.; Zhou, J.; Xiang, H. Wide distribution among halophilic archaea of a novel polyhydroxyalkanoate synthase subtype with homology to bacterial type III synthases. Appl. Environ. Microbiol. 2010, 76, 7811–7819. [Google Scholar] [CrossRef]
- Chen, G.-Q. Industrial production of PHA. In Plastics from Bacteria: Natural Functions and Applications; Chen, G.-Q., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 14, pp. 121–132. [Google Scholar]
- Chanprateep, S. Current trends in biodegradable polyhydroxyalkanoates. J. Biosci. Bioeng. 2010, 110, 621–632. [Google Scholar] [CrossRef]
- Poblete-Castro, I.; Becker, J.; Dohnt, K.; dos Santos, V.M.; Wittmann, C. Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 2012, 93, 2279–2290. [Google Scholar] [CrossRef]
- Greene, E.A.; Voordouw, G. Biodegradation of C5+ hydrocarbons by a mixed bacterial consortium from a C5+-contaminated site. Environ. Technol. 2004, 25, 355–363. [Google Scholar] [CrossRef]
- Jung, I.-G.; Park, C.-H. Characteristics of styrene degradation by Rhodococcus pyridinovorans isolated from a biofilter. Chemosphere 2005, 61, 451–456. [Google Scholar] [CrossRef]
- Gąszczak, A.; Bartelmus, G.; Greń, I. Kinetics of styrene biodegradation by Pseudomonas sp. E-93486. Appl. Microbiol. Biotechnol. 2012, 93, 565–573. [Google Scholar] [CrossRef]
- Ray, A.; Cot, M.; Puzo, G.; Gilleron, M.; Nigou, J. Bacterial cell wall macroamphiphiles: Pathogen-/microbe-associated molecular patterns detected by mammalian innate immune system. Biochimie 2013, 95, 33–42. [Google Scholar] [CrossRef]
- Chen, G.-Q.; Wu, Q. The application of polyhydroxyalkanoates as tissue engineering materials. Biomaterials 2005, 26, 6565–6578. [Google Scholar] [CrossRef]
- Wampfler, B.; Ramsauer, T.; Rezzonico, S.; Hischier, R.; Kohling, R.; Thony-Meyer, L.; Zinn, M. Isolation and purification of medium chain length poly(3-hydroxyalkanoates) (mcl-PHA) for medical applications using nonchlorinated solvents. Biomacromolecules 2010, 11, 2716–2723. [Google Scholar] [CrossRef]
- Karr, D.B.; Waters, J.K.; Emerich, D.W. Analysis of poly-β-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid chromatography and UV detection. Appl. Environ. Microbiol. 1983, 46, 1339–1344. [Google Scholar]
- Misaki, A.; Azuma, I.; Yamamura, Y. Structural and immunochemical studies on D-arabino-D-mannans and D-mannans of Mycobacterium tuberculosis and other Mycobacterium species. J. Biochem. 1977, 82, 1759–1770. [Google Scholar]
- Nigou, J.; Gilleron, M.; Puzo, G. Lipoarabinomannans: From structure to biosynthesis. Biochimie 2003, 85, 153–166. [Google Scholar] [CrossRef]
- Sutcliffe, I.; Brown, A.; Dover, L. The Rhodococcal cell envelope: Composition, organisation and biosynthesis. In Biology of Rhodococcus; Alvarez, H.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 16, pp. 29–71. [Google Scholar]
- Ruhland, G.J.; Fiedler, F. Occurrence and structure of lipoteichoic acids in the genus Staphylococcus. Arch. Microbiol. 1990, 154, 375–379. [Google Scholar]
- Sutcliffe, I.C. The lipoteichoic acids and lipoglycans of Gram-positive bacteria: A chemotaxonomic perspective. Syst. Appl. Microbiol. 1995, 17, 467–480. [Google Scholar] [CrossRef]
- Iwasaki, H.; Shimada, A.; Yokoyama, K.; Ito, E. Structure and glycosylation of lipoteichoic acids in Bacillus strains. J. Bacteriol. 1989, 171, 424–429. [Google Scholar]
- Sutcliffe, I.C.; Shaw, N. Atypical lipoteichoic acids of Gram-positive bacteria. J. Bacteriol. 1991, 173, 7065–7069. [Google Scholar]
- Danson, M.J.; Hough, D.W. The structural basis of protein halophilicity. Comp. Biochem. Physiol. Part. A Physiol. 1997, 117, 307–312. [Google Scholar] [CrossRef]
- Chen, G.-Q.; Zhang, G.; Park, S.; Lee, S. Industrial scale production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate). Appl. Microbiol. Biotechnol. 2001, 57, 50–55. [Google Scholar] [CrossRef]
- Choi, J.; Lee, S.Y. Factors affecting the economics of polyhydroxyalkanoate production by bacterial fermentation. Appl. Microbiol. Biotechnol. 1999, 51, 13–21. [Google Scholar] [CrossRef]
- Ganduri, V.; Ghosh, S.; Patnaik, P. Mixing control as a device to increase PHB production in batch fermentations with co-cultures of Lactobacillus delbrueckii and Ralstonia eutropha. Process Biochem. 2005, 40, 257–264. [Google Scholar] [CrossRef]
- Shi, H.; Shiraishi, M.; Shimizu, K. Metabolic flux analysis for biosynthesis of poly(β-hydroxybutyric acid) in Alcaligenes eutrophus from various carbon sources. J. Ferment. Bioeng. 1997, 84, 579–587. [Google Scholar] [CrossRef]
- Tanaka, K.; Katamune, K.; Ishizaki, A. Fermentative production of poly(β-hydroxybutyric acid) from xylose via L-lactate by a two-stage culture method employing Lactococcus lactis IO-1 and Alcaligenes eutrophus. Can. J. Microbiol. 1995, 41, 257–261. [Google Scholar] [CrossRef]
- van der Ha, D.; Nachtergaele, L.; Kerckhof, F.-M.; Rameiyanti, D.; Bossier, P.; Verstraete, W.; Boon, N. Conversion of biogas to bioproducts by algae and methane oxidizing bacteria. Environ. Sci. Technol. 2012, 46, 13425–13431. [Google Scholar] [CrossRef]
- Romo, D.M.R.; Grosso, M.V.; Solano, N.C.M.; Castaño, D.M. A most effective method for selecting a broad range of short and medium-chain-length polyhidroxyalcanoate producing microorganisms. Electron. J. Biotechnol. 2007, 10, 348–357. [Google Scholar]
- Solaiman, D.K.; Ashby, R.D. Rapid genetic characterization of poly(hydroxyalkanoate) synthase and its applications. Biomacromolecules 2005, 6, 532–537. [Google Scholar] [CrossRef]
- Spiekermann, P.; Rehm, B.H.; Kalscheuer, R.; Baumeister, D.; Steinbüchel, A. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 1999, 171, 73–80. [Google Scholar] [CrossRef]
- Melnicki, M.R.; Eroglu, E.; Melis, A. Changes in hydrogen production and polymer accumulation upon sulfur-deprivation in purple photosynthetic bacteria. Int. J. Hydrog. Energy 2009, 34, 6157–6170. [Google Scholar] [CrossRef]
- Wu, H.A.; Sheu, D.S.; Lee, C.Y. Rapid differentiation between short-chain-length and medium-chain-length polyhydroxyalkanoate-accumulating bacteria with spectrofluorometry. J. Microbiol. Meth. 2003, 53, 131–135. [Google Scholar] [CrossRef]
- Ostle, A.G.; Holt, J. Nile blue A as a fluorescent stain for poly-beta-hydroxybutyrate. Appl. Environ. Microbiol. 1982, 44, 238–241. [Google Scholar]
- Law, J.H.; Slepecky, R.A. Assay of poly-β-hydroxybutyric acid. J. Bacteriol. 1961, 82, 33–36. [Google Scholar]
- Hong, K.; Sun, S.; Tian, W.; Chen, G.-Q.; Huang, W. A rapid method for detecting bacterial polyhydroxyalkanoates in intact cells by fourier transform infrared spectroscopy. Appl. Microbiol. Biotechnol. 1999, 51, 523–526. [Google Scholar] [CrossRef]
- Gumel, A.; Annuar, M.; Heidelberg, T. Effects of carbon substrates on biodegradable polymer composition and stability produced by Delftia tsuruhatensis Bet002 isolated from palm oil mill effluent. Polym. Degrad. Stabil. 2012, 97, 1224–1231. [Google Scholar] [CrossRef]
- Hesselmann, R.P.; Fleischmann, T.; Hany, R.; Zehnder, A.J. Determination of polyhydroxyalkanoates in activated sludge by ion chromatographic and enzymatic methods. J. Microbiol. Meth. 1999, 35, 111–119. [Google Scholar] [CrossRef]
- Grubelnik, A.; Wiesli, L.; Furrer, P.; Rentsch, D.; Hany, R.; Meyer, V.R. A simple HPLC-MS method for the quantitative determination of the composition of bacterial medium chain-length polyhydroxyalkanoates. J. Sep. Sci. 2008, 31, 1739–1744. [Google Scholar] [CrossRef]
- Furrer, P.; Hany, R.; Rentsch, D.; Grubelnik, A.; Ruth, K.; Panke, S.; Zinn, M. Quantitative analysis of bacterial medium-chain-length poly([R]-3-hydroxyalkanoates) by gas chromatography. J. Chromatogr. A 2007, 1143, 199–206. [Google Scholar] [CrossRef]
- Tan, G.-Y.A.; Chen, C.-L.; Ge, L.; Li, L.; Wang, L.; Zhao, L.; Mo, Y.; Tan, S.N.; Wang, J.-Y. Enhanced gas chromatography-mass spectrometry method for bacterial polyhydroxyalkanoates (PHAs) analysis. J. Biosci. Bioeng. 2014, 117, 379–382. [Google Scholar] [CrossRef]
- Lee, E.Y.; Choi, C.Y. Gas chromatography-mass spectrometric analysis and its application to a screening procedure for novel bacterial polyhydroxyalkanoic acids containing long chain saturated and unsaturated monomers. J. Ferment. Bioeng. 1995, 80, 408–414. [Google Scholar] [CrossRef]
- Braunegg, G.; Sonnleitner, B.; Lafferty, R. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur. J. Appl. Microbiol. 1978, 6, 29–37. [Google Scholar] [CrossRef]
- Ward, A.C.; Dawes, E.A. A disk assay for poly-β-hydroxybutyrate. Anal. Biochem. 1973, 52, 607–613. [Google Scholar] [CrossRef]
- Slepecky, R.A.; Law, J.H. A rapid spectrophotometric assay of alpha, beta-unsaturated acids and beta-hydroxy acids. Anal. Chem. 1960, 32, 1697–1699. [Google Scholar] [CrossRef]
- Arcos-Hernandez, M.V.; Gurieff, N.; Pratt, S.; Magnusson, P.; Werker, A.; Vargas, A.; Lant, P. Rapid quantification of intracellular PHA using infrared spectroscopy: An application in mixed cultures. J. Biotechnol. 2010, 150, 372–379. [Google Scholar]
- de Rijk, T.C.; van de Meer, P.; Eggink, G.; Weusthuis, R.A. Methods for analysis of poly(3-hydroxyalkanoate) (PHA) composition. In Biopolymers Online; Doi, Y., Steinbüchel, A., Eds.; Wiley-VCH: Weinheim, Germany, 2005; Volume 3b, pp. 1–12. [Google Scholar]
- Korotkova, N.A.; Ashin, V.; Doronina, N.V.; Trotsenko, Y.A. A new method for quantitative determination of poly-3-hydroxybutyrate and 3-hydroxybutyrate-3-hydroxyvalerate copolymer in microbial biomass by reversed-phase high-performance liquid chromatography. Appl. Biochem. Microbiol. 1997, 33, 302–305. [Google Scholar]
- Zhang, S.; Norrlöw, O.; Wawrzynczyk, J.; Dey, E.S. Poly(3-hydroxybutyrate) biosynthesis in the biofilm of Alcaligenes eutrophus, using glucose enzymatically released from pulp fiber sludge. Appl. Environ. Microbiol. 2004, 70, 6776–6782. [Google Scholar] [CrossRef]
- Comeau, Y.; Hall, K.J.; Oldham, W.K. Determination of poly-β-hydroxybutyrate and poly-β-hydroxyvalerate in activated sludge by gas-liquid chromatography. Appl. Environ. Microbiol. 1988, 54, 2325–2327. [Google Scholar]
- Gumel, A.M.; Annuar, M.S.M.; Heidelberg, T. Biosynthesis and characterization of polyhydroxyalkanoates copolymers produced by Pseudomonas putida Bet001 isolated from palm oil mill effluent. PLoS One 2012, 7, e45214. [Google Scholar]
- Kunasundari, B.; Sudesh, K. Isolation and recovery of microbial polyhydroxyalkanoates. Express Polym. Lett. 2011, 5, 620–634. [Google Scholar] [CrossRef]
- Ramsay, J.A.; Berger, E.; Voyer, R.; Chavarie, C.; Ramsay, B.A. Extraction of poly-3-hydroxybutyrate using chlorinated solvents. Biotechnol. Tech. 1994, 8, 589–594. [Google Scholar] [CrossRef]
- Koller, M.; Bona, R.; Chiellini, E.; Braunegg, G. Extraction of short-chain-length poly-[(R)-hydroxyalkanoates] (scl-PHA) by the “anti-solvent” acetone under elevated temperature and pressure. Biotechnol. Lett. 2013, 35, 1023–1028. [Google Scholar] [CrossRef]
- Nonato, R.; Mantelatto, P.; Rossell, C. Integrated production of biodegradable plastic, sugar and ethanol. Appl. Microbiol. Biotechnol. 2001, 57, 1–5. [Google Scholar] [CrossRef]
- Berger, E.; Ramsay, B.A.; Ramsay, J.A.; Chavarie, C.; Braunegg, G. PHB recovery by hypochlorite digestion of non-PHB biomass. Biotechnol. Tech. 1989, 3, 227–232. [Google Scholar] [CrossRef]
- Hahn, S.K.; Chang, Y.K.; Kim, B.S.; Chang, H.N. Optimization of microbial poly(3-hydroxybutyrate) recover using dispersions of sodium hypochlorite solution and chloroform. Biotechnol. Bioeng. 1994, 44, 256–261. [Google Scholar] [CrossRef]
- Kapritchkoff, F.M.; Viotti, A.P.; Alli, R.C.P.; Zuccolo, M.; Pradella, J.G.C.; Maiorano, A.E.; Miranda, E.A.; Bonomi, A. Enzymatic recovery and purification of polyhydroxybutyrate produced by Ralstonia eutropha. J. Biotechnol. 2006, 122, 453–462. [Google Scholar] [CrossRef]
- Ashby, R.; Solaiman, D.; Foglia, T. Poly(ethylene glycol)-mediated molar mass control of short-chain- and medium-chain-length poly(hydroxyalkanoates) from Pseudomonas oleovorans. Appl. Microbiol. Biotechnol. 2002, 60, 154–159. [Google Scholar] [CrossRef]
- Durner, R.; Zinn, M.; Witholt, B.; Egli, T. Accumulation of poly[(R)-3-hydroxyalkanoates] in Pseudomonas oleovorans during growth in batch and chemostat culture with different carbon sources. Biotechnol. Bioeng. 2000, 72, 278–288. [Google Scholar]
- Jiang, X.; Ramsay, J.A.; Ramsay, B.A. Acetone extraction of mcl-PHA from Pseudomonas putida KT2440. J. Microbiol. Meth. 2006, 67, 212–219. [Google Scholar] [CrossRef]
- Hahn, S.K.; Chang, Y.K.; Lee, S.Y. Recovery and characterization of poly(3-hydroxybutyric acid) synthesized in Alcaligenes eutrophus and recombinant Escherichia coli. Appl. Environ. Microbiol. 1995, 61, 34–39. [Google Scholar]
- Cromwick, A.M.; Foglia, T.; Lenz, R.W. The microbial production of poly(hydroxyalkanoates) from tallow. Appl. Microbiol. Biotechnol. 1996, 46, 464–469. [Google Scholar] [CrossRef]
- Ashby, R.D.; Foglia, T.A. Poly(hydroxyalkanoate) biosynthesis from triglyceride substrates. Appl. Microbiol. Biotechnol. 1998, 49, 431–437. [Google Scholar] [CrossRef]
- Sánchez, R.J.; Schripsema, J.; da Silva, L.F.; Taciro, M.K.; Pradella, J.G.C.; Gomez, J.G.C. Medium-chain-length polyhydroxyalkanoic acids (PHAmcl) produced by Pseudomonas putida IPT 046 from renewable sources. European Polymer Journal 2003, 39, 1385–1394. [Google Scholar] [CrossRef]
- Fernández, D.; Rodríguez, E.; Bassas, M.; Viñas, M.; Solanas, A.M.; Llorens, J.; Marqués, A.M.; Manresa, A. Agro-industrial oily wastes as substrates for PHA production by the new strain Pseudomonas aeruginosa NCIB 40045: Effect of culture conditions. Biochem. Eng. J. 2005, 26, 159–167. [Google Scholar] [CrossRef]
- Yasotha, K.; Aroua, M.K.; Ramachandran, K.B.; Tan, I.K.P. Recovery of medium-chain-length polyhydroxyalkanoates (PHAs) through enzymatic digestion treatments and ultrafiltration. Biochem. Eng. J. 2006, 30, 260–268. [Google Scholar] [CrossRef]
- De Koning, G.J.M.; Kellerhals, M.; van Meurs, C.; Witholt, B. A process for the recovery of poly(hydroxyalkanoates) from Pseudomonads-Part 2. Bioprocess Eng. 1997, 17, 15–21. [Google Scholar] [CrossRef]
- De Koning, G.J.M.; Witholt, B. A process for the recovery of poly(hydroxyalkanoates) from Pseudomonads-Part 1. Bioprocess Eng. 1997, 17, 7–13. [Google Scholar] [CrossRef]
- Dai, Y.; Lambert, L.; Yuan, Z.; Keller, J. Characterisation of polyhydroxyalkanoate copolymers with controllable four-monomer composition. J. Biotechnol. 2008, 134, 137–145. [Google Scholar] [CrossRef]
- Saeed, K.A.; Ayorinde, F.O.; Eribo, B.E.; Gordon, M.; Collier, L. Characterization of partially transesterified poly(β-hydroxyalkanoate)s using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1999, 13, 1951–1957. [Google Scholar]
- Li, Z.; Yang, X.; Wu, L.; Chen, Z.; Lin, Y.; Xu, K.; Chen, G.-Q. Synthesis, characterization and biocompatibility of biodegradable elastomeric poly(ether-ester urethane)s based on poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) and poly(ethylene glycol) via melting polymerization. J. Biomater. Sci., Polym. Ed. 2009, 20, 1179–1202. [Google Scholar]
- Katime, I.; Cadenato, A. Compatibility of peo/poly(iso-butyl methacrylate) and peo/poly(tert-butyl methacrylate) blends by DTA. Mater. Lett. 1995, 22, 303–308. [Google Scholar] [CrossRef]
- Shamala, T.; Divyashree, M.; Davis, R.; Kumari, K.L.; Vijayendra, S.; Raj, B. Production and characterization of bacterial polyhydroxyalkanoate copolymers and evaluation of their blends by fourier transform infrared spectroscopy and scanning electron microscopy. Indian J. Microbiol. 2009, 49, 251–258. [Google Scholar] [CrossRef]
- Chen, S.; Liu, Q.; Wang, H.; Zhu, B.; Yu, F.; Chen, G.-Q.; Inoue, Y. Polymorphic crystallization of fractionated microbial medium-chain-length polyhydroxyalkanoates. Polymer 2009, 50, 4378–4388. [Google Scholar] [CrossRef]
- Wu, C.-S.; Liao, H.-T. The mechanical properties, biocompatibility and biodegradability of chestnut shell fibre and polyhydroxyalkanoate composites. Polym. Degrad. Stabil. 2014, 99, 274–282. [Google Scholar] [CrossRef]
- Teeka, J.; Imai, T.; Reungsang, A.; Cheng, X.; Yuliani, E.; Thiantanankul, J.; Poomipuk, N.; Yamaguchi, J.; Jeenanong, A.; Higuchi, T.; et al. Characterization of polyhydroxyalkanoates (PHAs) biosynthesis by isolated Novosphingobium sp. THA_AIK7 using crude glycerol. J. Ind. Microbiol. Biotechnol. 2012, 39, 749–758. [Google Scholar] [CrossRef]
- Meng, D.-C.; Shi, Z.-Y.; Wu, L.-P.; Zhou, Q.; Wu, Q.; Chen, J.-C.; Chen, G.-Q. Production and characterization of poly(3-hydroxypropionate-co-4-hydroxybutyrate) with fully controllable structures by recombinant Escherichia coli containing an engineered pathway. Metab. Eng. 2012, 14, 317–324. [Google Scholar] [CrossRef]
- Hartmann, R.; Hany, R.; Geiger, T.; Egli, T.; Witholt, B.; Zinn, M. Tailored biosynthesis of olefinic medium-chain-length poly[(R)-3-hydroxyalkanoates] in Pseudomonas putida GPo1 with improved thermal properties. Macromolecules 2004, 37, 6780–6785. [Google Scholar] [CrossRef]
- Hany, R.; Hartmann, R.; Böhlen, C.; Brandenberger, S.; Kawada, J.; Löwe, C.; Zinn, M.; Witholt, B.; Marchessault, R.H. Chemical synthesis and characterization of POSS-functionalized poly[3-hydroxyalkanoates]. Polymer 2005, 46, 5025–5031. [Google Scholar] [CrossRef]
- Wang, Q.; Tappel, R.C.; Zhu, C.; Nomura, C.T. Development of a new strategy for production of medium-chain-length polyhydroxyalkanoates by recombinant Escherichia coli via inexpensive non-fatty acid feedstocks. Appl. Environ. Microbiol. 2012, 78, 519–527. [Google Scholar] [CrossRef]
- De Waard, P.; van der Wal, H.; Huijberts, G.; Eggink, G. Heteronuclear NMR analysis of unsaturated fatty acids in poly(3-hydroxyalkanoates). Study of beta-oxidation in Pseudomonas putida. J. Biol. Chem. 1993, 268, 315–319. [Google Scholar]
- Eggink, G.; de Waard, P.; Huijberts, G.N. Formation of novel poly(hydroxyalkanoates) from long-chain fatty acids. Can. J. Microbiol. 1995, 41, 14–21. [Google Scholar] [CrossRef]
- Tripathi, L.; Wu, L.-P.; Dechuan, M.; Chen, J.; Wu, Q.; Chen, G.-Q. Pseudomonas putida KT2442 as a platform for the biosynthesis of polyhydroxyalkanoates with adjustable monomer contents and compositions. Bioresour. Technol. 2013, 142, 225–231. [Google Scholar] [CrossRef]
- Ballistreri, A.; Garozzo, D.; Giuffrida, M.; Impallomeni, G.; Montaudo, G. Sequencing bacterial poly(β-hydroxybutyrate-co-β-hydroxyvalerate) by partial methanolysis, HPLC fractionation, and fast-atom-bombardment mass spectrometry analysis. Macromolecules 1989, 22, 2107–2111. [Google Scholar] [CrossRef]
- Ballistreri, A.; Montaudo, G.; Garozzo, D.; Giuffrida, M.; Montaudo, M.S. Microstructure of bacterial poly(β-hydroxybutyrate-co-β-hydroxyvalerate) by fast atom bombardment mass spectrometry analysis of the partial pyrolysis products. Macromolecules 1991, 24, 1231–1236. [Google Scholar]
- Abate, R.; Ballistreri, A.; Montaudo, G.; Garozzo, D.; Impallomeni, G.; Critchley, G.; Tanaka, K. Quantitative applications of matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry: Determination of copolymer composition in bacterial copolyesters. Rapid Commun. Mass Spectrom. 1993, 7, 1033–1036. [Google Scholar] [CrossRef]
- Saeed, K.A.; Ayorinde, F.O.; Eribo, B.E.; Gordon, M.; Collier, L. Characterization of partially transesterified poly(beta-hydroxyalkanoate)s by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. AOAC Int. 2001, 84, 1109–11015. [Google Scholar]
- Chen, G.-Q. Introduction of bacterial plastics PHA, PLA, PBS, PE, PTT, and PPP. In Plastics from Bacteria: Natural Functions and Applications; Chen, G.-Q., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 14, pp. 1–16. [Google Scholar]
- Koller, M.; Bona, R.; Braunegg, G.; Hermann, C.; Horvat, P.; Kroutil, M.; Martinz, J.; Neto, J.; Pereira, L.; Varila, P. Production of polyhydroxyalkanoates from agricultural waste and surplus materials. Biomacromolecules 2005, 6, 561–565. [Google Scholar] [CrossRef]
- Li, L. MALDI-MS for polymer characterization. In MALDI MS: A Practical Guide to Instrumentation, Methods, and Applications, 2nd ed.; Hillenkamp, F., Peter-Katalinic, J., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2013; pp. 313–365. [Google Scholar]
- Aoyagi, Y.; Yamashita, K.; Doi, Y. Thermal degradation of poly[(R)-3-hydroxybutyrate], poly[ε-caprolactone], and poly[(S)-lactide]. Polym. Degrad. Stabil. 2001, 76, 53–59. [Google Scholar] [CrossRef]
- Bloembergen, S.; Holden, D.A.; Hamer, G.K.; Bluhm, T.L.; Marchessault, R.H. Studies of composition and crystallinity of bacterial poly(β-hydroxybutyrate-co-β-hydroxyvalerate). Macromolecules 1986, 19, 2865–2871. [Google Scholar] [CrossRef]
- Porter, M.; Yu, J. Monitoring the in situ crystallization of native biopolyester granules in Ralstonia eutropha via infrared spectroscopy. J. Microbiol. Meth. 2011, 87, 49–55. [Google Scholar] [CrossRef]
- Barham, P.J.; Keller, A.; Otun, E.L.; Holmes, P.A. Crystallization and morphology of a bacterial thermoplastic: Poly-3-hydroxybutyrate. J. Mater. Sci. 1984, 19, 2781–2794. [Google Scholar] [CrossRef]
- Simon-Colin, C.; Raguénès, G.; Crassous, P.; Moppert, X.; Guezennec, J. A novel mcl-PHA produced on coprah oil by Pseudomonas guezennei biovar. tikehau, isolated from a “kopara” mat of French Polynesia. Int. J. Biol. Macromol. 2008, 43, 176–181. [Google Scholar] [CrossRef]
- Cheng, S.-T.; Chen, Z.-F.; Chen, G.-Q. The expression of cross-linked elastin by rabbit blood vessel smooth muscle cells cultured in polyhydroxyalkanoate scaffolds. Biomaterials 2008, 29, 4187–4194. [Google Scholar] [CrossRef]
- Dufresne, A.; Kellerhas, M.B.; Witholt, B. Transcrystallization in mcl-PHAs: Cellulose whiskers composites. Macromolecules 1999, 32, 7396–7401. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tan, G.-Y.A.; Chen, C.-L.; Li, L.; Ge, L.; Wang, L.; Razaad, I.M.N.; Li, Y.; Zhao, L.; Mo, Y.; Wang, J.-Y. Start a Research on Biopolymer Polyhydroxyalkanoate (PHA): A Review. Polymers 2014, 6, 706-754. https://doi.org/10.3390/polym6030706
Tan G-YA, Chen C-L, Li L, Ge L, Wang L, Razaad IMN, Li Y, Zhao L, Mo Y, Wang J-Y. Start a Research on Biopolymer Polyhydroxyalkanoate (PHA): A Review. Polymers. 2014; 6(3):706-754. https://doi.org/10.3390/polym6030706
Chicago/Turabian StyleTan, Giin-Yu Amy, Chia-Lung Chen, Ling Li, Liya Ge, Lin Wang, Indah Mutiara Ningtyas Razaad, Yanhong Li, Lei Zhao, Yu Mo, and Jing-Yuan Wang. 2014. "Start a Research on Biopolymer Polyhydroxyalkanoate (PHA): A Review" Polymers 6, no. 3: 706-754. https://doi.org/10.3390/polym6030706
APA StyleTan, G.-Y. A., Chen, C.-L., Li, L., Ge, L., Wang, L., Razaad, I. M. N., Li, Y., Zhao, L., Mo, Y., & Wang, J.-Y. (2014). Start a Research on Biopolymer Polyhydroxyalkanoate (PHA): A Review. Polymers, 6(3), 706-754. https://doi.org/10.3390/polym6030706




