Low Molecular Weight pDMAEMA-block-pHEMA Block-Copolymers Synthesized via RAFT-Polymerization: Potential Non-Viral Gene Delivery Agents?
Abstract
: The aim of this study was to investigate non-viral pDNA carriers based on diblock-copolymers consisting of poly(2-(dimethyl amino)ethyl methacrylate) (pDMAEMA) and poly(2-hydroxyethyl methacrylate) (pHEMA). Specifically the block-lengths and molecular weights were varied to determine the minimal requirements for transfection. Such vectors should allow better transfection at acceptable toxicity levels and the entire diblock-copolymer should be suitable for renal clearance. For this purpose, a library of linear poly(2-(dimethyl amino)ethyl methacrylate-block-poly(2-hydroxyl methacrylate) (pDMAEMA-block-pHEMA) copolymers was synthesized via RAFT (reversible addition-fragmentation chain transfer) polymerization in a molecular weight (Mw) range of 17–35.7 kDa and analyzed using 1H and 13C NMR (nuclear magnetic resonance), ATR (attenuated total reflectance), GPC (gel permeation chromatography) and DSC (differential scanning calorimetry). Copolymers possessing short pDMAEMA-polycation chains were 1.4–9.7 times less toxic in vitro than polyethylenimine (PEI) 25 kDa, and complexed DNA into polyplexes of 100–170 nm, favorable for cellular uptake. The DNA-binding affinity and polyplex stability against competing polyanions was comparable with PEI 25 kDa. The zeta-potential of polyplexes of pDMAEMA-grafted copolymers remained positive (+15–30 mV). In comparison with earlier reported low molecular weight homo pDMAEMA vectors, these diblock-copolymers showed enhanced transfection efficacy under in vitro conditions due to their lower cytotoxicity, efficient cellular uptake and DNA packaging. The homo pDMAEMA115 (18.3 kDa) self-assembled with DNA into small positively charged polyplexes, but was not able to transfect cells. The grafting of 6 and 57 repeating units of pHEMA (0.8 and 7.4 kDa) to pDMAEMA115 increased the transfection efficacy significantly, implying a crucial impact of pHEMA on vector-cell interactions. The intracellular trafficking, in vivo transfection efficacy and kinetics of low molecular weight pDMAEMA-block-pHEMA are subject of ongoing studies.1. Introduction
The field of non-viral gene delivery remains a topic of intensive research efforts, and numerous polycations are under investigation [1,2]. One prominent vector in pDNA delivery is poly(ethylene imine) (PEI) 25 kDa, a branched polycation with a high charge density well known for its effective complexation, high transfection, but also significant toxicity [3]. To improve biocompatibility of PEI, several strategies were pursued such as a reduction of the molecular weight [4] or introduction of shielding component, e.g., PEGylation [5,6].
As the toxicity of a polymer, among other factors, is related to its charge density [3], polycations with more broadly spread charge distribution along the polymer chain, namely poly(2-(dimethyl amino)ethyl methacrylate (pDMAEMA) could be of interest. In comparison to PEI 25 kDa, pDMAEMA contains only tertiary amino groups, and the charge density is lower than in PEI. pDMAEMA shows an average pKa of 7.5 [7], and is thus sufficiently protonated at physiological pH for effective polyanion complexation. Previous reports described pDMAEMA to possess a decreased proton sponge effect compared to PEI 25 kDa [8] implying alternative mechanisms of membrane interaction, such as membrane destabilization [9]. This eventually led to reduced cell death, but the polymer retained the capacity to escape from endosomes [10].
A relationship between the polymer-DNA dissociation rate and transfection efficiency was established suggesting that pDMAEMA more readily releases DNA from polyplexes than polylysine [11]. The class of pDMAEMA was intensively investigated by Hennink and coworkers, demonstrating that homopolymers with molecular weights (Mw) of 300 kDa are effective DNA-carriers [12]. With increasing molecular weight, its transfection efficiency, complexation efficiency but also toxicity increased [10,13]. Scarce information on the performance of low molecular weight (<43 kDa) pDMAEMA is available from the literature, although such polymers could possibly be eliminated by renal excretion [9]. Therefore, a library of low molecular weight homo-pDMAEMAs with the chain length varying from 15 to 23.7 kDa was synthesized and grafted with pHEMA (poly(2-hydroxyethyl methacrylate)), resulting in water soluble linear diblock copolymers with a total Mw below 40 kDa. The number of tertiary amino groups of the charged copolymer-part exceeds 20 to 30-fold the minimal requirement of 4-6 groups reported to be necessary for efficient interaction with DNA [14].
Instead of applying ATRP (atom transfer radical polymerization) [10,15], in this study, the synthesis was performed via RAFT polymerization (reversible addition-fragmentation chain transfer) to avoid the use of toxic organometallic catalysts, and to attain narrow polydispersity indices (PDIs).
To improve biocompatibility and colloidal stability, uncharged blocks consisting of pHEMA were introduced in an attempt to reduce undesirable adverse interactions such as aggregation and cytotoxicity reported previously [16]. PEGylation of pDMAEMA has resulted in a decrease of zeta-potential and good steric stabilization of polycations at the cost of poor polyplex uptake into cells [17]. Another disadvantage of PEGylation could be seen in an “accelerated blood clearance” phenomenon (“ABC”) [18], occurring after repeated application of PEGylated vectors due to anti-PEG IgM-production and activation of the complement system. The hydrogel-forming pHEMA was earlier reported to be non-toxic and water soluble if not cross-linked [19], and biodegradable in linear form [20]. So far, no complement activation by pHEMA was reported despite longtime usage in medical applications [19].
Several pDMAEMA-pHEMA vector systems have been investigated recently showing their potential as gene delivery systems: blends of pDMAEMA and pHEMA were used to obtain nanoparticles [21], polymer with brush-like structures based on short chain pDMAEMA grafted on pHEMA backbones by cleavable carbonate ester linkage were reported [15]. Also star shaped pDMAEMAs with cleavable disulfide bonds synthesized via grafting with multiple short chain pDMAEMA-arms showed improved transfection efficiency [22].
Since high molecular weight homopolymers of pDMAEMA (300 kDa) were efficient transfection reagents, here, the molecular weight was systematically varied in the range from 290 to 15 kDa using RAFT to determine the minimal chain length required for transfection efficiency. Subsequently, such structures were modified using pHEMA to improve cytotoxicity and colloidal stability. These linear diblock copolymers, in contrast to previously studied pDMAEMA/pHEMA vectors, were designed to possess very narrow polydispersities, a less complicated steric structure and thus better accessibility for charge to charge interaction with genetic material, as well as low total molecular weight, allowing renal elimination without previous degradation.
2. Results and Discussion
2.1. Synthesis
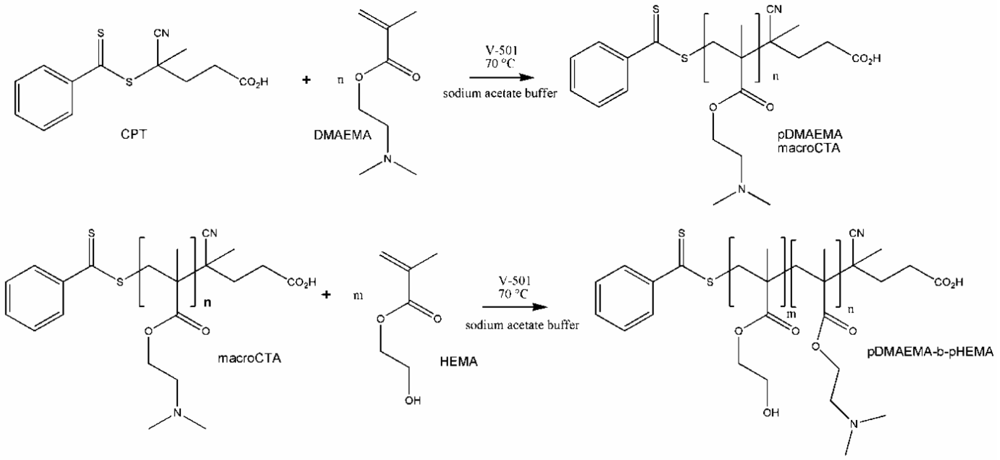
2.1.1. Synthesis of the Chain Transfer Agent CPT & the Macro Chain Transfer Agent (macroCTA) pDMAEMA
The 1H NMR (nuclear magnetic resonance) spectrum of 4-Cyanopentanoic acid dithiobenzoate (CPT) is displayed in Figure 1(A), for the 13C NMR spectrum, see Supplementary Information Figure S1. Strong agreement with the literature values was found in both cases [23-29].
For this study, four different pDMAEMA homo-polymers designated as pDMAEMA97, pDMAEMA105, pDMAEMA115 and pDMAEMA151 were synthesized. The index denotes the number of repeating units calculated by NMR measurements. For this calculation, the signal of the benzyl group at the end of each polymer strand (7.4–8.1 ppm) was correlated with the signal of the two methylene groups of the side chain (3.5–3.6 + 4.4–4.5 ppm). Shown here are the results from pDMAEMA115 (1) while the other homo-polymers behaved similarly. The NMR spectra shown in Figure 1(B) corresponds to that reported in [9]. In the attenuated total reflection (ATR) IR spectra, the two typical ester bands at 1,720 (C=O) and 1,150 (C–O), and the band of the backbone at 2,960 wavenumbers were found.
2.1.2. Synthesis of pDMAEMA-block-pHEMA Block Copolymers (pDMAEMA-block-pHEMA)
To calculate the number of HEMA repeating units in the polymer, the signal of the known number of methylene groups of the macroCTA was correlated with the signal of the methylene groups of HEMA (3.9–4.1 + 4.2–4.3 ppm). This was possible because the signals of the different parts do not overlap as displayed in Figure 1(C). Additionally, in the ATR IR spectra of the copolymers, the typical band for the hydroxyl group of pHEMA at 3,390 cm−1 was found. The ester band at 1,720 and 1,150 cm−1 belong to both blocks of the copolymer. The Tg of pDMAEMA [19] (−6 °C) and pHEMA [21] (∼70 °C) are known from the literature and decreased, as expected, to lower temperatures for the copolymers with increasing pDMAEMA part. The different block copolymers are shown in Table 1. For illustration of the synthesis of diblock copolymers, see Scheme 1.
2.1.3. Synthesis of pDMAEMA-pHEMA Random Copolymers (r-pDMAEMA-pHEMA)
The synthesis of the random polymers was achieved by simply mixing the monomers before starting the reaction. The weights were calculated as described for the macro CTA by correlating the signal of the benzyl group with the signal of the side chains of the different monomers. Here, the different ratios between the two monomers in the polymer were also calculated. The NMR spectra and the ATR spectra were comparable to the ones of the block copolymers, as expected. The ratio of the monomers is denoted by the last number in the abbreviated term of the polymers as shown in Table 1.
2.2. GPC (Gel Permeation Chromatography) Results of the Polymers
Comparing the values in Table 1, there was a discrepancy between the molecular weight calculated by NMR and those values determined by GPC. Since the results for molecular weight depend on the way of calculation, a discrepancy between NMR and GPC was expected for the copolymers with molecular weights of more than 20 kDa. However, the large differences for the polymers below 20 kDa containing a small amount of pHEMA were unexpected. The calculated refractive index was possibly not correct as these copolymers may interact with the column material leading to incomplete elution. All the polymers showed a PDI around 1, as expected for RAFT polymerization.
2.3. Nomenclature of the Synthesized pDMAEMA-block-pHEMA Copolymers
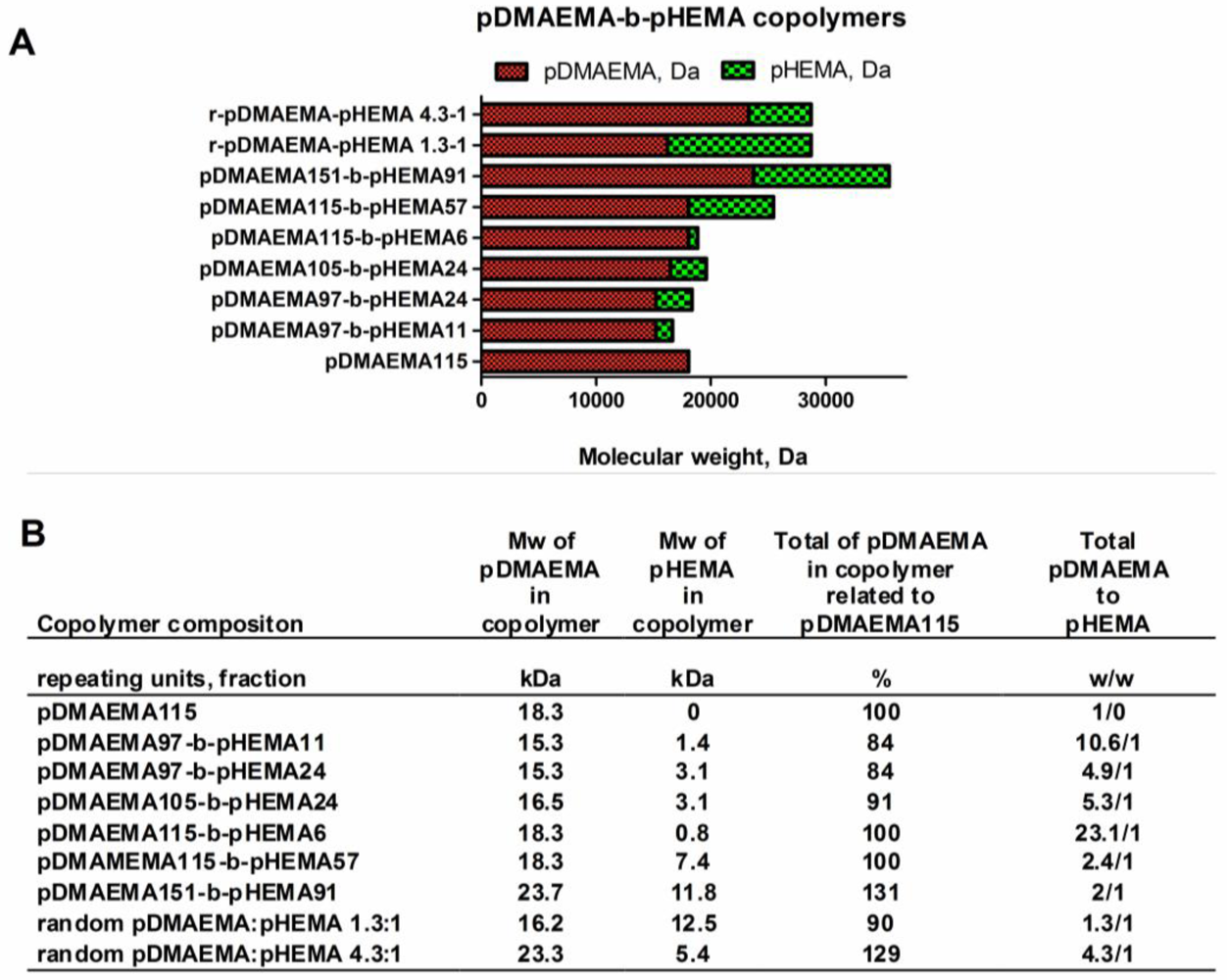
A library of copolymers containing different amounts of DMAEMA (2-(dimethyl amino)ethyl methacrylate and HEMA (2-hydroxyethyl methacrylate) repeating units was synthesized. The polymers were designated and classified as shown in Figure 2(A,B).
The synthesized polymers can be divided into three groups: homo-pDMAEMA, linear diblock pDMAEMA-block-pHEMA copolymers, and random copolymers with alternating sequence of pDMAEMA and pHEMA. The hypothesis for this work was that there would be a lower limit of the homo pDMAEMA chain length for effective transfection, and that the lack of transfection efficacy could be compensated for by grafting linear pHEMA. A pre-test with a panel of homo-polymers with subsequently decreasing pDMAEMA chain length was performed in form of in vitro transfections to point out the limit of transfection capacity of short chain polycations. Together with cytotoxicity data, the results from the transfection pre-test of homo-pDMAEMA 15–290 kDa are presented in Figure S2, Supplementary Information. It was found that a pDMAEMA95 homopolymer of 15 kDa possessing 95 repeating units was ineffective in gene delivery, whereas pDMAEMA406 with 64 kDa efficiently mediated transgene expression. These findings are in line with earlier studies, reporting that a 43 kDa homo-pDMAEMA [9] mediated no significant luciferase expression at N/P 8–20, but that the efficacy as vector could be increased at molecular weights above 112 kDa. Accordingly, in the present study it was found that the pDMAEMA115 homo-polymer of 18.3 kDa was inefficient and therefore chosen as low molecular weight “silent” reference vector. The copolymers pDMAEMA115-block-pHEMA57 & pDMAEMA115-block-pHEMA6 synthesized here contain the same polycationic part, but have increased molecular weights of 25.7 and 19.1 kDa, respectively, due to pHEMA grafting. Moreover, longer and shorter pDMAEMA chains were incorporated into other diblock construct to investigate the effect of pDMAEMA-block-pHEMA composition on DNA-complexation, charge, stability, toxicity, transfection efficacy and cellular uptake. The random pDMAEMA-pHEMA copolymers with larger and smaller polycationic parts were also included in the polymer library for investigations of structure function relationships (r-pDMAEMA-pHEMA 4.3–1 and r-pDMAEMA-pHEMA 1.3–1). The created copolymer library allowed for evaluation of the impact of the pDMAEMA/pHEMA proportion as well as the percentage of the polycationic part in relation to homo-pDMAEMA115 concerning vector efficacy in vitro.
2.4. DNA Complexation Efficiency
2.4.1. Agarose Gel Electrophoresis
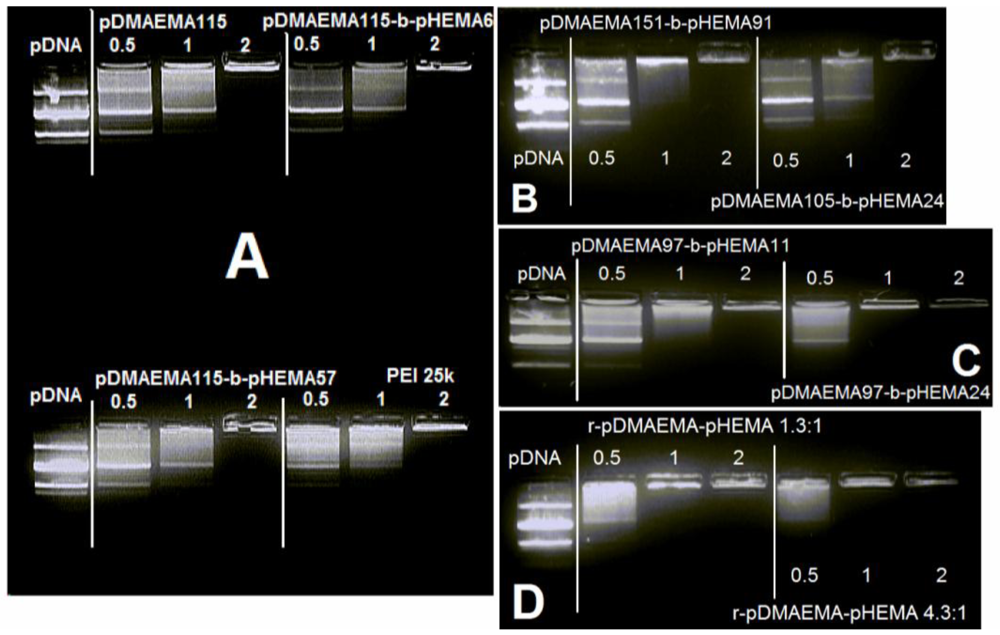
According to the gel electrophoresis performed with diblock and random pDMAEMA-pHEMA copolymers (Figure 3(A–D)), the N/P ratio of 2 was sufficient for complexing and immobilizing pDNA in case of all tested polymers. The N/P ratios of 10, 15 and 20 used for transfection experiments showed complete retardation of pDNA (data not shown). No differences were observed concerning the complexation efficiency of homo pDMAEMA115 compared to its diblock derivatives with 6 and 57 pHEMA units, and compared to pDMAEMA105-block-pHEMA24. Copolymer pDMAEMA151-block-pHEMA91 performed similarly as pDMAEMA115, but showed more complete retardation at N/P 1. Copolymer pDMAEMA97-block-pHEMA11 performed similarly to pDMAEMA151-block-pHEMA91. Yet, the random copolymers and pDMAEMA97-block-pHEMA24 were able to retard DNA at N/P 1 already, outreaching PEI 25k in its complexation properties. Based on the obtained gel electrophoresis data, all tested low molecular pDMAEMA-pHEMA constructs showed good pDNA-complexation even at low N/P ratios. The marginal differences of DNA-retardation at N/P 1 may be evidence of the impact of the polymer structure on interactions with pDNA. One of these slight differences can be seen in Figure 3(C), where pDMAEMA97-block-pHEMA24 shows a better complexation of DNA than pDMAEMA97-block-pHEMA11 at N/P ratio 1. The efficient complexation ability of the 15.3 kDa polycationic part of diblock copolymer pDMAEMA97-pHEMA24 was unexpected, as a 11.1 kDa homo-pDMAEMA was reported by Jiang et al. to be a poor complexer [15].
2.4.2. Heparin Assay — Stability against Competing Polyanions
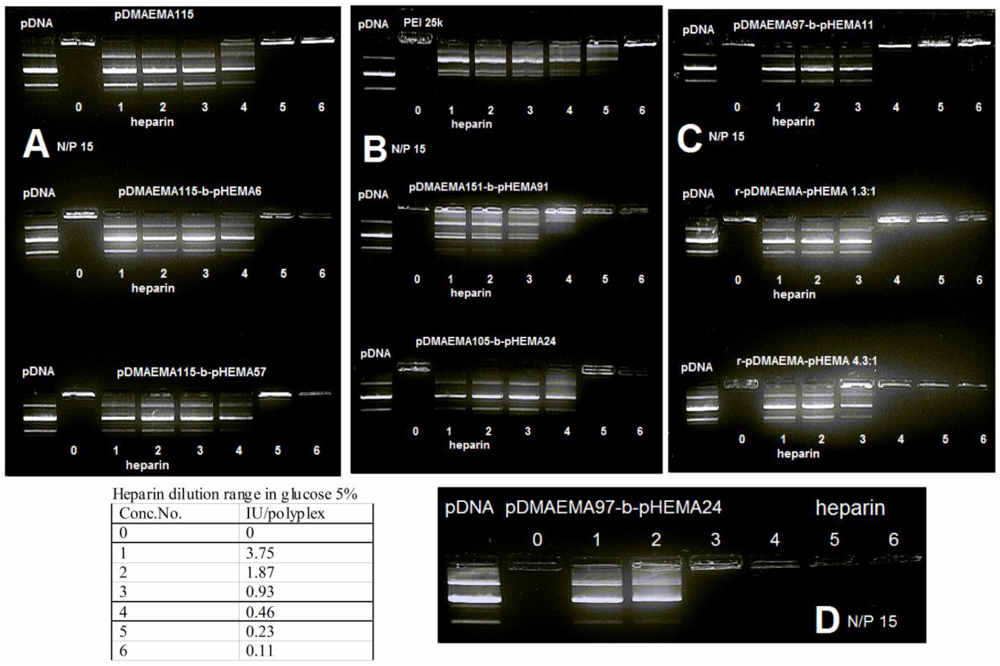
The trend observed in gel retardation experiments using polyplexes of lower N/P ratios corresponded well with the heparin competition assay of polyplexes at N/P 15 (Figure 4(A–D)). The copolymers with the same cationic block length of 115 units released DNA upon addition of the same heparin amount of 0.46 international units (IU). The copolymers pDMAEMA151-block-pHEMA91, pDMAEMA105-block-pHEMA24 as well as pDMAEMA115-derivatives appeared to be more resistant to competition with heparin than PEI 25k at 0.46 IU heparin, whereas pDMAEMA151-block-pHEMA91 had a less pronounced fluorescent band in lane 4 (0.46 IU). The random copolymers and pDMAEMA97-block-pHEMA11 were even more resistant to the presence of heparin releasing DNA only at higher concentrations of 0.93 IU. The highest resistance to competitive polyanions was observed in case of pDMAEMA97-block-pHEMA24, where 1.87 IU of heparin was needed to release DNA from the complex. Taken together, the most stable complexes were obtained using random copolymers and short chain diblock copolymers with only 97 pDMAEMA units. This may be due to a better interaction of copolymers with DNA thus providing a tighter polyplex structure. All tested copolymer complexes released DNA at heparin concentrations higher than the ones needed for destabilization of PEI 25k polyplexes. This fact can be interpreted as an advantage for protecting genetic material, but as well as a disadvantage, as far as DNA release is concerned at the site of action. Accordingly, decreased DNA release may cause reduced transfection efficiency of the most stable complexes.
2.5. Size and Zeta Potential
2.5.1. Size and Zeta Potential of Polyplexes in Isotonic Glucose
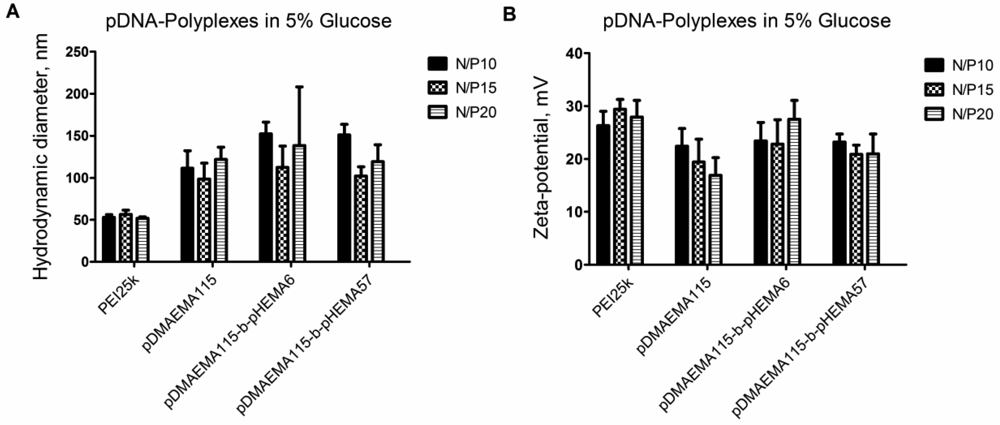
Isotonic glucose solution was used to formulate polyplexes with pDNA. For complete self-assembly of polyplexes, a 20 min incubation time was maintained after mixing. In measurements after shorter or no incubation, higher polydispersity indices and larger complex sizes were obtained (data not shown). PEI 25k yielded DNA-polyplexes in the size range <100 nm at all N/P ratios. At N/P ratios of 10, 15 and 20, the polyplex sizes of pDMAEMA-block-pHEMA diblock copolymers did not show statistically significant differences in hydrodynamic diameters compared to each other. All polymers were able to complex pDNA into polyplexes with hydrodynamic diameters of 100–170 nm. This is considered a size range suitable for cellular uptake [31,32]. Larger polyplexes were formed only in case of pDMAEMA105-block-pHEMA24 at N/P ratio 10, which may be due to complexation irregularities reflected in the broad standard deviation (Figure 5(A)). The hydrodynamic diameter of naked plasmid was earlier reported to be in the range of 300–400 nm [12].
The zeta potentials of polyplexes from diblock copolymers ranged from 10 to 30 mV depending on the N/P ratio (Figure 5(B)). A significant increase of the zeta potential at N/P 20 was only registered for pDMAEMA105-block-pHEMA24 and r-pDMAEMA-pHEMA 1.3:1. In general, no reduction in positive charge density could be observed for pHEMA-grafted copolymers in comparison to homo-pDMAEMA, suggesting that the pHEMA chains may not be localized on the polyplex surface but closer to the core of pDMAEMA and pDNA. The polyplex size also remained in the range of homo-pDMAEMA complexes with no obvious disturbance of complexation caused by grafting of pHEMA. Additionally, Figure 6(A,B) demonstrates similar trends in polyplex size and zeta potential concerning pDMAEMA115-derivatives.
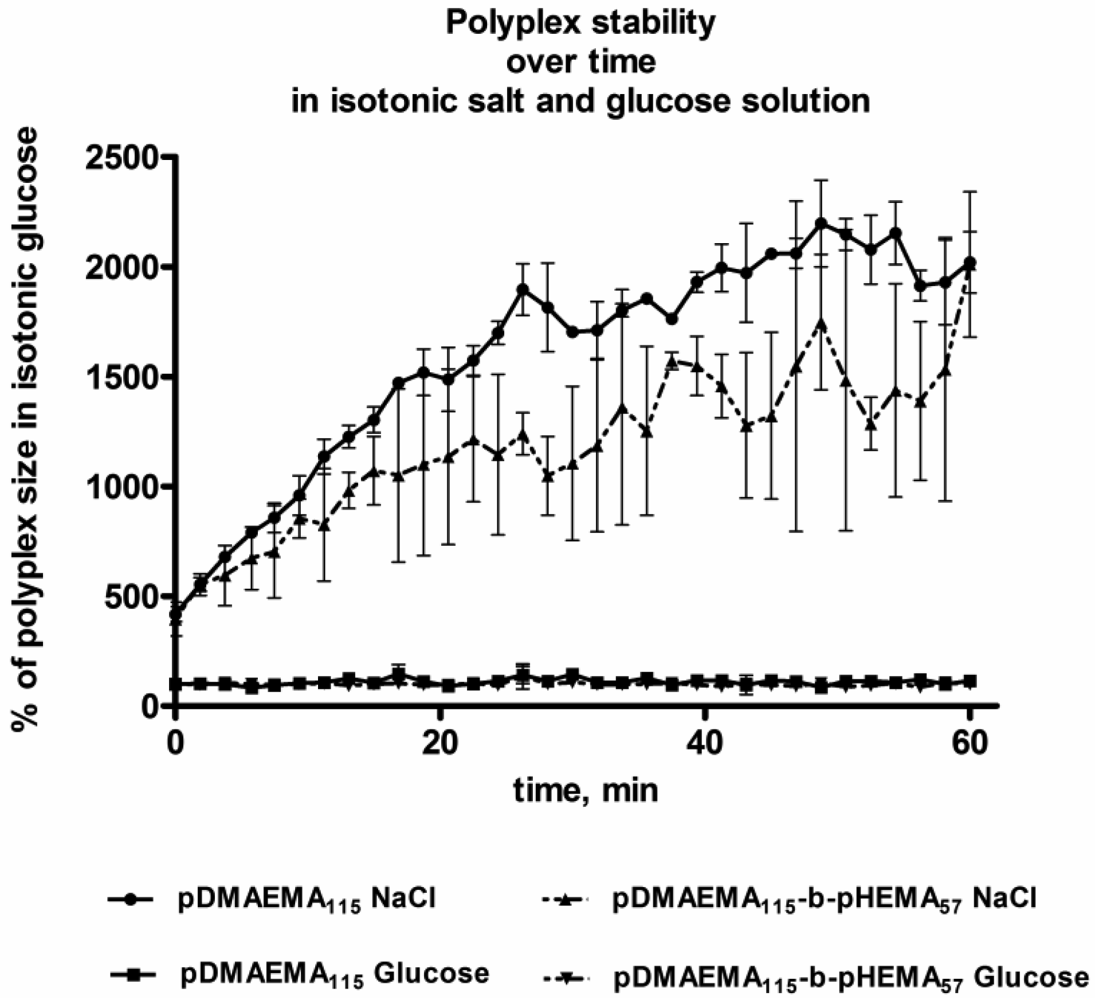
2.5.2. Polyplex Aggregation Behavior in Isotonic Salt and Glucose Solution over Time
According to the measurements of hydrodynamic diameters of polyplexes incubated in isotonic glucose and NaCl solution, similar trends were observed for pDMAEMA115 and pDMAEMA115-block-pHEMA57. In glucose, both polymer types showed good stability of their polyplexes with pDNA (Figure 7). In high ionic strength solution, on the contrary, significant polyplex aggregation could be observed and larger sized constructs appeared over time. From the aggregation experiment it may be expected that both types of polymers behaved similarly in cell experiments, as polyplexes of both types aggregated rapidly in the presence of high ionic strength medium. However, the Hennink group previously reported that the transfection efficiency of pDMAEMA polyplexes was not decreased in spite of an observed increase in size in the presence of serum [10,12].
2.6. Cytotoxicity of Polymers and Polyplexes in vitro (MTT, BCA)
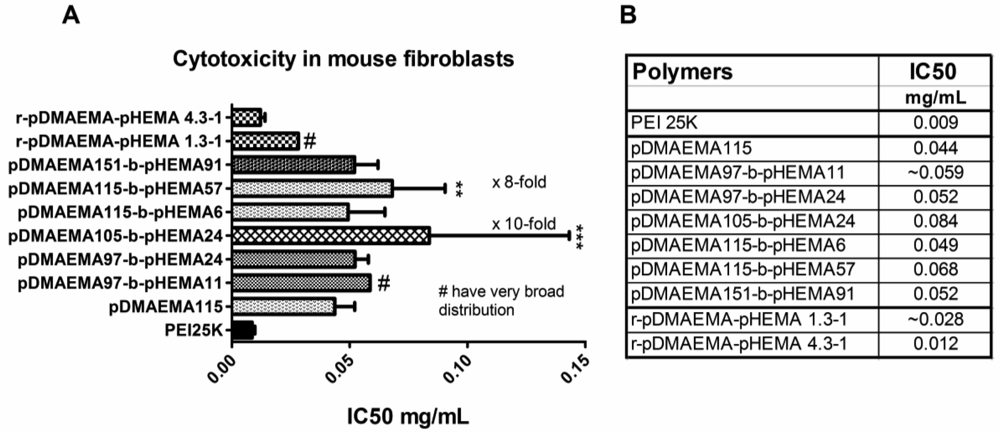
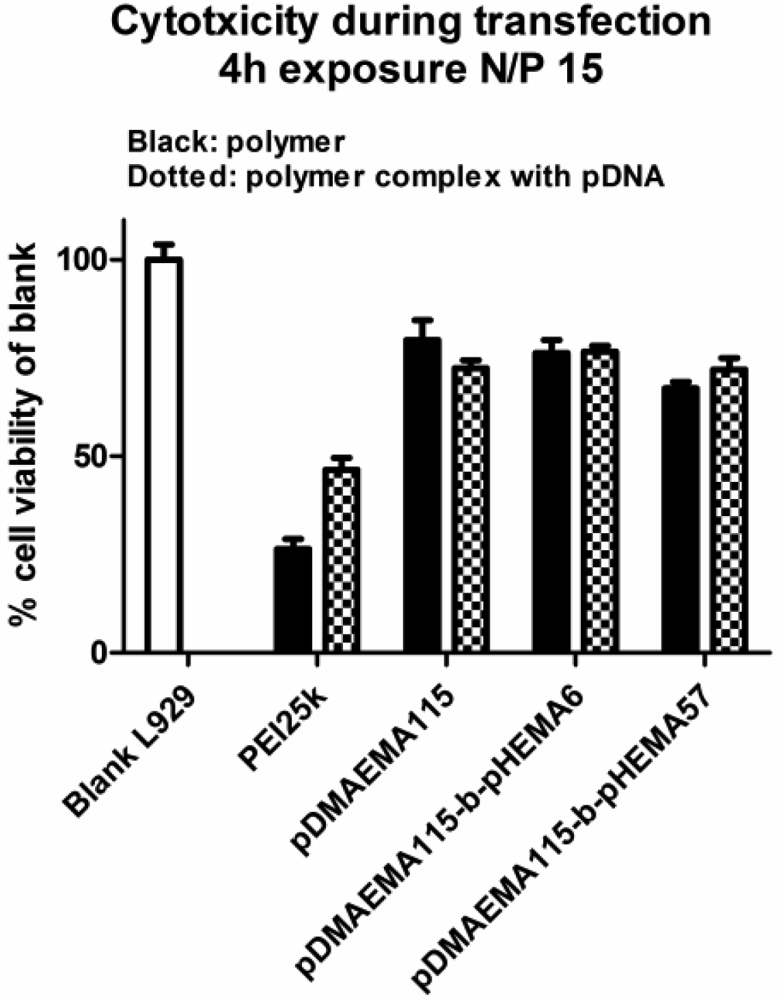
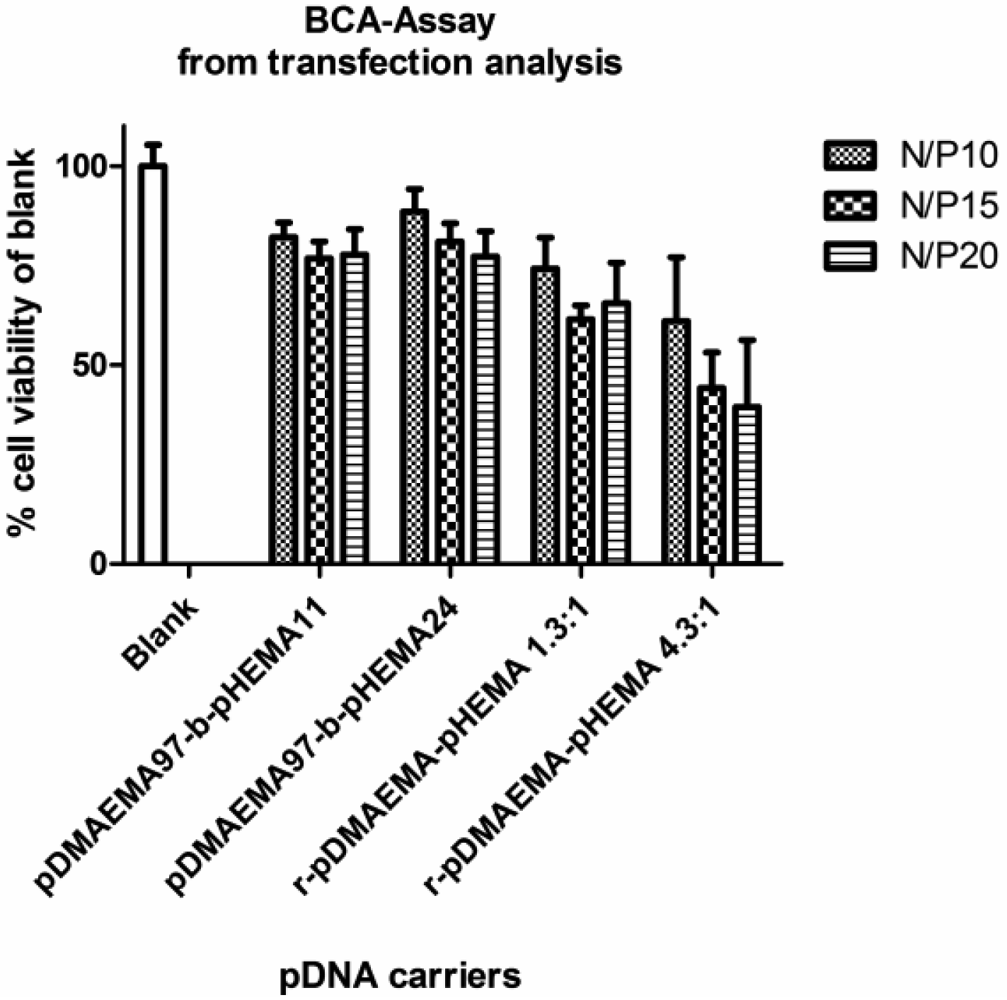
Cytotoxicity data obtained with MTT assays in L929 mouse fibroblasts indicated a better biocompatibility of pDMAEMA-pHEMA diblock copolymers with IC50 values 1.4–9.7 times higher than that of PEI 25k, depending on the polymer structure (Figure 8(A,B)). Comparing the IC50-values of the homo-polymers within the library synthesized here, the 18.3 kDa homo-pDMAEMA was nearly five times less toxic (IC50 = 0.043 mg/mL) than 290 kDa pDMAEMA (IC50 = 0.009 mg/mL) (Figure S2, Supplementary Information). These results were not unexpected as it has been described earlier that the toxicity of homo pDMAEMAs is a function of the molecular weight of the polycation [10]. Cherng et al. [12] reported an IC50-value of 0.03 mg/mL determined in XTT-assays for a pDMAEMA homo-polymer of Mw/Mn of 36 × 104/45 × 103 Da in COS-7 cells. Due to the very broad polydispersity (Mw/Mn) of the polymer, however, it cannot easily be compared with the RAFT polymers reported here which all feature PDI values below 1.20 (Table 1). Copolymer pDMAEMA105-block-pHEMA24 was 9.7-fold less toxic (***p < 0.001), and pDMAEMA115-block-pHEMA57 was 7.9-fold less toxic (**p < 0.01) than PEI 25k. The lowest IC50 values were obtained with r-pDMAEMA-pHEMA 4.3–1 being only 1.4 times less toxic than PEI 25k. The differences in toxicity between the different diblock-copolymers and homo-pDMAEMA solutions in the Mw range of 17–35.7 kDa (exposure for 24 h, Figure 8(A,B)), and between complexes with pDNA and solutions of pure pDMAEMA115 homo- or co-polymers (exposure for 4 h, Figure 9) were found to be not significant. The polymers pDMAEMA97-block-pHEMA11 and r-pDMAEMA-pHEMA 1.3–1 showed some irregularities in biocompatibility with mouse fibroblasts, resulting in very broad toxicity ranges. These irregularities could possibly be caused by residual impurities from the synthesis (see “Materials and Methods” Section 3.4.2. for more details). To further investigate toxicity issues, the protein analysis (BCA assay) following transfection experiments was conferred (see Figure 10). In this assay, a clear trend of decreasing protein values related to blank could be observed towards higher N/P ratios, correlating well with earlier described results [9,10]. As reported previously, cell membrane destabilization may be one of the reasons for cytotoxicity [9,10] and is connected with the charge density. The homopolymer pDMAEMA, however, has a lower charge density than PEI 25k. Additionally, grafting of uncharged pHEMA may contribute to the deviation from a linear trend of pDMAEMA-toxicity as a function of the chain length. The toxicity of random copolymers r-pDMAEMA-pHEMA 4.3–1 which was notably higher than that of diblock copolymers as observed in both MTT and BCA, correlated well with higher polycation content. Surprisingly, no significant differences were found concerning toxicity between pDMAEMA115-derivatives applied as polyplexes or pure polymers of the same concentration (Figure 9). However, Cherng et al. also observed only a partial “neutralizing effect” of polymer toxicity by plasmid for homo-pDMAEMA of Mw/Mn of 36 × 104/45 × 103 Da in COS-7 cells [12]. PEI 25k was much more toxic as pure polymer than its pDNA complexes (Figure 9), which is in line with earlier reports describing “charge neutralization” in polyplexes being able to decrease the toxicity of PEI [33].
The mechanism of interaction of diblock- and random-copolymers with cellular membranes as well as further optimization of the HEMA block-length for better biocompatibility is currently under investigation.
2.7. Transfection
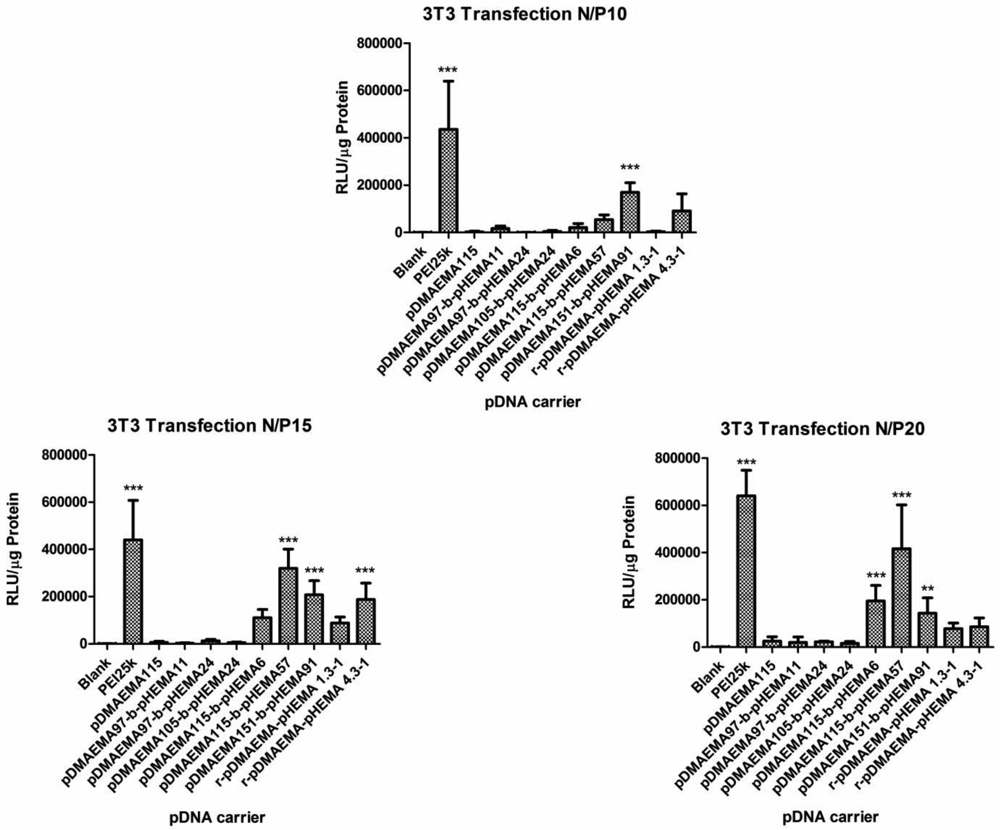
Based on the results of the physico-chemical characterization of pDMAEMA-pHEMA polyplexes concerning polyplex size distribution, zeta potential and complexation efficiency, none of the polymers from the tested panel could be excluded as candidate for transfection. However, the in vitro transfection experiments using pDNA encoding for luciferase in comparison to PEI 25k as standard presented a different picture. The obtained relative light units (RLU) related to μg protein were statistically evaluated using two-way ANOVA comparing them to blanks. Free DNA did not mediate significant luciferase expression compared to untreated cells (data not shown). Therefore, in further experiments, untreated cells were used as blanks. The p-values are indicated on the bar heads with stars (*p < 0.05; **p < 0.01;***p < 0.001). At N/P 10, only one efficient candidate (pDMAEMA151-block-pHEMA91) that caused significantly higher transgene expression than naked DNA was observed apart from PEI 25k (see Figure 11). At higher N/P ratios two additional candidates were identified mediating significant transgene expression (pDMAEMA115-block-pHEMA57, pDMAEMA115-block-pHEMA6). As expected from previous reports, where 43 kDa pDMAEMA was not successful in transfection [9], the homopolymer pDMAEMA consisting of 115 repeating units (18.3 kDa) was found to be an ineffective transfection agent at all tested N/P ratios. On the other hand, its derivatives pDMAEMA115-block-pHEMA57 and pDMAEMA115-block-pHEMA6 with Mw of 25.7 and 19.1 kDa, respectively, provided transfection efficiency at higher N/P ratios, whereas Jiang et al. only achieved moderate transfection results with brushed pDMAEMA-pHEMA copolymers far above 75 kDa at N/P 6 [15]. The diblock copolymers with slightly higher pDMAEMA content (151 units, total Mw of 35.7) were able to mediate transfection already at N/P 10. The random copolymer r-pDMAEMA-pHEMA 1.3–1 (29.0 kDa) yielded no statistically significant luciferase expression over the whole N/P-range tested. Thus, it can be assumed that one of the factors essential for transfection is the number of charged repeating units incorporated within the diblock copolymer, as a clear trend of increasing transfection efficacy towards higher homo-pDMAEMA molecular weights was already observed earlier [9,10]. The candidates possessing only 105 or even 97 pDMAEMA repeating units in a pDMAEMA-pHEMA diblock were ineffective, probably due to the lack of positive charge needed to perform transfection. This data strongly agree with the pre-test transfection experiment where homo-and diblock copolymers with 95 and 110 units pDMAEMA were tested and showed RLU values only at blank level (diblock copolymers data not shown, for homopolymers see Figure S2 Supplementary Information).
The unexpected observation that low molecular weight pDMEMA achieves transfection efficiency only after conjugation with pHEMA in a block structure points to the fact that interaction with cells is affected by this arrangement. It was reported that pHEMA possesses good tissue affinity [19] and may therefore have additional “fusogenic” interactions with cellular membranes enhancing the uptake of polyplexes. The candidates, pDMAEMA151-block-pHEMA91 and pDMAEMA115-block-pHEMA57, being successful in transfection carry rather long pHEMA blocks. The w/w proportion of pDMAEMA:pHEMA blocks for pDMAEMA151-block-pHEMA91 was 1.7:1 and for pDMAEMA115-block-pHEMA57 2:1, suggesting that a two-fold excess of the polycationic part is favorable for transfection. In case of pDMAEMA115-block-pHEMA6, the proportion was 19:1 and for r-pDMAEMA-pHEMA 4.3–1 4.3:1, leading to effective transfection only at one N/P ratio (N/P 20 and 15, respectively). Taken together, the most promising copolymers were found to contain at least 18.5 kDa of the polycationic part grafted to pHEMA in a pDMAEMA:pHEMA proportion of 2 to 1. The efficacy of random copolymers was observed to grow with increasing poylcationic part. The favorable impact of pHEMA on transfection efficacy is being further investigated.
2.8. Uptake Efficiency
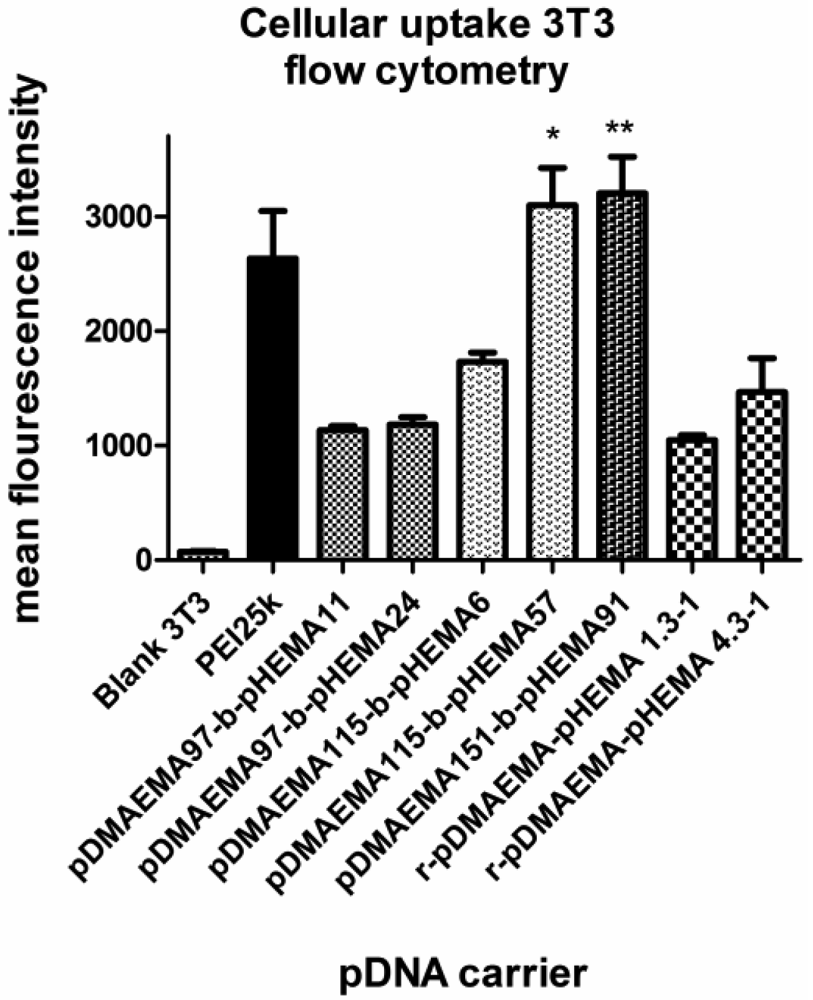
To gain a more detailed insight into the cell uptake of polyplexes from diblock copolymers, uptake efficiency was tested with fluorescently-labeled DNA in the same cell line as used for transfection at an N/P ratio of 15 (Figure 12). The geometric mean fluorescence intensity values (MFI) obtained in flow cytometry experiments were found to be in agreement with the transfection results. Layman et al. observed no influence of pDMAEMA length on cellular uptake in the molecular weight range from 43 to 915 kDa [9]. In this study, however, all pDMAEMA-pHEMAs successfully transfecting 3T3 cells were taken up to a higher degree than the ‘silent’ ones, suggesting that uptake is a critical step for these polyplexes. The structural analysis implies a favorable effect of pHEMA on cell affinity correlating well with previously reported findings [19], since the pDMAEMA115-derivative with a longer pHEMA-graft was taken up to a higher extent than the one with only 6 pHEMA units. Additionally, pDMAEMA151-block-pHEMA57 was also effectively taken up. PEI 25k showed a lower MFI due to its higher toxicity and thus higher percentage of dead cells not gated in the experiment. The same could be speculated regarding r-pDMAEMA-pHEMA 4.3:1.
2.9. Subcellular Distribution (CLSM, Confocal Laser Scanning Microscopy)
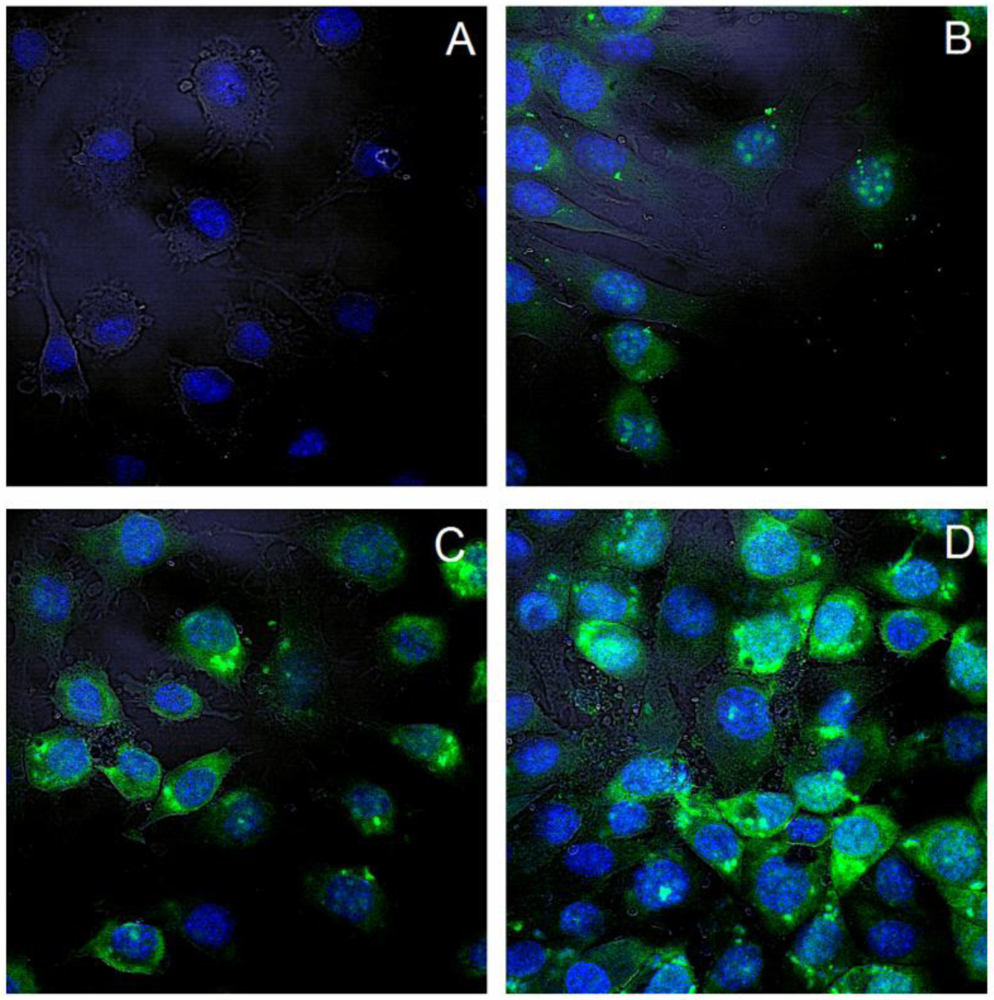
After internalization of polyplexes, the intracellular distribution is of importance. Confocal microscopy was performed to characterize the intracellular polyplex distribution 4 h post exposure to the cells. YoYo-1 labeled pDNA is displayed in green, and DAPI stained nuclei appear blue, the cell membranes are visualized with transmitted light. The images of all three channels are superimposed.
For this study, polyplexes of diblock copolymers with the most prominent transfection efficiency were depicted at an N/P ratio of 15. In all cases the green fluorescence was clearly observed in the perinuclear area, inside the nucleus as well as diffusely in the cytoplasm. This confirms an effective uptake within the period of 4 hours. Since microscopy does not allow for quantitative analysis, only representative images of transfected cells are shown in Figure 13. Cells treated with PEI-pDNA complexes appeared to be less intensively fluorescent. This may be related to the higher toxicity of PEI 25k at N/P 15 or to the fact that there was less fluorescently labeled DNA in the cytosol, but the DNA was translocated to the nucleus. Especially in cells treated with polyplexes of copolymers, broadly disseminated fluorescence as well as dot-formed green was present. This reflects the presence of polyplexes captured in endosomes as well as pDNA released into cytoplasm. Presence of fluorescence in the perinuclear area and inside the nucleus is a good predisposition for effective transfection. Layman et al. showed successful cellular uptake of polyplexes made of pDMAEMA 915 kDa as early as 30 min after application [9]. However, at that time point, only dotted increments were found, indicating endosomal capture of the polyplexes.
3. Materials and Methods
3.1. Materials
Benzyl chloride, Sulfur (80%), Potassium hexacyanoferrate(III), 2-Hydroxyethyl methacrylate (HEMA), 2-(Dimethyl amino) ethyl methacrylate (DMAEMA) and 4,4′-Azobis (4-cyanopentanoic acid) (V-501) were purchased from Sigma-Aldrich. Silica gel 60 (0.040–0.063 mm) for column chromatography (230–400 mesh ASTM) was ordered from Merck Chemicals. The dialysis membrane Spectra /Por 6 RC (MWCO 1 kDa) was delivered from VWR International GmbH. HEMA and DMAEMA were distilled under reduced pressure before use. Poly(ethylene imine) (Polymin™, 25 kDa, abbreviated as PEI 25k) was a gift from BASF (Ludwigshafen, Germany). pCMV-Luc (Lot: PF461-090623) was amplified by The Plasmid Factory (Bielefeld, Germany). Beetle Luciferin and heparin sodium salt were bought from Sigma-Aldrich Laborchemikalien GmbH (Seelze, Germany). SYBR® Gold was obtained from Invitrogen (Karlsruhe, Germany). The Pierce® BCA Protein Assay Kit was obtained from Thermo scientific (Schwerte, Germany). All other chemicals not listed were obtained from Sigma-Aldrich Chemie GmbH (Seelze, Germany) and used in the highest purity if not stated otherwise.
3.2. Synthesis
3.2.1. Synthesis of 4-Cyanopentanoic acid Dithiobenzoate (CPT) and Poly (2-(dimethyl amino)ethyl methacrylate) (pDMAEMA)
4-Cyanopentanoic acid dithiobenzoate (CPT), used as chain transfer agent (CTA), was synthesized as described in the literature [29]. The synthesis of pDMAEMA, which was used as a macroCTA, was performed similarly to the method of Scales et al. [28]. Briefly, 80 mmol of DMAEMA were mixed in an ice bath with sodium acetate buffer (pH 5.2) and adjusted to pH 5 with HCl. Subsequently, 0.8 mmol of CPT were dissolved in the mixture and either 0.1 or 0.2 mmol of 4,4′-Azobis (4-cyanopentanoic acid) (V-501) were added. The reaction was started by placing the reaction flask into the oil bath pre-heated to 70 °C. To stop the reaction, the mixture was frozen in liquid nitrogen after a predetermined time. The polymer was purified by dialysis in water, which was adjusted to pH 3–4 with acetic acid. The dialysate was changed three times a day during a period of 4 days. The slightly pink-colored polymer was dried by lyophilization.
3.2.2. Synthesis of Poly (2-(dimethyl amino)ethyl methacrylate)-block-Poly-2-Hydroxyethyl Methacrylate Block Copolymers (pDMAEMA-block-pHEMA)
To synthesize the block copolymer, 0.05 mmol of the macroCTA were dissolved in 5 mL sodium acetate buffer and 5 mmol HEMA were added. The amount of 0.125 mmol of V-501 was added by pipetting 0.25 mL of a freshly prepared stock solution (14 mg/mL) into the flask. The reaction was started by placing the reaction flask into in oil bath, which was pre-heated to 70 °C. To stop the reaction after a certain time, the mixture was frozen in liquid nitrogen. The purification and drying of the block copolymer was performed by dialysis and lyophilization as described above.
3.2.3. Synthesis of Poly(2-(dimethyl amino)ethyl methacrylate)-Poly-2-Hydroxyethyl Methacrylate Random Copolymers (r-pDMAEMA-pHEMA)
The random copolymers were prepared by mixing different ratios of DMAEMA and HEMA in a final volume of 10 mL sodium acetate buffer (pH 5.2) with 0.03 mmol V-501. The DMAEMA was therefore dissolved in 5 mL buffer first and adjusted to pH 5 with HCl. The reaction was started by placing the reaction flask in an oil bath, which was pre-heated to 70 °C. After 2.5 h, the reaction mixture was frozen in liquid nitrogen. The copolymer was precipitated in THF and dried over night at room temperature. Afterwards, the slightly pink-colored polymer was dissolved in water and dried by lyophilization.
3.2.4. Impurities
Residues of monomers could be a reason for toxicity of prepared polymers although all residues which may originate from the synthesis of the homo polymers and block copolymers were removed by dialysis of 40 mL of polymer solution in 10 L distilled water. The water was changed three times a day over a period of four days. Monomers left over in the polymer solution of random copolymers were separated from the polymer by precipitation of the polymer in THF. Residual THF was removed by lyophilization.
3.3. Characterization of Polymers
The CPT was characterized by 1H, 13C nuclear magnetic resonance (NMR) and attenuated total reflectance (ATR) spectroscopy whereas the polymers were also characterized by gel permeation chromatography (GPC) and differential scanning calorimetry (DSC). The NMR spectra were obtained with a JEOL GX 400D in either CDCl3, deuterated water, or methanol-d4. ATR measurements were performed on a Spectrometer of the Excalibur series from Digi-Lab containing an ATR unit with the WinIR Pro Version 3.3 software in a range between 600 and 4,000 cm−1. The GPC system contained a Duratec7505 degasser, Merck-Hitachi HPLC system (L6000 pump, AS-2000 auto sampler, T6300 column thermostat), Wyatt Dawn-EOS multi-angle laser light scattering detector (calibrated with toluene and normalized with 22 kDa pullulan standard) and an Optilab DSP refractometer. The columns used for sample separation were a PSS 10μ Novema pre-column and two PSS10μ Novema 30 columns of 8 × 300 mm. The measurements were performed at 35 ° C using 1% formic acid as eluent with a flow rate of 0.5 mL/min (laser wavelength: 690 nm, cell type: K5). The analysis and the calculation of the refractive indices of the copolymers were performed using the Wyatt Astra software V5.1.9. The refractive index of pDMAEMA was obtained from the literature [34]. The DSC measurements were performed using a DSC 7 unit with TAC7/DX controlling station from Perkin-Elmer in a range between −40 ° C and 120 ° C and a heat rate of 10 °C/min.
3.4. Polyplex Formation
For transfection, 0.5 μg plasmid DNA (pDNA) in a total of 25 μL polyplex solution was applied per well. The polyplexes were prepared by mixing equal volumes of pDNA and copolymer in isotonic glucose, calculated to afford the desired N/P ratios (positively charged polymer nitrogen atoms per phosphate groups in DNA) as described previously by Liu et al. [35]. After mixing by vigorous pipetting, the polyplexes were incubated for 20 min at room temperature for complete self-assembly and used freshly. The preparation was scaled up for several replicates, so that the total volume (max. 112.5 μL) was later divided into single aliquots of 25 μL after incubation, if not stated otherwise. The charge density involved in DNA-complexation defined as factor A (g polymer per mol nitrogen) was calculated to be 157 g/mol for homo-pDMAEMA and increased with larger percentage of the pHEMA part. PEI 25k polyplexes as control were prepared as stated above (factor A = 43 g/mol).
3.5. Retardation Efficiency on Agarose Gel & Heparin Competition Assay
To determine the efficacy of DNA packaging by polycationic polymer, gel electrophoresis was used as described previously [36]. For the experiments, 1% agarose gels containing 0.83 μg/mL ethidium bromide (EB) were run in TAE buffer (40 mM Tris/HCl, 1% acetic acid, 1 mM EDTA, pH 7.4) for 45 min at 80 V using an Electro-4 electrophoresis unit, Thermo Electron, Waltham, MA, USA. The gels were recorded after irradiation with UV-light using a gel documentation system (BioDocAnalyze, Biometra, Göttingen, Germany). The polyplexes of different N/P ratios (0.5, 1, 2, 5 for retardation assay, and 15 for heparin assay) were prepared as described above, and the volumes of 30 μL, containing 0.6 μg DNA were applied to each slot. To omit the soaring of polyplexes from the slots and to visualize the migration along the lane, 3 μL of loading buffer containing glycerol and bromophenole blue were added immediately prior to application to the gel.
To test the polyplex stability in the presence of competing anions (simulating serum components), they were exposed to heparin of different concentrations for 10 min after the usual incubation procedure of 20 min, then applied to the gel with loading buffer. As control for the visibility of DNA-EB intercalation, free DNA was run on the gel.
3.6. Polyplex size- and Charge-Determination via Dynamic Light Scattering (DLS) and Laser Doppler Anemometry (LDA)
3.6.1. Hydrodynamic Diameter and Zeta-Potential of pDNA-Polymer Complexes in Isotonic Glucose
Polyplexes used for this analysis were prepared and incubated as described above. The measurements were performed in triplicates for each N/P ratio and polymer. For each hydrodynamic size measurement (DLS), 50 μL polyplexes were applied directly into a disposable low volume cuvette (Eppendorf, Wesseling-Berzdorf, Germany) and analyzed with the Zetasizer Nano ZS (Malvern, Herrenberg, Germany) equipped with a 4 mW He-Ne laser at a wavelength of 633 nm at 25 °C. The light scattering was detected at a 173° backward scattering angle. The parameters of 5% glucose were applied (refractive index: 1.337, viscosity 1.0351 cP). For zeta potential measurements, the polyplexes were diluted with 750 μL isotonic glucose, applied into a clear zeta U-form cuvette (Malvern, Herrenberg, Germany) and analyzed with the Smoluchowski model, with a dielectric constant of 78.5. Each sample was measured in 5 runs. For both measurements, N/P ratios of 5, 10, 15, and 20 were tested and analyzed with the Zetasizer Nano ZS Software 6.20 (Malvern, Herrenberg, Germany) as reported previously [37].
3.6.2. Influence of Isotonic Salt and Glucose Solutions on Aggregation Behavior of Polyplexes (DLS)
To evaluate the impact of ionic strength of the continuous phase on the aggregation behavior of polyplexes, two types of polyplexes were chosen. Polyplexes of the homo-polymer (pDMAEMA115) and the diblock-copolymer (pDMAEMA115-block-pHEMA57) were measured concerning hydrodynamic diameters over the period of 1 h with 1 min interval using DLS. The volume and concentration of polyplexes were chosen according to conditions for transfection experiments: 25 μL complex suspension containing 0.5 μg pDNA at N/P 15 were added to 75 μL isotonic glucose or NaCl-solution after incubation for 20 min and measured as described under 3.6. The measurement of each sample was performed twice; the mean values and standard deviation are shown in Figure 7.
3.7. Cell Culture
For transfection experiments, confocal laser scanning microscopy (CLSM) and flow cytometry, mouse fibroblasts (3T3) were used that were cultured in DMEM high glucose (PAA Laboratories, Cölbe, Germany), supplemented with 10% fetal bovine serum (Cytogen, Sinn, Germany). For toxicity experiments, L929 mouse fibroblasts cultured in DMEM low glucose (PAA Laboratories, Cölbe, Germany), supplemented with 10% fetal bovine serum (Cytogen, Sinn, Germany) were used according to the USP (Unites States Pharmacopoeia) cytotoxicity test regulations and according to ISO 10993-5. The incubation of both cell lines was performed in a humidified atmosphere with 8% CO2 at 37 °C. For cell experiments, passages between 10 and 25 were applied.
3.8. Cytotoxicity Assays in vitro
3.8.1. MTT-Cytotoxicity Assay with Polymers
The toxicity of pDMAEMA-pHEMA diblock copolymers was tested using L929 murine fibroblasts in MTT-assays as previously described, using 2mg/mL MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide)-stock solution [38]. As pure polymers are considered to be more toxic than the polyplexes [12,35], the harsher conditions were tested by applying the pure polymer dilutions in a concentration range from 0.5 mg/mL to 0.00029 mg/mL to 96-well plates with 8,000 cells per well, seeded 24 h prior to the experiment. As 100% viability control, untreated cells were used. The IC50 values were compared with that of PEI 25k. For each polymer and dilution step, 4 replicates were used. After dissolving the metabolically formed formazane crystals in DMSO, the absorbance was measured using a plate reader (Titertek plus MS 212, ICN, Germany) at wavelengths of 570 and 690 nm. For data evaluation, GraphPadPrism 5.0 (Graph Pad Software, La Jolla, USA) software was used, the x-scale was plotted logarithmically and nonlinear fit with variable slope for data transformation was run to obtain the IC50 values.
3.8.2. MTT-Cytotoxicity Assay with pDNA-Polymer Complexes
To directly compare the toxicity of polyplexes to that of polymers only, three pDMAEMA115-derivatives were selected and applied to the cells at N/P 15 for 4 h at transfection conditions (see Section 3.9. Transfection efficiency). After 4 h of incubation, the viability evaluation was performed as described above. Each polymer complex was tested in replicates of four.
3.8.3. Toxicity via Protein Assay (Bicinchonic Acid, BCA)
To follow the toxic effect of polymer complexes during the whole transfection period (4 + 44 h), BCA assays were performed according to the manufacturer's protocol (Pierce® BCA Assay Kit) parallel to luciferase expression measurements as described below.
3.9. Transfection Efficiency
For transfection experiments, 3T3 cells were seeded at a density of 6,000 cells per well in 96-well-plates in 200 μL/well full medium 24 hours prior to transfection. After replacing the old medium with 75 μL of fresh full medium, 25 μL polyplexes per well, containing 0.5 μg DNA, were added to the cells and incubated for 4 h at normal cultivating conditions. The medium was changed 4 hours post transfection to 200 μL fresh medium, and cells were incubated for another 44 hours. After the incubation period, the cells were washed with PBS buffer, lysed with cell culture lysis reagent (CCLR, Promega, Mannheim, Germany) and assayed for luciferase expression with a 10 mM luciferin solution on a LumiSTAR plate reader (BMG Labtech, Offenburg, Germany). In parallel, the lysate samples were analyzed by BCA-Assay according to the manufacturer's protocol to determine the protein level. Transfections were performed in replicates of four, and the results are given as mean values +/− SD of relative light units (RLU) normalized to μ g protein according to the BCA assay.
3.10. Confocal Laser Scanning Microscopy (CLSM)
The intracellular distribution of polyplexes was studied by CLSM as described previously [36] using 3T3 cells and YoYo-1 labeled pDNA-polyplexes of N/P 15. After standard incubation with polyplexes for 4 h, cell fixation and DAPI-staining of cell nuclei followed.
3.11. Flow Cytometry
To quantify the intracellular uptake of copolymers transporting pDNA, flow cytometry was employed. 3T3 cells were seeded in 48-well plates at a density of 30,000 cells per well 24 h prior to transfection. For this experiment, polyplexes at N/P 15 were chosen as they were shown to be most effective for transfection and less toxic than polyplexes at N/P 20. To measure pDNA uptake, it was labeled using the intercalating dye YoYo-1 as described previously [36]. In short, polyplex preparation was performed as described for transfection experiments. 3T3 cells were incubated with the polyplexes for 4 h at 37 ° C and afterwards washed with PBS. PEI 25k was used as positive control, and as negative control, untreated cells were measured. Trypan blue 0.4% in PBS was used to quench the fluorescence on the cell surface, allowing for measurement of only internalized YoYo-1 fluorescence. After quenching, the cells were washed twice with PBS, trypsinized and fixed with a 1:1 mixture of FACSFlow (BD Biosciences, San Jose, CA) and 4% paraformaldehyde in PBS. The single cell solutions were measured using a FACSCantoII (BD Biosciences, San Jose, CA) with excitation at 488 nm and the emission filter set to a 530/30 bandpass. The gating was adjusted to evaluate 10,000 viable cells for each experiment. The results of three independent experiments were presented as geometric mean fluorescence intensity (MFI) +/− SD. Flow cytometry analysis was performed using FACSDiva™ software (BD Biosciences, San Jose, CA).
3.12. Statistics
All analytical tests were conducted in replicates of three or four if not otherwise stated. Results are given as mean values +/− standard deviation (SD). Two-way ANOVA and statistical evaluations were performed using Graph Pad Prism 5.0 (Graph Pad Software, La Jolla, USA) if not stated otherwise.
4. Conclusions
It was shown in this study that the in vitro transfection ability of low molecular weight pDMAEMA ≥18.3 kDa could be improved by grafting low molecular weight pHEMA. The high molecular weight homo-polymers were established as effective pDNA vectors by the Hennink group [10,12], with the lower limit for efficient homo-polymers being 43 kDa as reported by Layman et al. [9]. In agreement with these findings, no transfection for 18.3 kDa homo-pDMAEMA at N/P ratios up to 20 was observed in the experiments described here. Accordingly, this polymer was used as “negative control”, whereas its transfection efficiency was succesfully improved by grafting pHEMA. Efficient transfection could be achieved with homo-pDMAEMA of 64 kDa (see Supplementary Information Figure S2). The central idea of this project was on the one hand to create non-viral vectors with molecular weight below the kidney elimination limit suitable for excretion without previous degradation, and on the other hand decreasing the vector's toxicity by reducing the molecular weight of the polycationic part. This approach of improving biocompatibility was successfully performed by Jiang et al. [15], using a pHEMA backbone grafted with cleavable short pDMAEMA-chains, resulting in vectors of molecular weight over 75 kDa. In this present study, the molecular weight of efficient copolymers was successfully decreased to 30–35 kDa. The diblock pDMAEMA-block-pHEMA bearing only 115 pDMAEMA repeating units became an active transfectant after grafting with pHEMA to the total molecular weight of 19 kDa. The diblock copolymers bearing less than 115 polycation units could not be improved in transfection efficacy despite of pHEMA-grafting. Based only on physicochemical characteristics of the polyplexes (size, zeta potential, and complexation ability), no discrimination between ‘active’ and ‘inactive’ copolymers for DNA transfection could be made, all tested copolymers were potentially appropriate for gene delivery, possessing favorable complexation, cellular uptake and DNA-packaging. Only in transfection experiments, the potential as gene vectors could be discriminated. The cellular uptake and intracellular distribution strongly agree with transfection data, underlining the importance of the “charge” and “total Mw” balance. The pHEMA-grafting implies to be favorable for polyplexes in terms of cellular interaction and uptake. The toxicity of the homo-polymers was in good correlation with the pDMAEMA block length and thus with charge density. Concluding, the low molecular weight linear diblock pDMAEMA-block-pHEMA constructs starting from 19 kDa and possessing at least 18 kDa pDMAEMA, are promising candidates for further investigations in the field of gene delivery. Ongoing studies on in vivo transfection experiments, mechanistic aspects of complexation, and kinetic inside the cell are currently under investigation. For cell-type specific transfection, coupling of targeting ligands is being discussed.
Supplementary Materials
polymers-03-00693-s001.pdf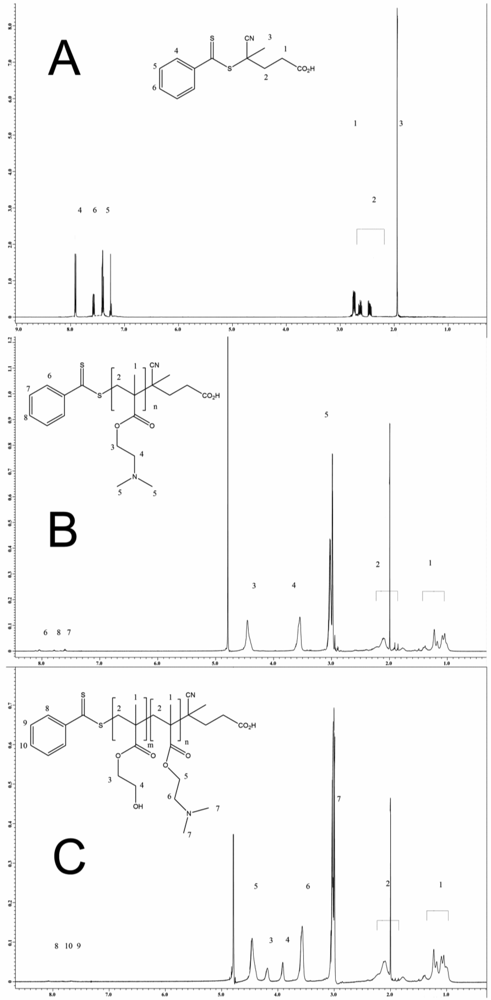

| Name | Polymer | M (NMR) (kDa) | Mn (kDa) | Mw (kDa) | Mw/Mn |
|---|---|---|---|---|---|
| 1 | pDMAEMA115 | 18.3 | 36.3 | 39.0 | 1.08 |
| 2 | pDMAEMA97-b-pHEMA11 | 17.0 | 43.9 | 45.5 | 1.04 |
| 3 | pDMAEMA97-b-pHEMA24 | 18.5 | 38.4 | 40.0 | 1.04 |
| 4 | pDMAEMA105-b-pHEMA24 | 19.9 | 31.2 | 31.5 | 1.01 |
| 5 | pDMAEMA115-b-pHEMA6 | 19.1 | 31.7 | 32.4 | 1.02 |
| 6 | pDMAEMA115-b-pHEMA57 | 25.7 | 25.9 | 26.5 | 1.02 |
| 7 | pDMAEMA151-b-pHEMA91 | 35.7 | 33.3 | 34.4 | 1.03 |
| 8 | r-pDMAEMA-pHEMA 1.3-1 | 29.0 | 26.6 | 27.4 | 1.03 |
| 9 | r-pDMAEMA-pHEMA 4.3-1 | 29.0 | 22.4 | 26.9 | 1.20 |
Acknowledgments
We are grateful to Eva Mohr (Department of Pharmaceutics and Biopharmacy) for technical support in cell culture work and Tanja Dicke for operating FACS-measurements.
References
- Morille, M.; Passirani, C.; Vonarbourg, A.; Clavreul, A.; Benoit, J.P. Progress in Developing Cationic Vectors for Non-Viral Systemic Gene Therapy against Cancer. Biomaterials 2008, 29, 3477–3496. [Google Scholar]
- Mintzer, M.A.; Simanek, E.E. Nonviral Vectors for Gene Delivery. Chem. Rev. 2009, 109, 259–302. [Google Scholar]
- Neu, M.; Fischer, D.; Kissel, T. Recent Advances in Rational Gene Transfer Vector Design Based on Poly(ethylene imine) and Its Derivatives. J. Gene Med. 2005, 7, 992–1009. [Google Scholar]
- Kunath, K.; von Harpe, A.; Fischer, D.; Petersen, H.; Bickel, U.; Voigt, K.; Kissel, T. Low-Molecular-Weight Polyethylenimine as a non-Viral Vector for DNA Delivery: Comparison of Physicochemical Properties, Transfection Efficiency and in vivo Distribution with High-Molecular-Weight Polyethylenimine. J. Control. Release 2003, 89, 113–125. [Google Scholar]
- Mao, S.; Neu, M.; Germershaus, O.; Merkel, O.; Sitterberg, J.; Bakowsky, U.; Kissel, T. Influence of Polyethylene Glycol Chain Length on the Physicochemical and Biological Properties of Poly(ethylene imine)-graft-Poly(ethylene glycol) Block Copolymer/SiRNA Polyplexes. Bioconjugate Chem. 2006, 17, 1209–1218. [Google Scholar]
- Neu, M.; Germershaus, O.; Behe, M.; Kissel, T. Bioreversibly Crosslinked Polyplexes of PEI and High Molecular Weight PEG Show Extended Circulation Times in vivo. J. Control. Release 2007, 124, 69–80. [Google Scholar]
- van de Wetering, P.; Zuidam, N.J.; van Steenbergen, M.J.; van der Houwen, O.A.G.J.; Underberg, W.J.M.; Hennink, W.E. A Mechanistic Study of the Hydrolytic Stability of Poly(2-(dimethylamino)ethyl methacrylate). Macromolecules 1998, 31, 8063–8068. [Google Scholar]
- Jones, R.A.; Poniris, M.H.; Wilson, M.R. pDMAEMA is internalised by endocytosis but does not physically disrupt endosomes. J. Control. Release 2004, 96, 379–391. [Google Scholar]
- Layman, J.M.; Ramirez, S.M.; Green, M.D.; Long, T.E. Influence of Polycation Molecular Weight on Poly(2-dimethylaminoethyl methacrylate)-Mediated DNA Delivery in vitro. Biomacromolecules 2009, 10, 1244–1252. [Google Scholar]
- van de Wetering, P.; Cherng, J.-Y.; Talsma, H.; Hennink, W.E. Relation between Transfection Efficiency and Cytotoxicity of Poly(2-(dimethylamino)ethyl methacrylate)/Plasmid Complexes. J. Control. Release 1997, 49, 59–69. [Google Scholar]
- Wink, T.; de Beer, J.; Hennink, W.E.; Bult, A.; van Bennekom, W.P. Interaction between Plasmid DNA and Cationic Polymers Studied by Surface Plasmon Resonance Spectrometry. Anal. Chem. 1999, 71, 801–805. [Google Scholar]
- Cherng, J.Y.; van de Wetering, P.; Talsma, H.; Crommelin, D.J.; Hennink, W.E. Effect of Size and Serum Proteins on Transfection Efficiency of Poly ((2-dimethylamino)ethyl methacrylate)-Plasmid Nanoparticles. Pharm. Res. 1996, 13, 1038–1042. [Google Scholar]
- Arigita, C.; Zuidam, N.J.; Crommelin, D.J.; Hennink, W.E. Association and Dissociation Characteristics of Polymer/DNA Complexes Used for Gene Delivery. Pharm. Res. 1999, 16, 1534–1541. [Google Scholar]
- Rungsardthong, U.; Ehtezazi, T.; Bailey, L.; Armes, S.P.; Garnett, M.C.; Stolnik, S. Effect of Polymer Ionization on the Interaction with DNA in Nonviral Gene Delivery Systems. Biomacromolecules 2003, 4, 683–690. [Google Scholar]
- Jiang, X.; Lok, M.C.; Hennink, W.E. Degradable-Brushed pHEMA-pDMAEMA Synthesized via ATRP and Click Chemistry for Gene Delivery. Bioconjugate Chem. 2007, 18, 2077–2084. [Google Scholar]
- Verbaan, F.; van Dam, I.; Takakura, Y.; Hashida, M.; Hennink, W.; Storm, G.; Oussoren, C. Intravenous Fate of Poly(2-(dimethylamino)ethyl methacrylate)-Based Polyplexes. Eur. J. Pharm. Sci. 2003, 20, 419–427. [Google Scholar]
- Verbaan, F.J.; Oussoren, C.; Snel, C.J.; Crommelin, D.J.; Hennink, W.E.; Storm, G. Steric Stabilization of Poly(2-(dimethylamino)ethyl methacrylate)-Based Polyplexes Mediates Prolonged Circulation and Tumor Targeting in Mice. J. Gene Med. 2004, 6, 64–75. [Google Scholar]
- Tagami, T.; Nakamura, K.; Shimizu, T.; Yamazaki, N.; Ishida, T.; Kiwada, H. CpG Motifs in pDNA-Sequences Increase anti-PEG IgM Production Induced by PEG-Coated pDNA-Lipoplexes. J. Control. Release 2010, 142, 160–166. [Google Scholar]
- Montheard, J.-P.; Chatzopoulos, M.; Chappard, D. 2-Hydroxyethyl Methacrylate (HEMA): Chemical Properties and Applications in Biomedical Fields. Polym. Rev. 1992, 32, 1–34. [Google Scholar]
- Mabilleau, G.; Moreau, M.F.; Filmon, R.; Basle, M.F.; Chappard, D. Biodegradability of Poly(2-hydroxyethyl methacrylate) in the Presence of the J774.2 Macrophage Cell Line. Biomaterials 2004, 25, 5155–5162. [Google Scholar]
- You, J.O.; Auguste, D.T. Nanocarrier Cross-Linking Density and pH Sensitivity Regulate Intracellular Gene Transfer. Nano Lett. 2009, 9, 4467–4473. [Google Scholar]
- Dai, F.; Sun, P.; Liu, Y.; Liu, W. Redox-Cleavable Star Cationic PDMAEMA by arm-First Approach of ATRP as a Nonviral Vector for Gene Delivery. Biomaterials 2010, 31, 559–569. [Google Scholar]
- Hosseini Nejad, E.; Castignolles, P.; Gilbert, R.G.; Guillaneuf, Y. Synthesis of Methacrylate Derivatives Oligomers by Dithiobenzoate-RAFT-Mediated Polymerization. J. Polym. Sci. A Polym. Chem. 2008, 46, 2277–2289. [Google Scholar]
- Patton, D.L.; Advincula, R.C. A Versatile Synthetic Route to Macromonomers via RAFT Polymerization. Macromolecules 2006, 39, 8674–8683. [Google Scholar]
- Xiong, Q.; Ni, P.; Zhang, F.; Yu, Z. Synthesis and Characterization of 2-(Dimethylamino)ethyl Methacrylate Homopolymers via aqueous RAFT Polymerization and Their Application in Miniemulsion Polymerization. Polym. Bull. 2004, 53, 1–8. [Google Scholar]
- Liu, L.; Wu, C.; Zhang, J.; Zhang, M.; Liu, Y.; Wang, X.; Fu, G. Controlled Polymerization of 2-(diethylamino)ethyl Methacrylate and Its Block Copolymer with N-Isopropylacrylamide by RAFT Polymerization. J. Polym. Sci. A Polym. Chem. 2008, 46, 3294–3305. [Google Scholar]
- York, A.W.; Kirkland, S.E.; McCormick, C.L. Advances in the Synthesis of Amphiphilic Block Copolymers via RAFT Polymerization: Stimuli-Responsive Drug and Gene Delivery. Adv. Drug Delivery Rev. 2008, 60, 1018–1036. [Google Scholar]
- Scales, C.W.; Huang, F.; Li, N.; Vasilieva, Y.A.; Ray, J.; Convertine, A.J.; McCormick, C.L. Corona-Stabilized Interpolyelectrolyte Complexes of SiRNA with Nonimmunogenic, Hydrophilic/Cationic Block Copolymers Prepared by Aqueous RAFT Polymerization. Macromolecules 2006, 39, 6871–6881. [Google Scholar]
- Mitsukami, Y.; Donovan, M.S.; Lowe, A.B.; McCormick, C.L. Water-Soluble Polymers. 81. Direct Synthesis of Hydrophilic Styrenic-Based Homopolymers and Block Copolymers in Aqueous Solution via RAFT. Macromolecules 2001, 34, 2248–2256. [Google Scholar]
- van de Wetering, P.; Cherng, J.Y.; Talsma, H.; Crommelin, D.J.A.; Hennink, W.E. 2-(dimethylamino)ethyl Methacrylate Based (co)polymers as Gene Transfer Agents. J. Control. Release 1998, 53, 145–153. [Google Scholar]
- Chen, L.; Li, H.; Zhao, R.; Zhu, J. Study Progress of Cell Endocytosis. Chin.-Ger. J. Clin. Oncol. 2009, 8, 360–365. [Google Scholar]
- Prokop, A.; Davidson, J.M. Nanovehicular Intracellular Delivery Systems. J. Pharm. Sci. 2008, 97, 3518–3590. [Google Scholar]
- Merkel, O.M.; Beyerle, A.; Beckmann, B.M.; Zheng, M.; Hartmann, R.K.; Stoger, T.; Kissel, T.H. Polymer-Related Off-Target Effects in Non-Viral siRNA Delivery. Biomaterials 2011, 32, 2388–2398. [Google Scholar]
- Sahnoun, M.; Charreyre, M.-T.; Veron, L.; Delair, T.; D'Agosto, F. Synthetic and Characterization Aspects of Dimethylaminoethyl Methacrylate Reversible Addition Fragmentation Chain Transfer (RAFT) Polymerization. J. Polym. Sci. A Polym. Chem. 2005, 43, 3551–3565. [Google Scholar]
- Liu, Y.; Nguyen, J.; Steele, T.; Merkel, O.; Kissel, T. A New Synthesis Method and Degradation of Hyper-Branched Polyethylenimine Grafted Polycaprolactone Block Mono-Methoxyl Poly (ethylene glycol) Copolymers (hy-PEI-g-PCL-block-mPEG) as Potential DNA Delivery Vectors. Polymer 2009, 50, 3895–3904. [Google Scholar]
- Germershaus, O.; Mao, S.; Sitterberg, J.; Bakowsky, U.; Kissel, T. Gene Delivery Using Chitosan, Trimethyl Chitosan or Polyethylenglycol-graft-Trimethyl Chitosan Block Copolymers: Establishment of Structure-Activity Relationships in vitro. J. Control. Release 2008, 125, 145–154. [Google Scholar]
- Merkel, O.M.; Mintzer, M.A.; Librizzi, D.; Samsonova, O.; Dicke, T.; Sproat, B.; Garn, H.; Barth, P.J.; Simanek, E.E.; Kissel, T. Triazine Dendrimers as Nonviral Vectors for in vitro and in vivo RNAi: The Effects of Peripheral Groups and Core Structure on Biological Activity. Mol. Pharm. 2010, 7, 969–983. [Google Scholar]
- Nguyen, J.; Reul, R.; Roesler, S.; Dayyoub, E.; Schmehl, T.; Gessler, T.; Seeger, W.; Kissel, T. Amine-Modified Poly(Vinyl Alcohol)s as Non-viral Vectors for siRNA Delivery: Effects of the Degree of Amine Substitution on Physicochemical Properties and Knockdown Efficiency. Pharma. Res. 2010, 27, 2670–2682. [Google Scholar]
©2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Samsonova, O.; Pfeiffer, C.; Hellmund, M.; Merkel, O.M.; Kissel, T. Low Molecular Weight pDMAEMA-block-pHEMA Block-Copolymers Synthesized via RAFT-Polymerization: Potential Non-Viral Gene Delivery Agents? Polymers 2011, 3, 693-718. https://doi.org/10.3390/polym3020693
Samsonova O, Pfeiffer C, Hellmund M, Merkel OM, Kissel T. Low Molecular Weight pDMAEMA-block-pHEMA Block-Copolymers Synthesized via RAFT-Polymerization: Potential Non-Viral Gene Delivery Agents? Polymers. 2011; 3(2):693-718. https://doi.org/10.3390/polym3020693
Chicago/Turabian StyleSamsonova, Olga, Christian Pfeiffer, Markus Hellmund, Olivia M. Merkel, and Thomas Kissel. 2011. "Low Molecular Weight pDMAEMA-block-pHEMA Block-Copolymers Synthesized via RAFT-Polymerization: Potential Non-Viral Gene Delivery Agents?" Polymers 3, no. 2: 693-718. https://doi.org/10.3390/polym3020693
APA StyleSamsonova, O., Pfeiffer, C., Hellmund, M., Merkel, O. M., & Kissel, T. (2011). Low Molecular Weight pDMAEMA-block-pHEMA Block-Copolymers Synthesized via RAFT-Polymerization: Potential Non-Viral Gene Delivery Agents? Polymers, 3(2), 693-718. https://doi.org/10.3390/polym3020693





