A Novel Three-Colour Fluorescence in Situ Hybridization Approach for the Detection of t(7;12)(q36;p13) in Acute Myeloid Leukaemia Reveals New Cryptic Three Way Translocation t(7;12;16)
Abstract
:1. Introduction
| No. | Disease | Karyotype | Pt no. | Ref. |
|---|---|---|---|---|
| 1 | T-ALL | 48,XX,t(7;12)(q36;p13),+8,+19 | 116 | [20] |
| 2 | AML-M0 | 47,XX,t(7;12)(q36;p13),+19 | 1 | [14] |
| 3 | AML | 48,XX,t(7;12)(q36;p13),+8,+19 | 2 | [14] |
| 4 | AML | 46,XY,t(7;12)(q36;p13)/47,idem,+8 | 13 | [21] |
| 5 | AML-M4 | 47,XY,t(7;12)(q36;p13),+8 | 46 | [22] |
| 6 | AML-M2 | 47,XX,t(7;12)(q36;p13),+19 | 1 | [23] |
| 7 | AML-M5a | 49,XY,t(5;7;12)(q31;q36;p13),+8,+19,+del(22)(q13) | 1 | [15] |
| 8 | ABL | 48,XY,t(7;12)(q36;p13),+19,+22 | 2 | [15] |
| 9 | AML-M0 | 48,XY,t(1;7;12)(q25;q36;p13),+8,+19 | 3 | [15] |
| 10 | AML-M2 | 47,XX,t(7;12)(q36;p13),+19/49,idem,+X,+8 | 63 | [24] |
| 11 | AML-M2 | 47,XX,t(7;12)(q36;p13.1),+19 | 64 | [24] |
| 12 | AML-M6 | 46,XY,der(7)t(7;12)(q32;p13)del(12)(p13)/ 47,idem,+19/47,idem,+8 | 26 | [25] |
| 13 | AML-M2 | 48,XX,t(7;12)(q32;p13),+13,+19 | 27 | [25] |
| 14 | AML | 47,XY,t(7;12)(q36;p13),+19 | 1 | [7] |
| 15 | AML | 48,XY,ins(12;7)(p13;q36;q11.1),+13,+19 | 2 | [7] |
| 16 | AML | 46,XY,t(7;12)(q36;p13) | 1 | [11] |
| 17 | AML-M1 | 47,XY,der(7)t(7;12)(q36;p13)del(12)(p13p13),der(12)t(7;12)(q36;p13),+19 | 2 | [11] |
| 18 | AML-M3v | 47,XY,t(7;12)(q36;p13),+19 | 3 | [11] |
| 19 | AML | 47,XX,t(7;12)(q36;p13),+19/48,idem,+19 | 4 | [11] |
| 20 | AML | 47,XX,t(7;12)(q36;p13),+19 | 6 | [11] |
| 21 | AML | 46,XX,t(7;12)(q36;p13) | 9 | [11] |
| 22 | AML | 46,XX,t(7;12)(q32;p13)/47,idem,+19 | 10 | [11] |
| 23 | AMKL | 46,XX,add(7)(q22),del(12)(p12p13) | 1 | [26] |
| 24 | MDS | 46,XX,der(7)t(7;12)(q22;p13)del(7)(q22q36),der(12)t(7;12)(q36;p13) | 1 | [8] |
| 25 | AML-M5 | 47,XY,del(7)(q32q35-36),t(7;12)(q36;p13),+19 | 2 | [8] |
| 26 | AML-M1 | 47,XX,t(7;12)(q36;p13),+19 | 3 | [9] |
| 27 | T-ALL | 50,XX,+6,del(12)(p13),+18,+19,+22 | 4 | [9] |
| 28 | AML-M0 | 47,XY,t(7;12)(q36;p13),+der(19) | 5 | [9] |
| 29 | AML-M4 | 48,XY,t(7;12)(q36;p13),+8,+19 | 6 | [9] |
| 30 | ALL | 47,XY,t(7;12)(q36;p13),+19 | 7 | [9] |
| 31 | AML | 47,XX,t(7;12)(q36;p13),+8/48,idem,+19/ 50,idem,+X,+19,+19/51,idem,+X,+8,+19,+19 | 17 | [9] |
| 32 | AML-M0 | 47,XY,t(7;12)(q36;p13),+19 | 6 | [10] |
| 33 | AML | 48,XY,t(7;12)(q36;p13),+8,+19 | 7 | [10] |
| 34 | AML-M0 | 46,XX,t(7;12)(q32;p13)/47,idem,+19 | 1 | [12] |
| 35 | AML-M2 | 47,XX,t(7;12)(q36p13),+19 | 4 | [12] |
| 36 | ALL | 47,XX,del(7)(q31),del(12(p13) | 5 | [12] |
| 37 | AML-M0 | 47,XX,+19 | 6 | [12] |
| 38 | AML-M5 | 47,XX,t(7;12)(q36p13),+19 | 7 | [12] |
| 39 | AML | 48,XY,t(7;12)(q36;p13),+8,+19 | 2 | [19] |
| 40 | AML-M2 | 47,XX,t(7;12)(q36;p13),+19 | 3 | [19] |
| 41 | AML-M0 | 47,XX,t(7;12)(q36;p13),+19 | 4 | [19] |
| 42 | AML-M0 | 47,XX,del(7)(q11.2~21),del(12)(p13),+mar | 5 | [19] |
| 43 | AML-M2 | 47,XX,del(12)(q12),+19 | 6 | [19] |
| 44 | AML | 46,XY,inv(2)(p11p13),t(7;12)(q36;p13),der(16)t(1;16)(q22;p13),add(21)(q22) | 5 | [27] |
2. Experimental Section
2.1. Patients Samples
| pt | Age/sex | Disease | Karyotype | Ref. |
|---|---|---|---|---|
| 1 | 7 mo/F | AML | 46,XX,der(7)t(7;12)(q22;p13)del(7)(q22q36) | [8,10] |
| Revised karyotype: | ||||
| 46,XX,der(7)t(7;12)(q36;p13)del(7)(q22q36) | ||||
| 2 | 3 mo/M | AML-M0 | 47,XY,t(7;12)(q36;p13),+der(19) | [10] |
| 3 | 5 mo/F | AML-M1 | 47,XX,t(7;12)(q36;p13),+19 | [10] |
| 4 | 8 mo/F | AML | 47,XX,t(7;12),+19 | [23] |
| 5 | 4 mo/F | AML-M2 | 47,XX,t(7;16)(q36;q12),+19 | [19] |
| Revised karyotype: | ||||
| 47,XX,der(16)t(7;12;16)(q36;p13;q12)inv(16)(p11.2q12),+19 | ||||
| 6 | 10 mo/F | AML-M4 | 48,XX,+19+22,inv(16)(p13q22),del(12p)(p13) | this study |
| Revised karyotype: | ||||
| 48,XX,+19+22,t(7;12)(q36;p13),inv(16)(p13q22) | ||||
| 7 | 6 mo/F | AML-M7 | 47,XX,+19/idem,t(7;12)(q36;p13),+mar | this study |
| 8 | 5 mo/F | MPAL | 46,XX,del(7)(q11),del(12)(p13) | this study |
| Revised karyotype: | ||||
| 46,XX, der(7)t(7;12)(q11;p13)del(7)(q11q36) |
2.2. Probes
2.3. Fluorescence in Situ Hybridization
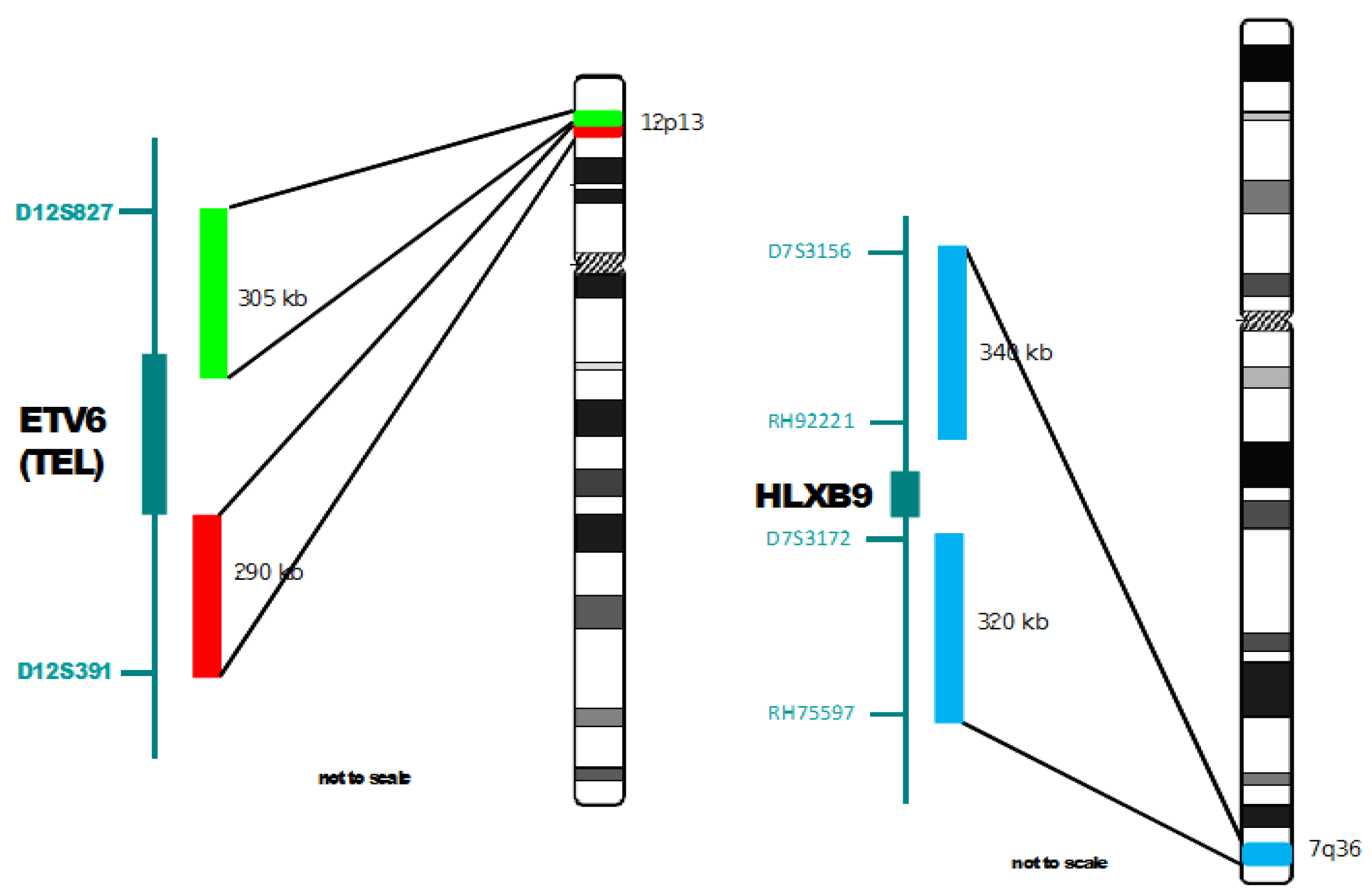
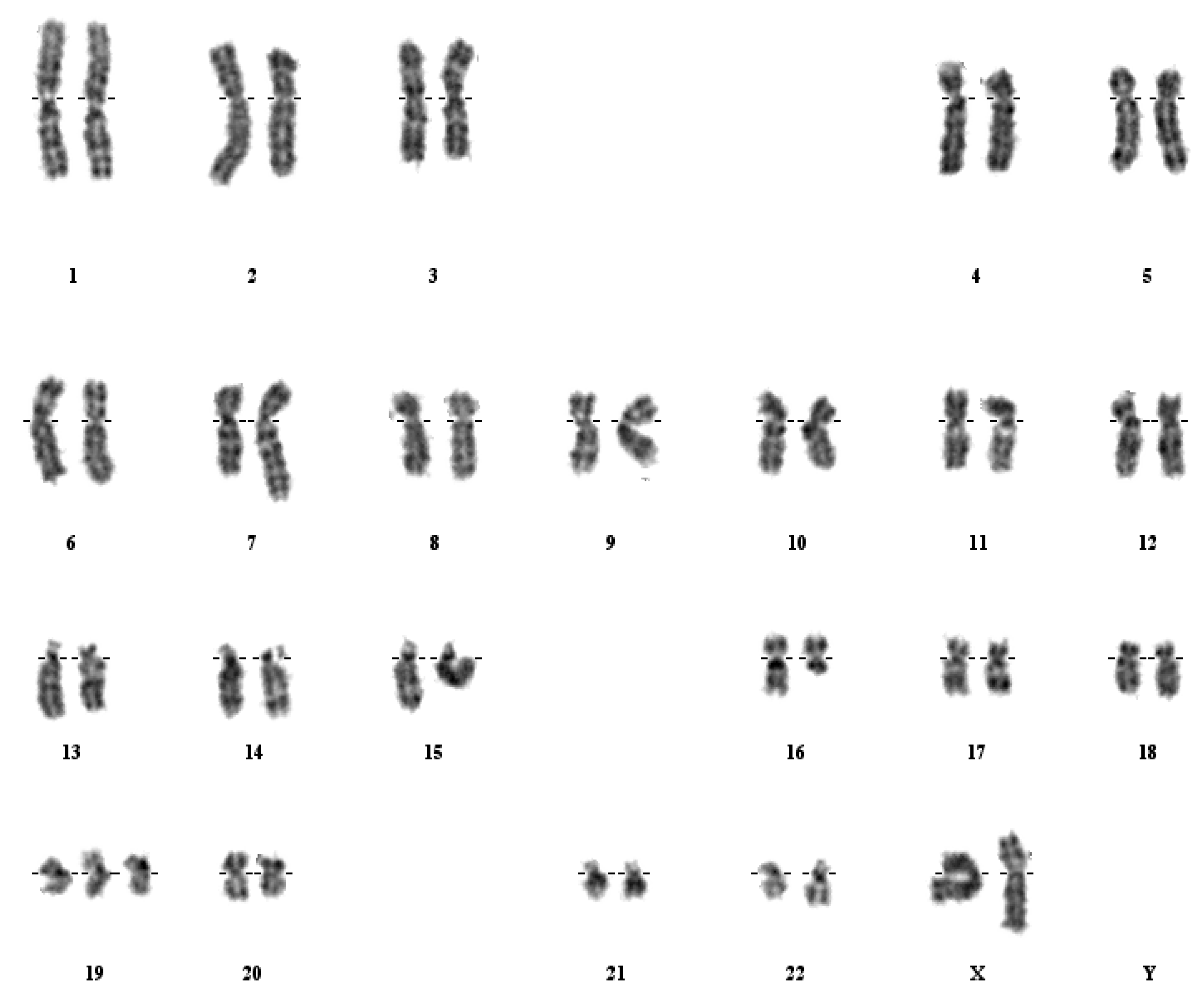
3. Results

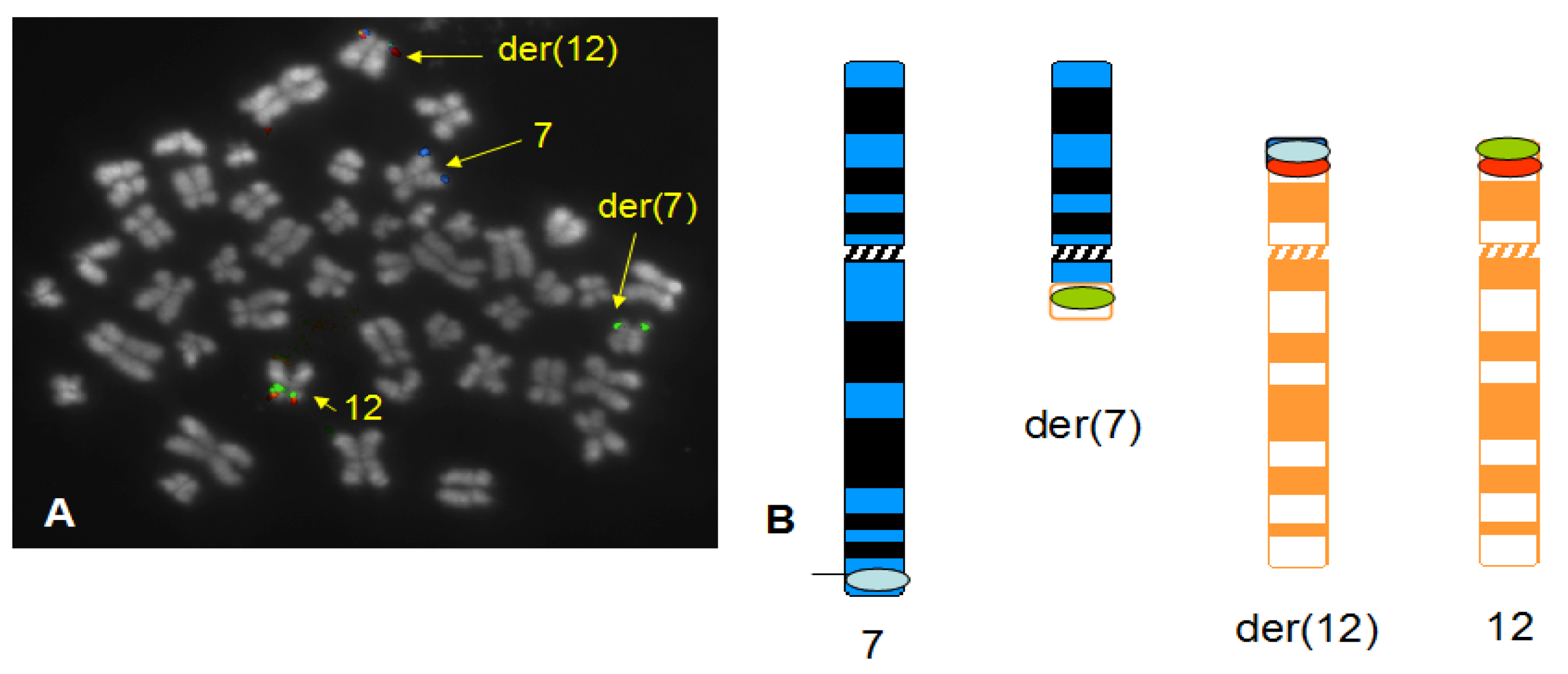
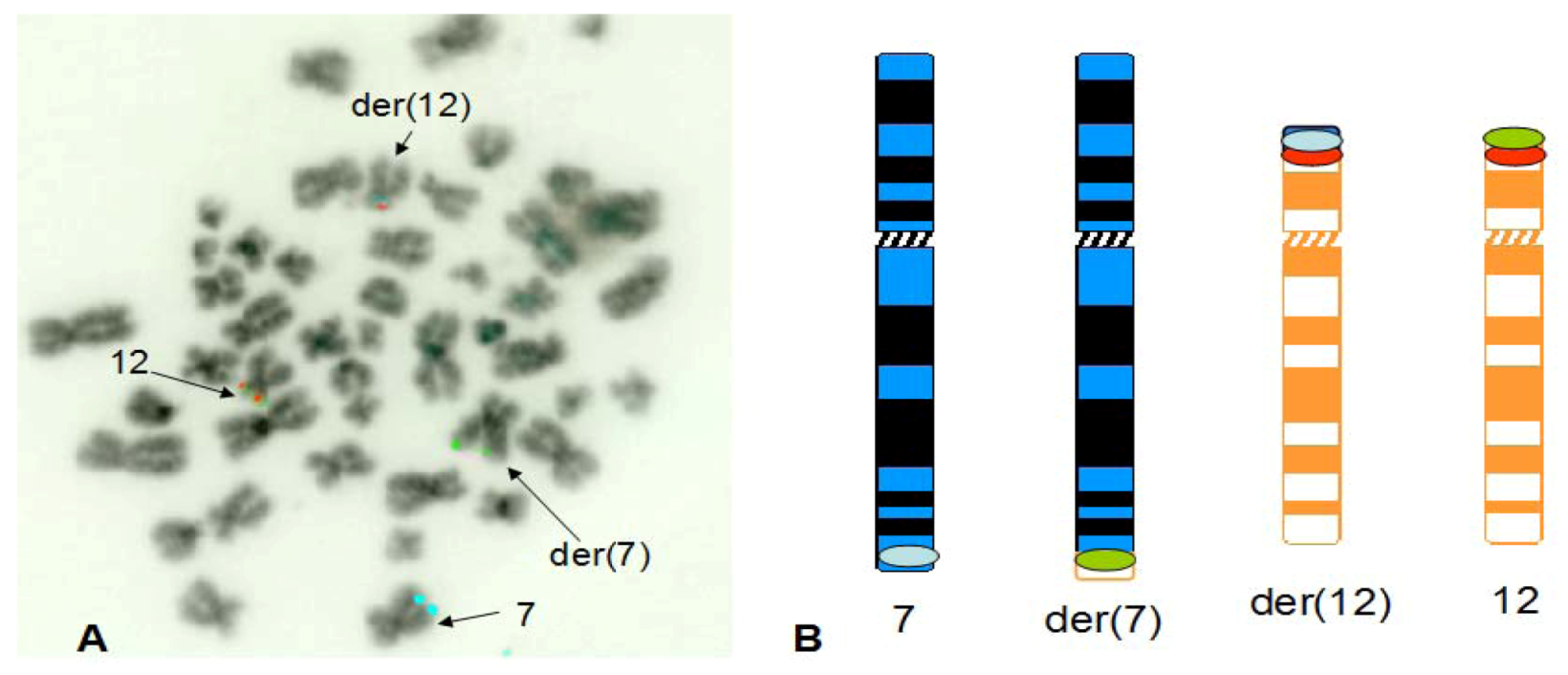
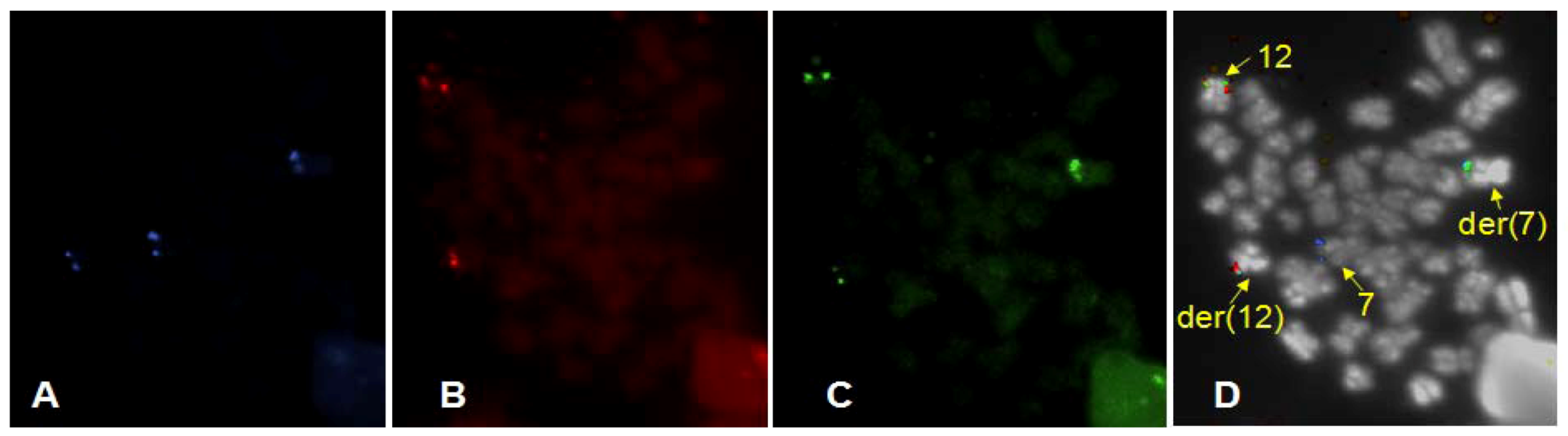

4. Discussion
4.1. Summary of Results on Chromosome 7 and 12 Breakpoints
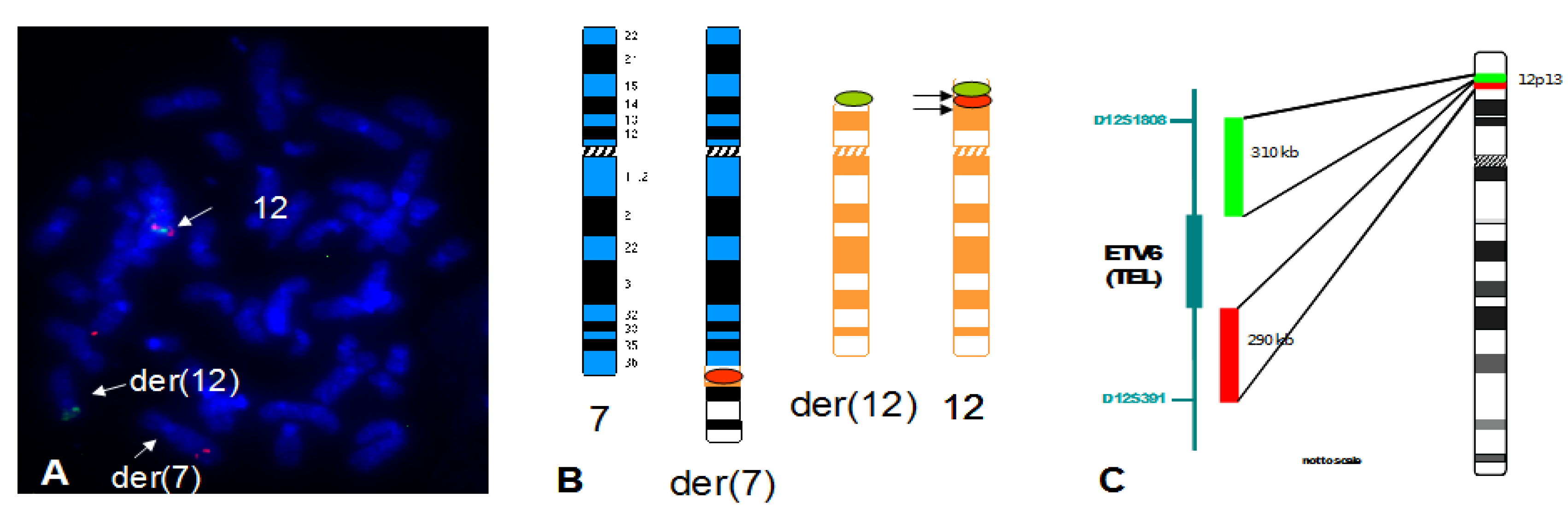
4.2. Finding of a New Cryptic t(7;12) Rearrangement
5. Conclusions
References
- Keen-Kim, D.; Nooraie, F.; Raio, P.N. Cytogenetics biomarkers for human cancer. Front. Biosci. 2008, 13, 5928–5949. [Google Scholar] [CrossRef]
- Lin, C.; Yang, L.; Rosenfeld, M.G. Molecular logic underlying chromosomal translocations, random or non-random? Adv. Cancer Res. 2012, 113, 241–279. [Google Scholar] [CrossRef]
- Bohlander, S.K. Fusion genes in leukemia: An emerging network. Cytogenet. Cell Genet. 2000, 91, 52–56. [Google Scholar] [CrossRef]
- Gough, S.M.; Slape, C.I.; Aplan, P.D. NUP98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef]
- Marschalek, R. Mechanisms of leukemogenesis by MLL fusion proteins. Br. J. Haematol. 2011, 152, 141–154. [Google Scholar] [CrossRef]
- De Braekeleer, E.; Douet-Guilbert, N.; Morel, F.; Le Bris, M.J.; Basinko, A.; de Braekeleer, M. ETV6 fusion genes in haematological malignancies: A review. Leuk. Res. 2012, 36, 945–961. [Google Scholar] [CrossRef]
- Simmons, H.M.; Oseth, L.; Nguyen, P.; O’Leary, M.; Conklin, K.F.; Hirsch, B. Cytogenetic and molecular heterogeneity of 7q36/12p13 rearrangements in childhood AML. Leukemia 2002, 16, 2408–2416. [Google Scholar] [CrossRef]
- Tosi, S.; Giudici, G.; Mosna, G.; Harbott, J.; Specchia, G.; Grosveld, G.; Privitera, E.; Kearney, L.; Biondi, A.; Cazzaniga, G. Identification of new partner chromosomes involved in fusions with the ETV6 (TEL) gene in hematologic malignancies. Genes Chromosomes Cancer 1998, 21, 223–229. [Google Scholar] [CrossRef]
- Tosi, S.; Harbott, J.; Teigler-Schlegel, A.; Haas, O.A.; Pirc-Danoewinata, H.; Harrison, C.J.; Biondi, A.; Cazzaniga, G.; Kempski, H.; Scherer, S.W.; et al. t(7;12)(q36;p13), a new recurrent translocation involving ETV6 in infant leukemia. Genes Chromosomes Cancer 2000, 29, 325–332. [Google Scholar] [CrossRef]
- Tosi, S.; Hughes, J.; Scherer, S.W.; Nakabayashi, K.; Harbott, J.; Haas, O.A.; Cazzaniga, G.; Biondi, A.; Kempski, H.; Kearney, L. Heterogeneity of the 7q36 breakpoints in the t(7;12) involving ETV6 in infant leukemia. Genes Chromosomes Cancer 2003, 38, 191–200. [Google Scholar] [CrossRef]
- Slater, R.M.; van Drunen, E.; Kroes, W.G.; Weghuis, D.O.; van den Berg, E.; Smit, E.M.; van der Does-van den Berg, A.; van Wering, E.; Hahlen, K.; Carroll, A.J.; et al. t(7;12)(q36;p13) and t(7;12)(q32;p13)—Translocations involving ETV6 in children 18 months of age or younger with myeloid disorders. Leukemia 2001, 15, 915–920. [Google Scholar] [CrossRef]
- Von Bergh, A.R.; van Drunen, E.; van Wering, E.R.; van Zutven, L.J.; Hainmann, I.; Lonnerholm, G.; Meijerink, J.P.; Pieters, R.; Beverloo, H.B. High incidence of t(7;12)(q36;p13) in infant AML but not in infant ALL, with a dismal outcome and ectopic expression of HLXB9. Genes Chromosomes Cancer 2006, 45, 731–739. [Google Scholar] [CrossRef]
- Beverloo, H.B.; Panagopoulos, I.; Isaksson, M.; van Wering, E.; van Drunen, E.; de Klein, A.; Johansson, B.; Slater, R. Fusion of the homeobox gene HLXB9 and the ETV6 gene in infant acute myeloid leukemias with the t(7;12)(q36;p13). Cancer Res. 2001, 61, 5374–5377. [Google Scholar]
- Ballabio, E.; Cantarella, C.D.; Federico, C.; di Mare, P.; Hall, G.; Harbott, J.; Hughes, J.; Saccone, S.; Tosi, S. Ectopic expression of the HLXB9 gene is associated with an altered nuclear position in t(7;12) leukemias. Leukemia 2009, 23, 1179–1182. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Lim, J.; Kim, Y.; Han, K.; Lee, J.; Chung, N.G.; Cho, B.; Kim, H.K. Three-way complex translocations in infant acute myeloid leukemia with t(7;12)(q36;p13): The incidence and correlation of a HLXB9 overexpression. Cancer Genet. Cytogenet. 2009, 191, 102–105. [Google Scholar] [CrossRef]
- Hollington, P.; Neufing, P.; Kalionis, B.; Waring, P.; Bentel, J.; Wattchow, D.; Tilley, W.D. Expression and localization of homeodomain proteins DLX4, HB9, and HB24 in malignant and benign human colorectal tissues. Anticancer Res. 2004, 24, 955–962. [Google Scholar]
- Wilkens, L.; Jaggi, R.; Hammer, C.; Inderbitzin, D.; Giger, O.; von Neuhoff, N. The homeobox gene HLXB9 is upregulated in a morphological subset of poorly differentiated hepatocellular carcinoma. Virchows Arch. 2011, 458, 697–708. [Google Scholar] [CrossRef]
- Wildenhain, S.; Ingenhag, D.; Ruckert, C.; Degistirici, O.; Dugas, M.; Meisel, R.; Hauer, J.; Borkhardt, A. Homeobox protein HB9 binds to the prostaglandin E receptor 2 promoter and inhibits intracellular cAMP mobilization in leukemic cells. J. Biol. Chem. 2012, 287, 40703–40712. [Google Scholar]
- Wildenhain, S.; Ruckert, C.; Rottgers, S.; Harbott, J.; Ludwig, W.D.; Schuster, F.R.; Beldjord, K.; Binder, V.; Slany, R.; Hauer, J.; et al. Expression of cell-cell intracting genes distinguishes HLXB9/TEL from MLL-positive childhood acute myeloid leukemia. Leukemia 2010, 24, 1657–1660. [Google Scholar] [CrossRef]
- Andreasson, P.; Hoglund, M.; Bekassy, A.N.; Garwicz, S.; Heldrup, J.; Mitelman, F.; Johansson, B. Cytogenetic and FISH studies of a single center consecutive series of 152 childhood acute lymphoblastic leukemias. Eur. J. Haematol. 2000, 65, 40–51. [Google Scholar]
- Hagemeijer, A.; van Zanen, G.E.; Smit, E.M.; Hahlen, K. Bone marrow karyotypes of children with non-lymphocytic leukemia. Pediatr. Res. 1979, 13, 1247–1254. [Google Scholar] [CrossRef]
- Hagemeijer, A.; Hahlen, K.; Abels, J. Cytogenetic follow-up of patients with nonlymphocytic leukemia. II. Acute nonlymphocytic leukemia. Cancer Genet. Cytogenet. 1981, 3, 109–124. [Google Scholar] [CrossRef]
- Hauer, J.; Tosi, S.; Schuster, F.R.; Harbott, J.; Kolb, H.J.; Borkhardt, A. Graft versus leukemia effect after haploidentical HSCT in a MLL negative infant AML with HLXB9/ETV6 rearrangement. Pediatr. Blood Cancer 2008, 50, 921–923. [Google Scholar] [CrossRef]
- Raimondi, S.C.; Chang, M.N.; Ravindranath, Y.; Behm, F.G.; Gresik, M.V.; Steuber, C.P.; Weinstein, H.J.; Carroll, A.J. Chromosomal abnormalities in 478 children with acute myeloid leukemia: Clinical characteristics and treatment outcome in a co-operative Pediatric Oncology Group Study POG 8821. Blood 1999, 94, 3707–3716. [Google Scholar]
- Satake, N.; Maseki, N.; Nishiyama, M.; Kobayashi, H.; Sakurai, M.; Inaba, H.; Katano, N.; Horikoshi, Y.; Eguchi, H.; Miyake, M.; et al. Chromosome abnormalities and MLL rearrangements in acute myeloid leukemia of infants. Leukemia 1999, 13, 1013–1017. [Google Scholar] [CrossRef]
- Taketani, T.; Taki, T.; Sako, M.; Ishii, T.; Yamaguchi, S.; Hayashi, Y. MNX1-ETV6 fusion gene in an megakaryoblastic leukemia and expression of the MNX1 gene in leukemia and normal B cell lines. Cancer Genet. Cytogenet. 2008, 186, 115–119. [Google Scholar] [CrossRef]
- Wlodarska, I.; La Starza, R.; Baens, M.; Dierlamm, J.; Uyttebroeck, A.; Selleslag, D.; Francine, A.; Mecucci, C.; Hagemeijer, A.; van den Berghe, H.; et al. Fluorescence in situ hybridization characterization of new translocations involving TEL (ETV6) in a wide spectrum of hematologic malignancies. Blood 1998, 91, 1399–1406. [Google Scholar]
- Vieira, L.; Marques, B.; Cavaleiro, C.; Ambrosio, A.P.; Jorge, M.; Neto, A.; Costa, J.M.; Junior, E.C.; Boavida, M.G. Molecular cytogenetic characterization of rearrangements involving 12p in leukaemia. Cancer Genet. Cytogenet. 2005, 157, 134–139. [Google Scholar] [CrossRef]
- Johansson, B.; Floretos, T.; Mitelman, F. Cytogenetic and molecular genetic evolution of chronic myeloid leukaemia. Acta Haematol. 2002, 107, 76–94. [Google Scholar] [CrossRef]
- Tosi, S.; Cabot, G.; Giudici, G.; Attuati, V.; Morandi, P.; Rambaldi, A.; Dohner, H.; Biondi, A. Detection of the breakpoint cluster region-ABL fusion in chronic myeloid leukaemia and variant Philadelphia chromosome translocations by in situ hybridisation. Cancer Genet. Cytogenet. 1996, 89, 153–156. [Google Scholar] [CrossRef]
- Cho, S.R.; Park, S.J.; Kim, H.J.; Park, I.J.; Choi, J.R.; Jung, H.J.; Park, J.E. Acute promyelocytic leukemia with complex translocation t(5;17;15)(q35;q21;q22): Case report and review of the literature. J. Pediatr. Hematol. Oncol. 2011, 33, e326–e329. [Google Scholar]
- Liu, S.; Li, Q.; Pang, W.; Bo, L.; Qin, S.; Liu, X.; Teng, Q.; Qain, L.; Wang, J. A new complex variant t(4;15;17) in acute promyelocytic leukaemia: Fluorescence in situ hybridization confirmation and literature review. Cancer Genet. Cytogenet. 2001, 130, 33–37. [Google Scholar] [CrossRef]
- Belloni, E.; Trubia, M.; Mancini, M.; Derme, V.; Nanni, M.; Lahortiga, I.; Riccioni, R.; Confalonieri, S.; Lo Coco, F.; di Fiore, P.P.; et al. A new complex rearrangement involving the ETV6, LOC115548, and MN1 genes in a case of acute myeloid leukaemia. Genes Chromosomes Cancer 2004, 41, 272–277. [Google Scholar] [CrossRef]
- Crescenzi, B.; La Starza, R.; Nozzoli, C.; Ciolli, S.; Matteucci, C.; Romoli, S.; Rigacci, L.; Gorello, P.; Bosi, A.; Martelli, M.F.; et al. Molecular cytogenetic findings in a four-way t(1;12;5;12)(p36;p13;q33;q24) underlying the ETV6-PDGFRB fusion gene in chronic myelomonocytic leukemia. Cancer Genet. Cytogenet. 2007, 176, 67–71. [Google Scholar] [CrossRef]
- Mathew, S.; Shurtleff, S.A.; Raimondi, S.C. Novel cryptic, complex rearrangements involving ETV6-CBFA (TEL-AML1) genes identified by fluorescence in situ hybridization in pediatric patients with acute lymphoblastic leukemia. Genes Chromosomes Cancer 2001, 32, 188–193. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Naiel, A.; Vetter, M.; Plekhanova, O.; Fleischman, E.; Sokova, O.; Tsaur, G.; Harbott, J.; Tosi, S. A Novel Three-Colour Fluorescence in Situ Hybridization Approach for the Detection of t(7;12)(q36;p13) in Acute Myeloid Leukaemia Reveals New Cryptic Three Way Translocation t(7;12;16). Cancers 2013, 5, 281-295. https://doi.org/10.3390/cancers5010281
Naiel A, Vetter M, Plekhanova O, Fleischman E, Sokova O, Tsaur G, Harbott J, Tosi S. A Novel Three-Colour Fluorescence in Situ Hybridization Approach for the Detection of t(7;12)(q36;p13) in Acute Myeloid Leukaemia Reveals New Cryptic Three Way Translocation t(7;12;16). Cancers. 2013; 5(1):281-295. https://doi.org/10.3390/cancers5010281
Chicago/Turabian StyleNaiel, Abdulbasit, Michael Vetter, Olga Plekhanova, Elena Fleischman, Olga Sokova, Grigory Tsaur, Jochen Harbott, and Sabrina Tosi. 2013. "A Novel Three-Colour Fluorescence in Situ Hybridization Approach for the Detection of t(7;12)(q36;p13) in Acute Myeloid Leukaemia Reveals New Cryptic Three Way Translocation t(7;12;16)" Cancers 5, no. 1: 281-295. https://doi.org/10.3390/cancers5010281





