Cancer Biomarkers: Are We Ready for the Prime Time?
Abstract
:1. Introduction: Defining Biomarkers
2. Historical Perspective
3. Classification of Cancer Biomarkers
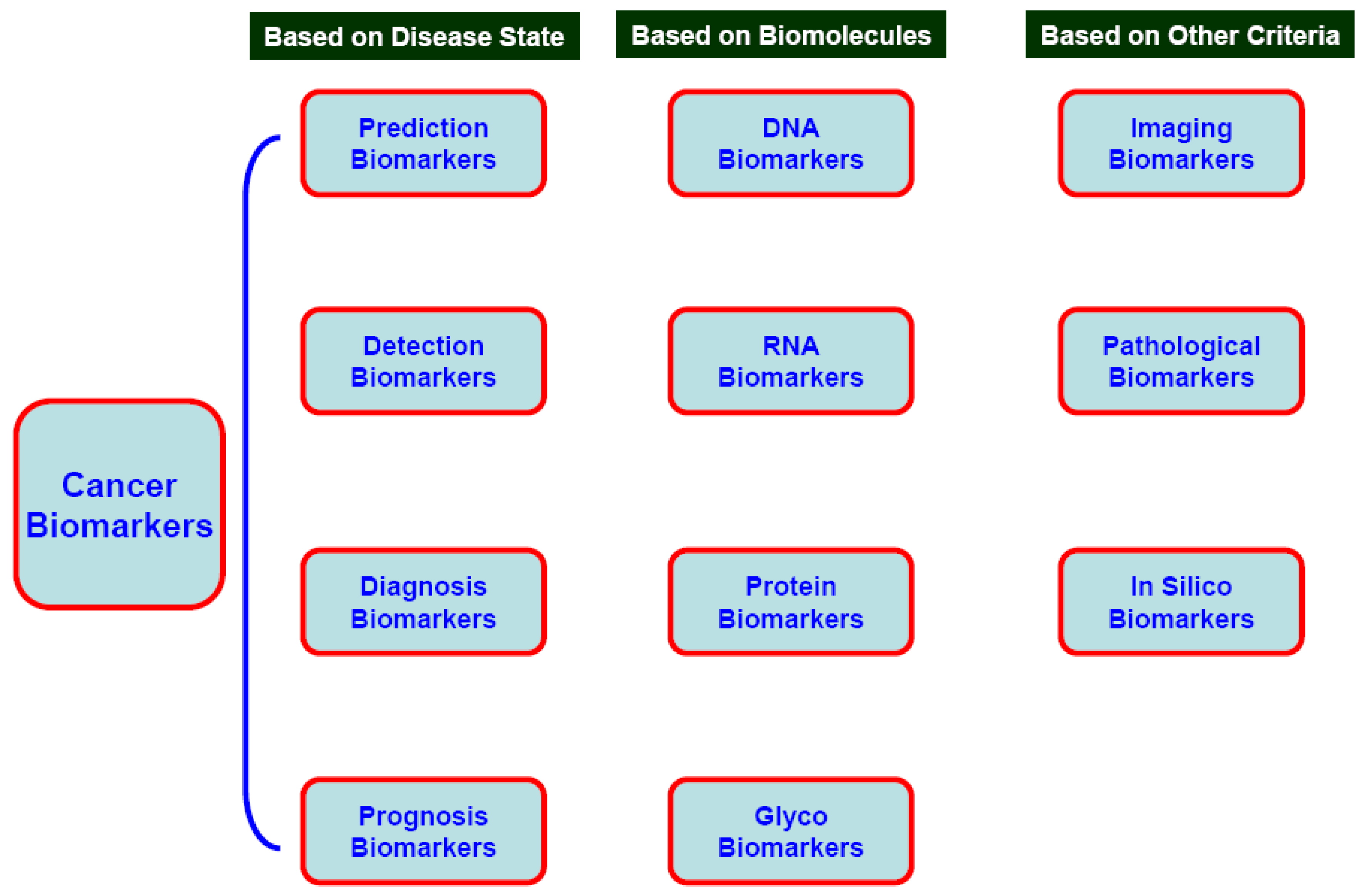
3.1. Prediction, Detection, Diagnostic, Prognostic, and Pharmacodynamics Cancer Biomarkers
3.2. Cancer Biomarkers on the Basis of Biomolecules
3.2.1. DNA
3.2.2. RNA and Micro RNA (miRNA)
3.2.3. Protein Markers
3.2.4. Carbohydrate Biomarkers
3.3. Pathogenic Cancer Markers
3.3.1. Viral markers
3.3.2. Bacterial Markers
3.3.3. Imaging Markers
4. Bioinformatics and Cancer Biomarkers
5. Cancer Biomarkers for Selected Organ Sites
5.1. Lung
5.2. Uterine and Cervical Cancers
5.3. Breast Cancers
5.4. Liver Cancer
5.5. Prostate Cancer
5.6. Head and Neck Cancers
6. Concluding Remarks
6.1. Challenges in the Field and Potential Solutions
Acknowledgements
References
- Gold, P.; Freedman, S.O. Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and absorption techniques. J. Exp. Med. 1965, 121, 439–462. [Google Scholar] [CrossRef]
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; Bloomfield, C.D.; Lander, E.S. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef]
- Barry, M.J. PSA screening for prostate cancer: the current controversy–a viewpoint. Patient outcomes research team for prostatic diseases. Ann. Oncol. 1998, 9, 1279–1282. [Google Scholar] [CrossRef]
- Barry, M.J. The PSA Conundrum. Arch. Intern. Med. 2006, 9, 7–8. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Couper, M.P.; Zikmund-Fisher, B.J.; Levin, C.A.; McNaughton-Collins, M.; Helitzer, D.L.; VanHoewyk, J.; Barry, M.J. Prostate cancer screening decisions: results from the National Survey of Medical Decisions (DECISIONS study). Arch. Intern. Med. 2009, 169, 1611–1618. [Google Scholar] [CrossRef]
- Lippi, G.; Plebani, M.; Franchini, M.; Guidi, G.C.; Favaloro, E.J. Prostate-specific antigen, prostate cancer, and disorders of hemostasis. Semin. Thromb. Hemost. 2009, 35, 654–564. [Google Scholar] [CrossRef]
- Chatterjee, S.K.; Zetter, B.R. Cancer biomarkers: knowing the present and predicting the future. Future Oncol. 2005, 1, 37–50. [Google Scholar] [CrossRef]
- Ludwig, J.A.; Weinstein, J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer 2005, 5, 584–586. [Google Scholar]
- Negm, R.; Verma, M.; Srivastava, S. The promise of biomarkers in cancer screening and detection. Trends Mol. Med. 2002, 8, 288–293. [Google Scholar] [CrossRef]
- Sawyers, C.L. The cancer biomarker problem. Nature 2008, 452, 548–552. [Google Scholar] [CrossRef]
- Verma, M.; Maruvada, P.; Srivastava, S. Current approaches in mitochondrial proteomics. Mito. Matters 2004, 3, 3–5. [Google Scholar] [CrossRef]
- Verma, M. Biomarkers for risk assessment in molecular epidemiology of cancer. Tech. Cancer Res. Treatment. 2004, 3, 505–514. [Google Scholar]
- Verma, M. Pancreatic cancer epidemiology. Tech. Cancer Res. Treatment 2005, 4, 295–302. [Google Scholar]
- Verma, M.; Manne, U. Genetic and epigenetic biomarkers in cancer diagnosis and identifying high risk populations. Crit. Rev. Hematol. Oncol. 2006, 60, 9–18. [Google Scholar] [CrossRef]
- Verma, M.; Kumar, D. Application of mitochondrial genome information in cancer epidemiology. Clin. Chimica. Acta 2007, 383, 41–50. [Google Scholar] [CrossRef]
- Verma, M.; Naviaux, R.; Tanaka, M.; Kumar, D.; Franceschi, C.; Singh, K. Mitochondrial DNA and cancer epidemiology. Cancer Res. 2007, 67, 437–439. [Google Scholar] [CrossRef]
- Verma, M.; Seminara, D.; Arena, J.F.; John, C.; Iwamoto, K.; Hartmuller, V. (2006). Genetic and epigenetic biomarkers in cancer: improving diagnosis and risk assessment. Mol. Diagn. Ther. 2006, 10, 1–15. [Google Scholar] [CrossRef]
- Verma, M.; Kumar, D. Application of mitochondrial genome information in cancer epidemiology. Clin. Chimica. Acta 2007, 383, 41–50. [Google Scholar] [CrossRef]
- Verma, M. Human epigenome and cancer. In Human Genome Epidemiology, 2nd ed.; Khoury, M., Ed.; Oxford University Press: London, UK, 2009; pp. 551–558. [Google Scholar]
- Mishra, A.; Bharti, A.C.; Varghese, P.; Saluja, D.; Das, B.C. Differential expression and activation of NF-kappaB family proteins during oral carcinogenesis: Role of high risk human papillomavirus infection. Int. J. Cancer. 2006, 119, 2840–2850. [Google Scholar] [CrossRef]
- Roses, R.E.; Paulson, E.C.; Sharma, A.; Schueller, J.E.; Nisenbaum, H.; Weinstein, S.; Fox, K.R.; Zhang, P.J.; Czerniecki, B.J. HER-2/neu overexpression as a predictor for the transition from in situ to invasive breast cancer. Cancer Epidemiol. Biomarker Prev. 2009, 18, 1386–1389. [Google Scholar]
- Sharma, S.V.; Bell, D.W.; Settleman, J.; Haber, D.A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 2007, 7, 169–181. [Google Scholar]
- Milone, J.H.; Enrico, A. Treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia. Leuk. Lymphoma 2009, 50, 9–15. [Google Scholar] [CrossRef]
- Habis, A.H.; Vernon, S.D.; Lee, D.R.; Verma, M.; Srivastava, S.; Unger, U. Molecular quality of exfoliated cervical cells: implications for molecular epidemiology and biomarker discovery. Cancer Epidemiol. Biomarkers Pre. 2004, 13, 492–496. [Google Scholar]
- Lau, P.; Chin, J.L.; Pautler, S.; Razvi, H.; Izawa, J.I. NMP22 is predictive of recurrence in high-risk superficial bladder cancer patients. Can. Urol. Assoc. J. 2009, 3, 454–458. [Google Scholar]
- Kageyama, S.; Isono, T.; Matsuda, S.; Ushio, Y.; Satomura, S.; Terai, A.; Arai, Y.; Kawakita, M.; Okada, Y.; Yoshiki, T. Urinary calreticulin in the diagnosis of bladder urothelial carcinoma. Int. J. Urol. 2009, 6, 481–486. [Google Scholar]
- Shariat, S.F.; Karakiewicz, P.I.; Godoy, G.; Karam, J.A.; Ashfaq, R.; Fradet, Y.; Isbarn, H.; Montorsi, F.; Jeldres, C.; Bastian, P.J.; Nielsen, M.E.; Müller, S.C.; Sagalowsky, A.I.; Lotan, Y. Survivin as a prognostic marker for urothelial carcinoma of the bladder: a multicenter external validation study. Clin. Cancer Res. 2009, 15, 7012–7019. [Google Scholar] [CrossRef]
- Sidransky, D. Emerging molecular markers of cancer. Nat. Reviews. Cancer. 2002, 2, 210–219. [Google Scholar] [CrossRef]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Verma, M.; Srivastava, S. Epigenetics in Cancer: Implications for Early Detection and Prevention. Lancet Oncol. 2002, 3, 755–763. [Google Scholar] [CrossRef]
- Verma, M.; Kagan, J.; Sidransky, S.; Srivasatava, S. Proteomic analysis of the cancer cell mitochondria. Nat. Rev. Cancer 2003, 3, 789–795. [Google Scholar] [CrossRef]
- Enokida, H.; Shiina, H.; Igawa, M.; Ogishima, T.; Kawakami, T.; Bassett, W.W.; Anast, J.W.; Li, L.C.; Urakami, S.; Terashima, M.; Verma, M.; Kawahara, M.; Nakagawa, M.; Kane, C.J.; Carroll, P.R.; Dahiya, R. CpG hypermethylation of MDR1 gene contributes to the pathogenesis and progression of human prostate cancer. Cancer Res. 2004, 64, 5956–5962. [Google Scholar] [CrossRef]
- Kaneuchi, M.; Sasaki, M.; Tanaka, Y.; Shiina, H.; Verma, M.; Ebina, Y.; Nomura, E.; Yamamoto, R.; Sakuragi, N.; Dahiya, R. Expression and methylation status of 14-3-3 sigma gene can characterize the different histological features of ovarian cancer. Biochem. Biophys. Res. Commun. 2004, 316, 1156–62. [Google Scholar] [CrossRef]
- Velculescu, V.E.; Zhang, L.; Vogelstein, B.; Kinzler, K.W. Serial analysis of gene expression. Science 1995, 270, 484–487. [Google Scholar]
- Gray, J.W.; Collins, C. Genome changes and gene expression in human solid tumors. Carcinogenesis 2000, 21, 443–452. [Google Scholar] [CrossRef]
- Mischel, P.S.; Cloughesy, T.F.; Nelson, S.F. DNA-microarray analysis of brain cancer: molecular classification for therapy. Nat. Rev. Neurosci. 2004, 5, 782–792. [Google Scholar] [CrossRef]
- Rajeevan, M.S.; Dimulescu, I.M.; Vernon, S.D.; Verma, M.; and Unger, E. Global amplification of sense RNA: a novel method to replicate and archive mRNA for gene expression analysis. Genomics 2003, 82, 491–497. [Google Scholar] [CrossRef]
- Weigl, B.H.; Bardell, R.L.; Cabrera, C.R. Lab-on-a-chip for drug development. Adv. Drug. Deliv. Rev. 2003, 55, 349–377. [Google Scholar] [CrossRef]
- Bartels, C.L.; Tsongalis, G.J. MicroRNAs: novel biomarkers for human cancer. Clin. Chem. 2009, 55, 623–631. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006, 11, 857–866. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; Downing, J.R.; Jacks, T.; Horvitz, H.R.; Golub, T.R. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Hurst, D.R.; Edmonds, M.D.; Welch, D.R. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009, 69, 7495–7498. [Google Scholar] [CrossRef]
- Shenouda, S.K.; Alahari, S.K. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009, 28, 369–378. [Google Scholar] [CrossRef]
- Srivastava, S.; Verma, M.; Gopal-Srivastava, R. Proteomic maps of the cancer-associated infectious agents. J. Proteome Res. 2005, 4, 1171–1180. [Google Scholar] [CrossRef]
- Ravichandran, R.; Vasquez, G.B.; Srivastava, S.; Verma, M.; Petricoin, E.; Lubell, J.; Sriram, R.D.; Barker, P.E.; Gilliland, G.L. Data Standards for Proteomics: Mitochondrial two-dimensional Polyacrylamide Gel Electrophoresis Data as a Model System. Mitochondrion 2004, 3, 327–336. [Google Scholar] [CrossRef]
- Everley, P.A.; Krijgsveld, J.; Zetter, B.R.; Gygi, S.P. Quantitative cancer proteomics: stable isotope labeling with amino acids in cell culture (SILAC) as a tool for prostate cancer research. Mol. Cell Proteomics 2004, 3, 729–735. [Google Scholar] [CrossRef]
- Paweletz, C.P.; Charboneau, L.; Bichsel, V.E.; Simone, N.L.; Chen, T.; Gillespie, J.W.; Emmert-Buck, M.R.; Roth, M.J.; Petricoin III, E.F.; Liotta, L.A. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 2001, 20, 1981–1989. [Google Scholar] [CrossRef]
- Zhou, G.; Li, H.; DeCamp, D.; Chen, S.; Shu, H.; Gong, Y.; Flaig, M.; Gillespie, J.W.; Hu, N.; Taylor, P.R.; Emmert-Buck, M.R.; Liotta, L.A.; Petricoin, E.F., 3rd.; Zhao, Y. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol. Cell Proteomics 2002, 2, 117–124. [Google Scholar]
- Seydel, C. Quantum dots get wet. Science 2003, 300, 80–81. [Google Scholar] [CrossRef]
- Seshi, B. An integrated approach to mapping the proteome of the human bone marrow stromal cell. Proteomics 2006, 6, 5169–5182. [Google Scholar] [CrossRef]
- Shafer, M.W.; Mangold, L.; Partin, A.W.; Haab, B.B. Antibody array profiling reveals serum TSP-1 as a marker to distinguish benign from malignant prostatic disease. Prostate 2007, 67, 255–267. [Google Scholar] [CrossRef]
- Grizzle, E.; Adam, B.; Bigbee, W.L.; Conrads, T.P.; Carroll, C.; Feng, Z.; Izbicka, E.; Jendoubi, M.; Johnsey, D.; Kagan, J.; Verma, M.; Leach, R.; McCarthy, D. B.; Semmes, O. J.; Srivastava, S.; Srivastava, S.; Thompson, I. M.; Thornquist, M.D.; Zhen, Z.; Zhiqiang, Z. Serum Protein Expression Profiling for Cancer Detection: Validation of a SELDI-based Approach for Prostate Cancer. Dis. Markers 2004, 19, 185–195. [Google Scholar] [CrossRef]
- Srinivas, P.; Verma, M.; Zhao, Y.; Srivastava, S. Proteomics for cancer biomarkers discovery. Clin. Chem. 2002, 48, 1160–1169. [Google Scholar]
- Abbott, K.L.; Nairn, A.V.; Hall, E.M.; Horton, M.B.; McDonald, J.F.; Moremen, K.W.; Dinulescu, D.M.; Pierce, M. Focused glycomic analysis of the N-linked glycan biosynthetic pathway in ovarian cancer. Proteomics 2008, 8, 3210–3220. [Google Scholar] [CrossRef]
- Orntoft, T.F.; Vestergaard, E.M. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis 1999, 20, 362–371. [Google Scholar] [CrossRef]
- Saldova, R.; Wormald, M.R.; Dwek, R.A.; Rudd, P.M. Glycosylation changes on serum glycoproteins in ovarian cancer may contribute to disease pathogenesis. Dis. Markers 2008, 25, 219–232. [Google Scholar] [CrossRef]
- Misonou, Y.; Shida, K.; Korekane, H.; Seki, Y.; Noura, S.; Ohue, M.; Miyamoto, Y. Comprehensive clinico-glycomic study of 16 colorectal cancer specimens: elucidation of aberrant glycosylation and its mechanistic causes in colorectal cancer cells. J. Proteome Res. 2009, 8, 2990–3005. [Google Scholar] [CrossRef]
- Powlesland, A.S.; Hitchen, P.G.; Parry, S.; Graham, S.A.; Barrio, M.M.; Elola, M.T.; Mordoh, J.; Dell, A.; Drickamer, K.; Taylor, M.E. Targeted glycoproteomic identification of cancer cell glycosylation. Glycobiology 2009, 19, 899–909. [Google Scholar] [CrossRef]
- Mechref, Y.; Hussein, A.; Bekesova, S.; Pungpapong, V.; Zhang, M.; Dobrolecki, L.E.; Hickey, R.J.; Hammoud, Z.T.; Novotny, M.V. Quantitative serum glycomics of esophageal adenocarcinoma and other esophageal disease onsets. J. Proteome Res. 2009, 8, 2656–2666. [Google Scholar] [CrossRef]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. [Google Scholar] [CrossRef]
- An, H.J.; Miyamoto, S.; Lancaster, K.S.; Kirmiz, C.; Li, B.; Lam, K.S.; Leiserowitz, G.S.; Lebrilla, C.B. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. J. Proteome Res. 2006, 5, 1626–1635. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Munger, K. Viruses associated with human cancer. Biochem. Biophys. Acta 2008, 1782, 127–150. [Google Scholar]
- Taneja, S.; Sen, S.; Gupta, V.K.; Aggarwal, R.; Jameel, S. Plasma and urine biomarkers in acute viral hepatitis E. Proteome Sci. 2009, 7, 39–40. [Google Scholar] [CrossRef]
- Boxus, M.; Willems, L. Mechanisms of HTLV-1 persistence and transformation. Br. J. Cancer. 2009, 101, 1497–501. [Google Scholar] [CrossRef]
- Yin, M.; Hu, Z.; Tan, D.; Ajani, J.A.; Wei, Q. Molecular epidemiology of genetic susceptibility to gastric cancer: focus on single nucleotide polymorphisms in gastric carcinogenesis. Am. J. Transl. Res. 2009, 1, 44–54. [Google Scholar]
- Suerbaum, S.; Michetti, P. Helicobacter pylori infection. N Engl J Med. 2002, 347, 1175–1186. [Google Scholar] [CrossRef]
- Moriyama, E.H.; Zheng, G.; Wilson, B.C. Optical molecular imaging: from single cell to patient. Clin. Pharmacol. Ther. 2008, 84, 267–271. [Google Scholar] [CrossRef]
- Giovacchini, G.; Picchio, M.; Coradeschi, E.; Scattoni, V.; Bettinardi, V.; Cozzarini, C.; Freschi, M.; Fazio, F.; Messa, C. [(11)C]choline uptake with PET/CT for the initial diagnosis of prostate cancer: relation to PSA levels, tumour stage and anti-androgenic therapy. Eur. J. Nucl. Med. Mol. Imaging. 2008, 35, 1065–1073. [Google Scholar] [CrossRef]
- Andrechek, E.R.; Cardiff, R.D.; Chang, J.T.; Gatza, M.L.; Acharya, C.R.; Potti, A.; Nevins, J.R. Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc. Natl. Acad. Sci. USA. 2009, 106, 16387–16392. [Google Scholar]
- Buccheri, G.; Ferrigno, D. Lung tumour markers in oncology practice: a study of TPA and CA125. Br. J. Cancer 2002, 87, 1112–1118. [Google Scholar] [CrossRef]
- Slebos, R.J.; Kibbelaar, R.E.; Dalesio, O.; Kooistra, A.; Stam, J.; Meijer, C.J.; Wagenaar, S.S.; Vanderschueren, R.G.; van Zandwijk, N.; Mooi, W.J. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N. Engl. J. Med. 1990, 323, 561–565. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Varella-Garcia, M.; Bunn, P.A, Jr.; Di Maria, M.V.; Veve, R.; Bremmes, R.M.; Barón, A.E.; Zeng, C.; Franklin, W.A. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol. 2003, 21, 3798–3807. [Google Scholar] [CrossRef]
- Christensen, J.G.; Burrows, J.; Salgia, R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005, 225, 1–26. [Google Scholar] [CrossRef]
- Steels, E.; Paesmans, M.; Berghmans, T.; Branle, F.; Lemaitre, F.; Mascaux, C.; Meert, A.P.; Vallot, F.; Lafitte, J.J.; Sculier, J.P. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur. Respir. J. 2001, 18, 705–719. [Google Scholar] [CrossRef]
- Salgia, R.; Skarin, A.T. Molecular abnormalities in lung cancer. J. Clin. Oncol. 1998, 16, 1207–1217. [Google Scholar]
- Belinsky, S.A.; Klinge, D.M.; Dekker, J.D.; Smith, M.W.; Bocklage, T.J.; Gilliland, F.D.; Crowell, R.E.; Karp, D.D.; Stidley, C.A.; Picchi, M.A. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin.Cancer Res. 2005, 11, 6505–6511. [Google Scholar] [CrossRef]
- zur Hausen, H. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2002, 2, 342–350. [Google Scholar] [CrossRef]
- Ishimi, Y.; Okayasu, I.; Kato, C.; Kwon, H.J.; Kimura, H.; Yamada, K.; Song, S.Y. Enhanced expression of Mcm proteins in cancer cells derived from uterine cervix. Eur. J. Biochem. 2003, 270, 1089–1101. [Google Scholar] [CrossRef]
- Murphy, N.; Ring, M.; Heffron, C.C.; King, B.; Killalea, A.G.; Hughes, C.; Martin, C.M.; McGuinness, E.; Sheils, O.; O'Leary, J.J. p16INK4A, CDC6, and MCM5: predictive biomarkers in cervical preinvasive neoplasia and cervical cancer. J. Clin. Pathol. 2005, 58, 525–534. [Google Scholar]
- Murphy, N.; Ring, M.; Killalea, A.G.; Uhlmann, V.; O'Donovan, M.; Mulcahy, F.; Turner, M.; McGuinness, E.; Griffin, M.; Martin, C.; Sheils, O.; O'Leary, J.J. p16INK4A as a marker for cervical dyskaryosis: CIN and cGIN in cervical biopsies and ThinPrep smears. J. Clin. Pathol. 2003, 56, 56–63. [Google Scholar] [CrossRef]
- Kumar, D.; Verma, M. Molecular markers of cervical squamous cell carcinoma. CME J. Gynecol. Oncol. 2006, 11, 41–60. [Google Scholar]
- Cheng, Q.; Lau, W.M.; Chew, S.H.; Ho, T.H.; Tay, S.K.; Hui, K.M. Identification of molecular markers for the early detection of human squamous cell carcinoma of the uterine cervix. Br. J. Cancer 2002, 86, 274–281. [Google Scholar] [CrossRef]
- Nakano, K.; Watney, E.; McDougall, J.K. Telomerase activity and expression of telomerase RNA component and telomerase catalytic subunit gene in cervical cancer. Am. J. Pathol. 1998, 153, 857–864. [Google Scholar] [CrossRef]
- Harris, L.; Fritsche, H.; Mennel, R.; Norton, L.; Ravdin, P.; Taube, S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr. American Society of Clinical Oncology. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. 2007, 25, 5287–5312. [Google Scholar] [CrossRef]
- Borgoño, C.A.; Grass, L.; Soosaipillai, A.; Yousef, G.M.; Petraki, C.D.; Howarth, D.H.; Fracchioli, S.; Katsaros, D.; Diamandis, E.P. Human kallikrein 14: a new potential biomarker for ovarian and breast cancer. Cancer Res. 2003, 63, 9032–9041. [Google Scholar]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; Ménard, S.; Palazzo, J.P.; Rosenberg, A.; Musiani, P.; Volinia, S.; Nenci, I.; Calin, G.A.; Querzoli, P.; Negrini, M.; Croce, C.M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Sotiriou, C.; Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef]
- Jing, F.; Zhang, J.; Tao, J.; Zhou, Y.; Jun, L.; Tang, X.; Wang, Y.; Hai, H. Hypermethylation of tumor suppressor genes BRCA1, p16 and 14-3-3sigma in serum of sporadic breast cancer patients. Onkologie 2007, 30, 14–19. [Google Scholar] [CrossRef]
- Banerjee, H.N.; Verma, M. Use of nanotechnology for the development of novel cancer biomarkers. Expert Rev. Mol. Diagn. 2006, 6, 679–683. [Google Scholar] [CrossRef]
- Gupta, S.; Bent, S.; Kohlwes, J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann. Intern. Med. 2003, 139, 46–50. [Google Scholar] [CrossRef]
- Liebman, H.A.; Furie, B.C.; Tong, M.J.; Blanchard, R.A.; Lo, K.J.; Lee, S.D.; Coleman, M.S.; Furie, B. Des-gamma-carboxy (abnormal) prothrombin as a serum marker of primary hepatocellular carcinoma. N. Engl. J. Med. 1984, 310, 1427–1431. [Google Scholar] [CrossRef]
- Wang, S.S.; Lu, R.H.; Lee, F.Y.; Chao, Y.; Huang, Y.S.; Chen, C.C.; Lee, S.D. Utility of lentil lectin affinity of alpha-fetoprotein in the diagnosis of hepatocellular carcinoma. J. Hepatol. 1996, 25, 66–71. [Google Scholar]
- Block, T.M.; Comunale, M.A.; Lowman, M.; Steel, L.F.; Romano, P.R.; Fimmel, C.; Tennant, B.C.; London, W.T.; Evans, A.A.; Blumberg, B.S.; Dwek, R.A.; Mattu, T.S.; Mehta, A.S. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc. Natl. Acad. Sci. USA. 2005, 102, 779–784. [Google Scholar] [CrossRef]
- Zhu, Z.W.; Friess, H.; Wang, L.; Abou-Shady, M.; Zimmermann, A.; Lander, A.D.; Korc, M.; Kleeff, J.; Büchler, M.W. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut 2001, 48, 558–564. [Google Scholar] [CrossRef]
- Catalona, W.J.; Partin, A.W.; Slawin, K.M.; Brawer, M.K.; Flanigan, R.C.; Patel, A.; Richie, J.P.; deKernion, J.B.; Walsh, P.C.; Scardino, P.T.; Lange, P.H.; Subong, E.N.; Parson, R.E.; Gasior, G.H.; Loveland, K.G.; Southwick, P.C. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: a prospective multicenter clinical trial. JAMA 1998, 279, 1542–1547. [Google Scholar] [CrossRef]
- Gaylis, F.D.; Keer, H.N.; Wilson, M.J.; Kwaan, H.C.; Sinha, A.A.; Kozlowski, J.M. Plasminogen activators in human prostate cancer cell lines and tumors: correlation with the aggressive phenotype. J. Urol. 1989, 142, 193–198. [Google Scholar]
- Ivanovic, V.; Melman, A.; Davis-Joseph, B.; Valcic, M.; Geliebter, J. Elevated plasma levels of TGF-beta 1 in patients with invasive prostate cancer. Nat. Med. 1995, 1, 282–284. [Google Scholar] [CrossRef]
- Kattan, M.W.; Shariat, S.F.; Andrews, B.; Zhu, K.; Canto, E.; Matsumoto, K.; Muramoto, M.; Scardino, P.T.; Ohori, M.; Wheeler, T.M.; Slawin, K.M. The addition of interleukin-6 soluble receptor and transforming growth factor beta1 improves a preoperative nomogram for predicting biochemical progression in patients with clinically localized prostate cancer. J. Clin. Oncol. 2003, 21, 3573–3579. [Google Scholar] [CrossRef]
- Paul, B.; Dhir, R.; Landsittel, D.; Hitchens, M.R.; Getzenberg, R.H. Detection of prostate cancer with a blood-based assay for early prostate cancer antigen. Cancer Res. 2005, 65, 4097–4100. [Google Scholar] [CrossRef]
- Reiter, R.E.; Gu, Z.; Watabe, T.; Thomas, G.; Szigeti, K.; Davis, E.; Wahl, M.; Nisitani, S.; Yamashiro, J.; Le Beau, M.M.; Loda, M.; Witte, O.N. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc. Natl. Acad. Sci. USA 1998, 95, 1735–1740. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Barrette, T.R.; Rubin, M.A.; Ghosh, D.; Chinnaiyan, A.M. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002, 62, 4427–4433. [Google Scholar]
- Shariat, S.F.; Lamb, D.J.; Kattan, M.W.; Nguyen, C.; Kim, J.; Beck, J.; Wheeler, T.M.; Slawin, K.M. Association of preoperative plasma levels of insulin-like growth factor I and insulin-like growth factor binding proteins-2 and -3 with prostate cancer invasion, progression, and metastasis. J. Clin. Oncol. 2002, 20, 833–841. [Google Scholar] [CrossRef]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.; Otte, A.P.; Rubin, M.A.; Chinnaiyan, A.M. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef]
- van Houten, V.M.; Tabor, M.P.; van den Brekel, M.W.; Denkers, F.; Wishaupt, R.G.; Kummer, J.A.; Snow, G.B.; Brakenhoff, R.H. Molecular assays for the diagnosis of minimal residual head-and-neck cancer: methods, reliability, pitfalls, and solutions. Clin. Cancer Res. 2000, 6, 3803–3816. [Google Scholar]
- Nunes, D.N.; Kowalski, L.P.; Simpson, A.J. Detection of oral and oropharyngeal cancer by microsatellite analysis in mouth washes and lesion brushings. Oral. Oncol. 2000, 36, 525–528. [Google Scholar] [CrossRef]
- Shores, C.G.; Yin, X.; Funkhouser, W.; Yarbrough, W. Clinical evaluation of a new molecular method for detection of micrometastases in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 2004, 130, 937–942. [Google Scholar] [CrossRef]
- Moll, R.; Franke, W.W.; Schiller, D.L.; Geiger, B.; Krepler, R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell 1982, 31, 11–24. [Google Scholar] [CrossRef]
- Franchi, A.; Santucci, M.; Masini, E.; Sardi, I.; Paglierani, M.; Gallo, O. Expression of matrix metalloproteinase 1, matrix metalloproteinase 2, and matrix metalloproteinase 9 in carcinoma of the head and neck. Cancer 2002, 95, 1902–1910. [Google Scholar] [CrossRef]
- Chen, Z.; Lee, T.L.; Yang, X.P.; Dong, G.; Loercher, A.; Van Waes, C. cDNA microarray and bioinformatic analysis of nuclear factor-kappaB related genes in squamous cell carcinoma. Methods Mol. Biol. 2007, 383, 81–99. [Google Scholar]
- Srivastava, S.; Gray, J.W.; Reid, B.J.; Grad, O.; Greenwood, A.; Hawk, E.T. Translational Research Working Group. Translational Research Working Group developmental pathway for biospecimen-based assessment modalities. Clin. Cancer Res. 2008, 14, 5672–5677. [Google Scholar] [CrossRef]
- Pepe, M.S.; Etzioni, R.; Feng, Z.; Potter, J.D.; Thompson, M.L.; Thornquist, M.; Winget, M.; Yasui, Y. Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst. 2001, 93, 1054–1061. [Google Scholar] [CrossRef]
- Bosman, F.T.; Yan, P.; Tejpar, S.; Fiocca, R.; Van Cutsem, E.; Kennedy, R.D.; Dietrich, D.; Roth, A. Tissue biomarker development in a multicentre trial context: a feasibility study on the PETACC3 stage II and III colon cancer adjuvant treatment trial. Clin. Cancer Res. 2009, 15, 5528–5533. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mishra, A.; Verma, M. Cancer Biomarkers: Are We Ready for the Prime Time? Cancers 2010, 2, 190-208. https://doi.org/10.3390/cancers2010190
Mishra A, Verma M. Cancer Biomarkers: Are We Ready for the Prime Time? Cancers. 2010; 2(1):190-208. https://doi.org/10.3390/cancers2010190
Chicago/Turabian StyleMishra, Alok, and Mukesh Verma. 2010. "Cancer Biomarkers: Are We Ready for the Prime Time?" Cancers 2, no. 1: 190-208. https://doi.org/10.3390/cancers2010190
APA StyleMishra, A., & Verma, M. (2010). Cancer Biomarkers: Are We Ready for the Prime Time? Cancers, 2(1), 190-208. https://doi.org/10.3390/cancers2010190



