Evaluation of Ochratoxin Recognition by Peptides Using Explicit Solvent Molecular Dynamics
Abstract
:1. Introduction
2. Results and Discussion
2.1. Conformational Preferences of Peptide in Solution
2.1.1. Hexamer (SNLHPK)
2.1.2. Octamer (CSIVEDGK)
2.1.3. NFO4 (VYMNRKYYKCCK)
2.1.4. 13-mer (GPAGIDGPAGIRC)
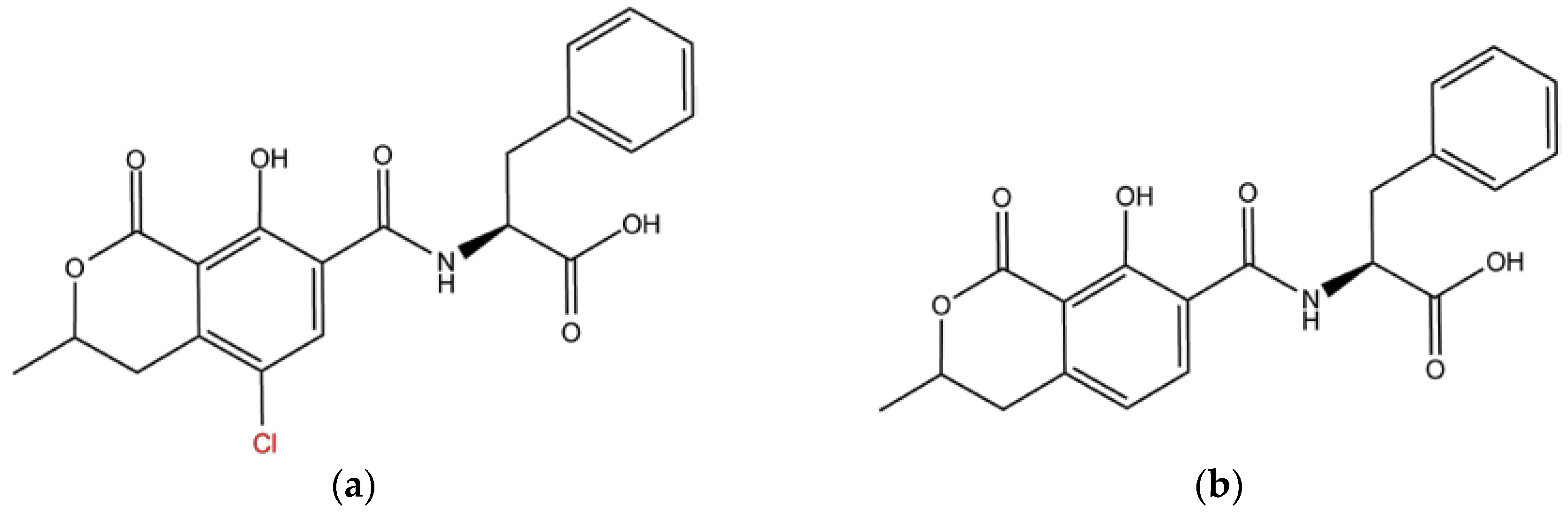
2.2. Structural and Energetic Characteristics Influencing the Peptide Affinity and Selectivity to Ochratoxins
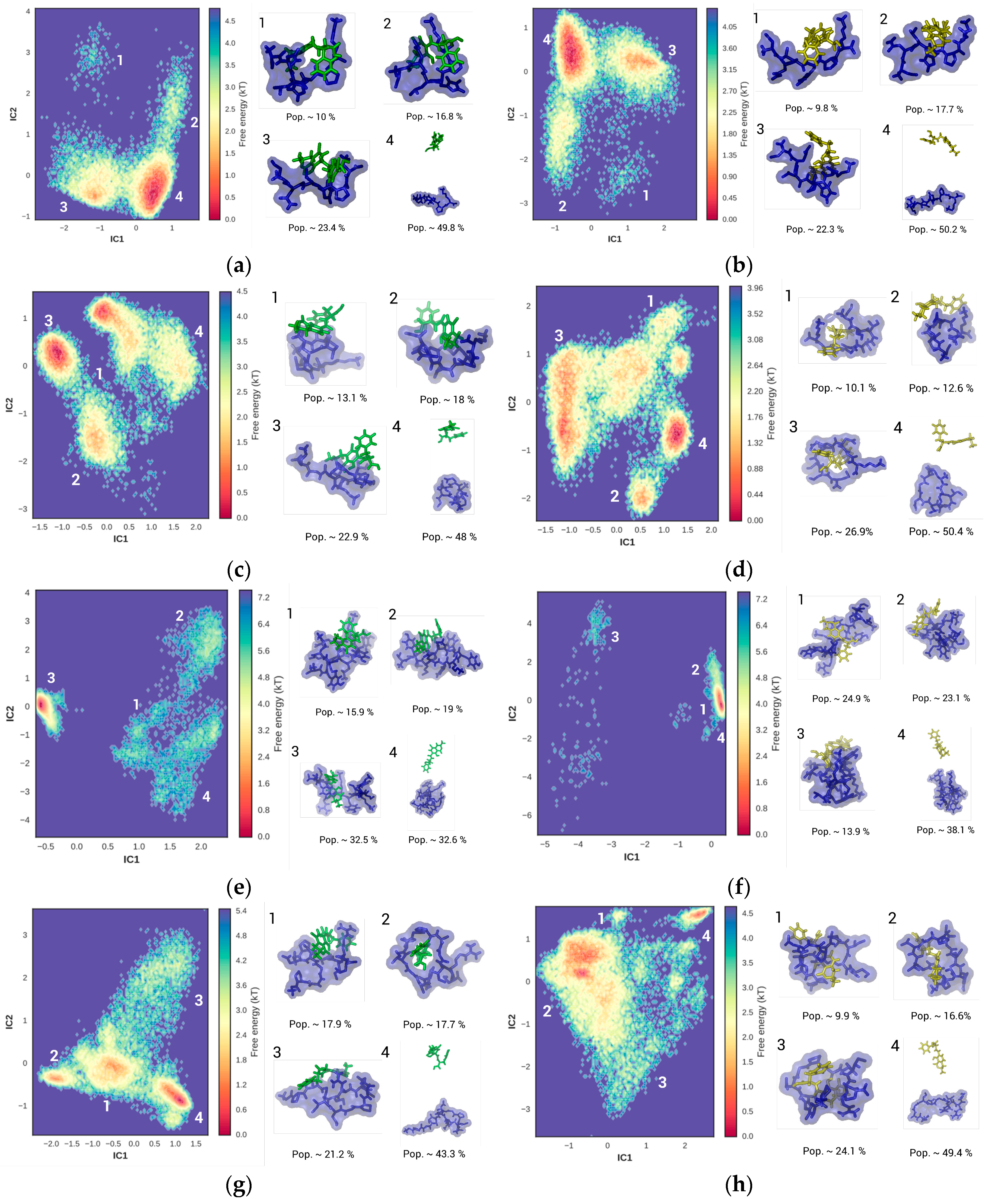
2.2.1. Hexamer–OTA/OTB
2.2.2. Octamer–OTA/OTB
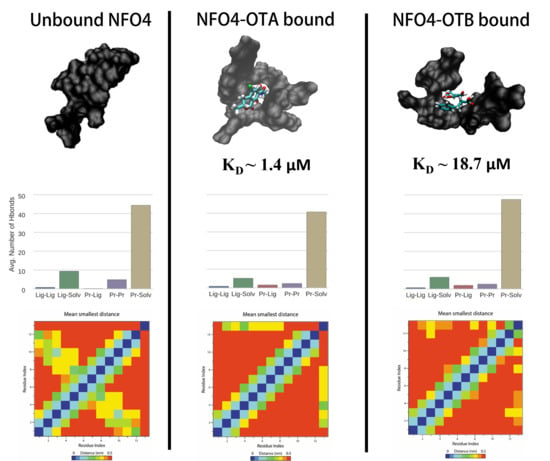
2.2.3. NFO4–OTA/OTB
2.2.4. 13-mer–OTA/OTB
2.3. Validation of Results and Implications for Computer-Aided Design of Biosensing Platforms
3. Conclusions
4. Materials and Methods
4.1. Simulation Protocol
4.2. Sampling of Kinetically Relevant Peptide and Peptide–Hapten Configurations
4.3. Predictive Modeling of the Peptide Folding and Binding Kinetics
4.3.1. MSM Construction and Iteration
4.3.2. Validation of MSM Macrostates
4.3.3. Estimation of Equilibrium Population Distribution and Binding Free Energy
4.4. Other Trajectory Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Malir, F.; Ostry, V.; Pfohl-Leszkowicz, A.; Malir, J.; Toman, J. Ochratoxin A: 50 years of research. Toxins 2016, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Kőszegi, T.; Poór, M. Ochratoxin A: Molecular interactions, mechanisms of toxicity and prevention at the molecular level. Toxins 2016, 8, 111. [Google Scholar]
- Heussner, A.; Bingle, L. Comparative ochratoxin toxicity: A review of the available data. Toxins 2015, 7, 4253. [Google Scholar] [CrossRef] [PubMed]
- Malir, F.; Ostry, V.; Pfohl-Leszkowicz, A.; Novotna, E. Ochratoxin A: Developmental and reproductive toxicity—An overview. Birth Defects Res. Part B Dev. Reprod. Toxicol. 2013, 98, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Ha, T. Recent advances for the detection of ochratoxin A. Toxins 2015, 7, 5276–5300. [Google Scholar] [CrossRef] [PubMed]
- Bazin, I.; Tria, S.A.; Hayat, A.; Marty, J.-L. New biorecognition molecules in biosensors for the detection of toxins. Biosens. Bioelectron. 2017, 87, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.; Arya, S.K.; Vasudev, A.; Bhansali, S. Recent advances in detection of ochratoxin-A. Open J. Appl. Biosens. 2013, 2, 1–11. [Google Scholar] [CrossRef]
- European Union Commission Regulation (EC). Commission Regulation: Setting Maximum Levels for Certain Contaminants in Foodstuffs; European Union Commission Regulation (EC), 1 June 2015; Brussels, Belgium. [Google Scholar]
- Tria, S.A.; Lopez-Ferber, D.; Gonzalez, C.; Bazin, I.; Guiseppi-Elie, A. Microfabricated biosensor for the simultaneous amperometric and luminescence detection and monitoring of ochratoxin A. Biosens. Bioelectron. 2016, 79, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Rhouati, A.; Yang, C.; Hayat, A.; Marty, J.-L. Aptamers: A promising tool for ochratoxin A detection in food analysis. Toxins 2013, 5, 1988. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Velu, R.; De Girolamo, A.; Valenzano, S.; Pascale, M.; Smith, M.; DeRosa, M. Comparison of in-solution biorecognition properties of aptamers against ochratoxin A. Toxins 2016, 8, 336. [Google Scholar] [CrossRef] [PubMed]
- Vidal, J.C.; Bonel, L.; Ezquerra, A.; Hernández, S.; Bertolín, J.R.; Cubel, C.; Castillo, J.R. Electrochemical affinity biosensors for detection of mycotoxins: A review. Biosens. Bioelectron. 2013, 49, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Giraudi, G.; Ferrero, V.E.; Anfossi, L.; Baggiani, C.; Giovannoli, C.; Tozzi, C. Solid-phase extraction of ochratoxin A from wine based on a binding hexapeptide prepared by combinatorial synthesis. J. Chromatogr. A 2007, 1175, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Heurich, M.; Altintas, Z.; Tothill, I.E. Computational design of peptide ligands for ochratoxin A. Toxins (Basel) 2013, 5, 1202–1218. [Google Scholar] [CrossRef] [PubMed]
- Bazin, I.; Andreotti, N.; Hassine, A.I.H.; De Waard, M.; Sabatier, J.M.; Gonzalez, C. Peptide binding to ochratoxin a mycotoxin: A new approach in conception of biosensors. Biosens. Bioelectron. 2013, 40, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Siantar, D.P.; Halverson, C.A.; Kirmiz, C.; Peterson, G.F.; Hill, N.R.; Dugar, S.M. Ochratoxin A in wine: Survey by antibody- and polymeric-based SPE columns using HPLC/fluorescence detection. Am. J. Enol. Vitic. 2003, 54, 170–177. [Google Scholar]
- Giovannoli, C.; Passini, C.; Volpi, G.; Di Nardo, F.; Anfossi, L.; Baggiani, C. Peptide-based affinity media for solid-phase extraction of ochratoxin A from wine samples: Effect of the solid support on binding properties. Talanta 2015, 144, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Oliveberg, M.; Wolynes, P.G. The experimental survey of protein-folding energy landscapes. Q. Rev. Biophys. 2005, 38, 245–288. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.J.; Shukla, D.; Beauchamp, K.A.; Pande, V.S. To milliseconds and beyond: Challenges in the simulation of protein folding. Curr. Opin. Struct. Biol. 2013, 23, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Boehr, D.D.; Nussinov, R.; Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009, 5, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Mobley, D.L.; Dill, K.A. Binding of small-molecule ligands to proteins: ”What you see” is not always “what you get”. Structure 2009, 17, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Plattner, N.; Noe, F. Protein conformational plasticity and complex ligand-binding kinetics explored by atomistic simulations and markov models. Nat. Commun. 2015, 6, 7653. [Google Scholar] [CrossRef] [PubMed]
- Maximova, T.; Moffatt, R.; Ma, B.; Nussinov, R.; Shehu, A. Principles and overview of sampling methods for modeling macromolecular structure and dynamics. PLoS Comput. Biol. 2016, 12, e1004619. [Google Scholar] [CrossRef] [PubMed]
- Abrams, C.; Bussi, G. Enhanced sampling in molecular dynamics using metadynamics, replica-exchange, and temperature-acceleration. Entropy 2014, 16, 163. [Google Scholar] [CrossRef]
- Cossio, P.; Marinelli, F.; Laio, A.; Pietrucci, F. Optimizing the performance of bias-exchange metadynamics: Folding a 48-residue lysm domain using a coarse-grained model. J. Phys. Chem. B 2010, 114, 3259–3265. [Google Scholar] [CrossRef] [PubMed]
- Fiorin, G.; Klein, M.L.; Hénin, J. Using collective variables to drive molecular dynamics simulations. Mol. Phys. 2013, 111, 3345–3362. [Google Scholar] [CrossRef]
- Piana, S.; Laio, A. A bias-exchange approach to protein folding. J. Phys. Chem. B 2007, 111, 4553–4559. [Google Scholar] [CrossRef] [PubMed]
- Zerze, G.H.; Miller, C.M.; Granata, D.; Mittal, J. Free energy surface of an intrinsically disordered protein: Comparison between temperature replica exchange molecular dynamics and bias-exchange metadynamics. J. Chem. Theory Comput. 2015, 11, 2776–2782. [Google Scholar] [CrossRef] [PubMed]
- Pande, V.S.; Beauchamp, K.; Bowman, G.R. Everything you wanted to know about Markov State Models but were afraid to ask. Methods 2010, 52, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Chodera, J.D.; Noe, F. Markov State Models of biomolecular conformational dynamics. Curr. Opin. Struct. Biol. 2014, 25, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Silva, D.A.; Meng, L.; Yue, A.; Huang, X. Quantitatively characterizing the ligand binding mechanisms of choline binding protein using Markov State Model analysis. PLoS Comput. Biol. 2014, 10, e1003767. [Google Scholar] [CrossRef] [PubMed]
- Trendelkamp-Schroer, B.; Wu, H.; Paul, F.; Noe, F. Estimation and uncertainty of reversible Markov Models. J. Chem. Phys. 2015, 143, 174101. [Google Scholar] [CrossRef] [PubMed]
- Scherer, M.K.; Trendelkamp-Schroer, B.; Paul, F.; Pérez-Hernández, G.; Hoffmann, M.; Plattner, N.; Wehmeyer, C.; Prinz, J.-H.; Noé, F. Pyemma 2: A software package for estimation, validation, and analysis of Markov Models. J. Chem. Theory Comput. 2015, 11, 5525–5542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, M.; Kang, Y.; Xie, H.; Wang, X.; Song, H.; Li, X.; Fang, W. Identification of a high-affinity monoclonal antibody against ochratoxin A and its application in enzyme-linked immunosorbent assay. Toxicon 2015, 106, 89–96. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Velu, R.; Hill, K.; Bardóczy, V.; Mészáros, T.; DeRosa, M. Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins 2014, 6, 2435. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Aguado, J.A.; Penner, G. Determination of ochratoxin A with a DNA aptamer. J. Agric. Food Chem. 2008, 56, 10456–10461. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. Zinc: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Koziara, K.B.; Stroet, M.; Malde, A.K.; Mark, A.E. Testing and validation of the automated topology builder (ATB) version 2.0: Prediction of hydration free enthalpies. J. Comput. Aided Mol. Des. 2014, 28, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lin, Z.; van Gunsteren, W.F. Validation of the GROMOS 54a7 force field with respect to β-peptide folding. J. Chem. Theory Comput. 2011, 7, 1237–1243. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Freitag, F.; Gattin, Z.; Haberkern, H.; Jaun, B.; Siwko, M.; Vyas, R.; van Gunsteren, W.F.; Dolenc, J. Validation of the GROMOS 54a7 force field regarding mixed α/β-peptide molecules. Helv. Chim. Acta 2012, 95, 2562–2577. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Bonomi, M.; Branduardi, D.; Bussi, G.; Camilloni, C.; Provasi, D.; Raiteri, P.; Donadio, D.; Marinelli, F.; Pietrucci, F.; Broglia, R.A.; et al. PLUMED: A portable plugin for free-energy calculations with molecular dynamics. Comput. Phys. Commun. 2009, 180, 1961–1972. [Google Scholar] [CrossRef]
- Pietrucci, F.; Marinelli, F.; Carloni, P.; Laio, A. Substrate binding mechanism of HIV-1 protease from explicit-solvent atomistic simulations. J. Am. Chem. Soc. 2009, 131, 11811–11818. [Google Scholar] [CrossRef] [PubMed]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.-P.; Lane, T.J.; Pande, V.S. MDTraj: A modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]


| Peptide Name | Num. of Residues 1 | Preferred Macrostate 2 | Basin Depth 3 | 2° Structure Preference 4 | Pr.-Pr. H-Bonds 5 | Pr.-Solv. H-Bonds 6 |
|---|---|---|---|---|---|---|
| Hexamer(SNLHPK) | 6 | Model #3 | 2.7 kT | Random/Coil | 2 | 24 |
| Octamer (CSIVEDGK) | 8 | Model #4 | 4.9 kT | Random/Coil | 8 | 20 |
| NFO4 (VYMNRKYYKCCK) | 12 | Model #1 | 2.9 kT | Random/Coil | 2 | 44 |
| 13-mer (GPAGIDGPAGIRC) | 13 | Model #3 | 5.0 kT | Random/Coil | 6 | 18 |
| System Name | Complex Dist. 1 (%) | Pref. State 2 | Well Depth 3 | Str. Shifts 4 | Cont. Freq. 5 | RMSF Shift 6 | ΔG° (kJ/mol) 7 |
|---|---|---|---|---|---|---|---|
| Hexamer-OTA | 50.2 | #3 (47%) | <3.0 kT | No | 2/6 | Decreased | −15.43 (1.92) |
| Hexamer-OTB | 49.8 | #3 (45%) | <3.0 kT | No | 5/6 | No Shift | −15.31 (1.42) |
| Octamer-OTA | 52.0 | #3 (44%) | ~4.5 kT | No | 3/8 | Increased | −15.61 (1.92) |
| Octamer-OTB | 49.6 | #3 (54%) | ~3.0 kT | No | 8/8 | Decreased | −14.52 (1.26) |
| NFO4-OTA | 67.4 | #3 (48%) | >7.0 kT | Yes | 2/12 | Increased | −33.34 (3.43) |
| NFO4-OTB | 61.9 | #1 (40%) | >7.0 kT | Yes | 1/12 | Increased | −26.99 (3.22) |
| 13mer-OTA | 56.7 | #3 (37%) | <3.0 kT | No | 5/13 | Decreased | −14.81 (1.46) |
| 13mer-OTB | 50.6 | #3 (48%) | <3.0 kT | No | 2/13 | Decreased | −13.64 (2.59) |
| Recognition Molecule | ΔG°OTA-PRE (kJ/mol) | ΔG°OTB-PRE (kJ/mol) | KD-OTA(μM) Pred. 1 | Selectivity Pred. 1 | KD-OTA(μM)-Expt |
|---|---|---|---|---|---|
| Albumin | - | - | - | - | 0.019–1 2 |
| Antibody | - | - | - | - | 0.00001–0.083 3 |
| DNA Aptamer | - | - | - | - | 0.096–0.370 4 |
| Hexamer | −15.43 (1.92) | −15.31 (1.42) | 1991 | ~1 | 29.4 5 |
| Octamer | −15.61 (1.42) | −14.52 (1.26) | 1861 | ~2 | 11.8 6 |
| NFO4 | −33.34 (3.43) | −26.99 (3.22) | 1.47 | ~13 | 0.079 7 |
| 13-mer | −14.81 (1.46) | −13.64 (2.59) | 2563 | ~2 | 15.7 6 |
| Parameter | Rg | Nhb | Φcorr | d1 |
|---|---|---|---|---|
| Sigma (kJ/mol) | 0.020 | 0.500 | 0.400 | 0.025 |
| Interval range | 0.50–1.20 | 4–40 | 5–25 | 0.10–0.50 |
| Bias factor | 8 | 8 | 8 | 8 |
| Deposition Frequency (ps) | 1 | 1 | 1 | 1 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thyparambil, A.A.; Bazin, I.; Guiseppi-Elie, A. Evaluation of Ochratoxin Recognition by Peptides Using Explicit Solvent Molecular Dynamics. Toxins 2017, 9, 164. https://doi.org/10.3390/toxins9050164
Thyparambil AA, Bazin I, Guiseppi-Elie A. Evaluation of Ochratoxin Recognition by Peptides Using Explicit Solvent Molecular Dynamics. Toxins. 2017; 9(5):164. https://doi.org/10.3390/toxins9050164
Chicago/Turabian StyleThyparambil, Aby A., Ingrid Bazin, and Anthony Guiseppi-Elie. 2017. "Evaluation of Ochratoxin Recognition by Peptides Using Explicit Solvent Molecular Dynamics" Toxins 9, no. 5: 164. https://doi.org/10.3390/toxins9050164