Quantitative Proteomic Analysis of Venoms from Russian Vipers of Pelias Group: Phospholipases A2 are the Main Venom Components
Abstract
:1. Introduction
2. Results
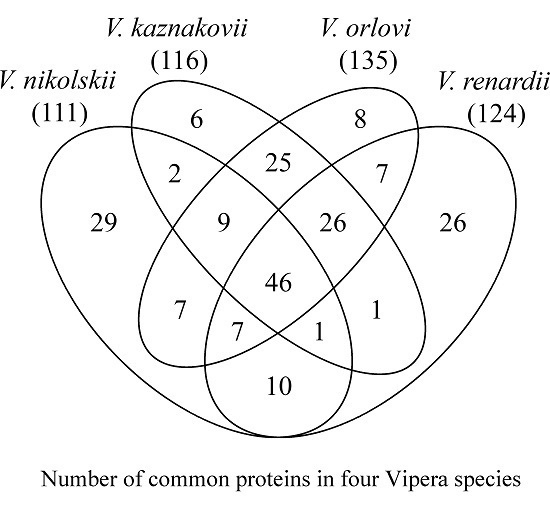
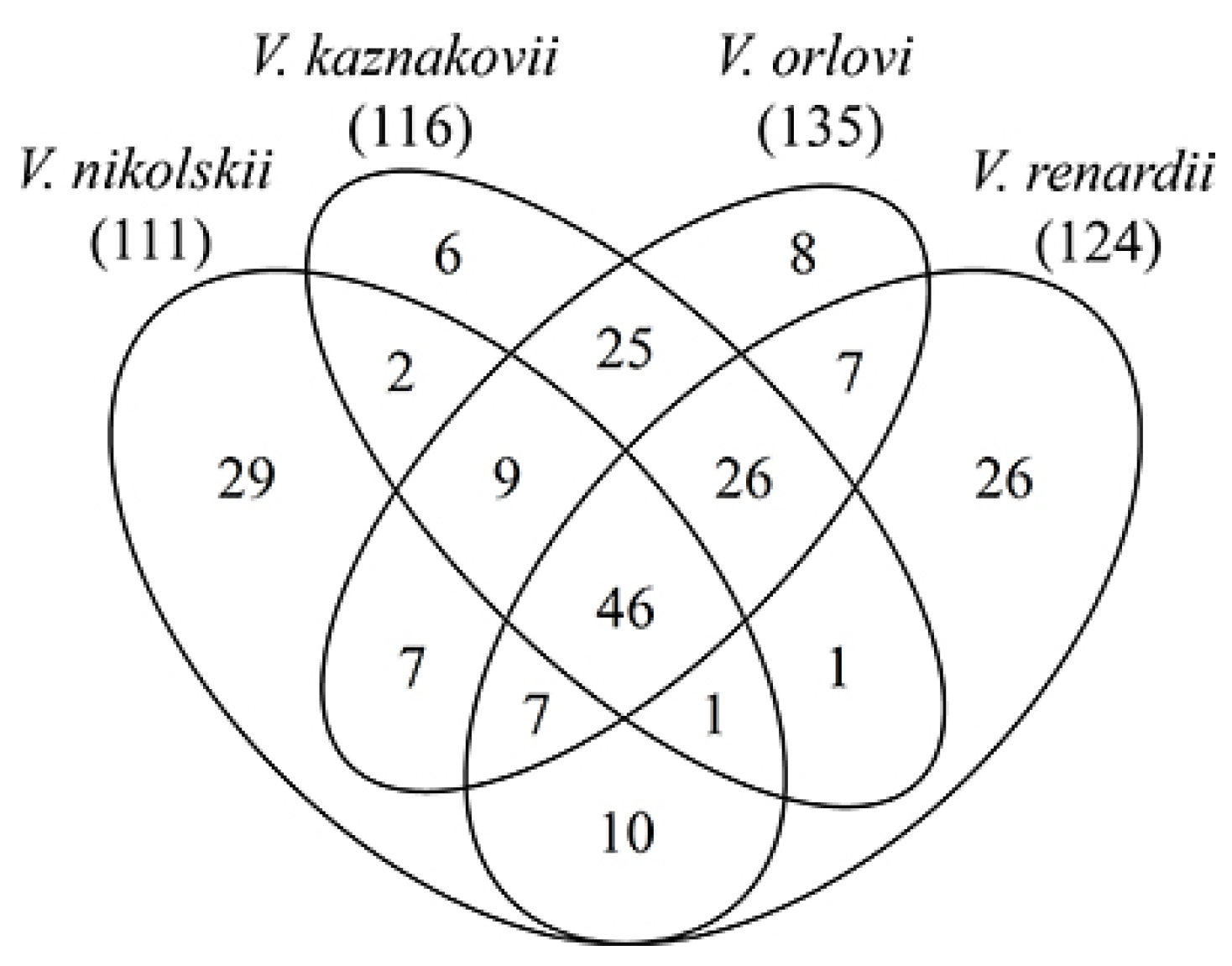
2.1. Venom Proteins Identification
2.2. Composition of Russian Viper Venoms
2.3. Identification of Endogenous Peptides in the Venoms
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. In-Solution Trypsin Digestion of Venom Samples
5.2. Endogenous Venom Peptides Isolation
5.3. LC-MS/MS Analysis
5.4. LC-MS/MS Data Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| FC | Fold Changes |
| FDR | False Discovery Rate |
| B-NAP | Bradykinin potentiating and C-type natriuretic peptides |
| BP | Blood protein |
| CP | Cysteine Proteases |
| CRISP | Cysteine-rich secretory protein |
| CTL | C-type lectin like |
| Dis | Disintegrin |
| Hya | Hyaluronidase |
| Kunitz | Kunitz type proteinase inhibitor |
| LFQ | label-free quantification |
| LAAO | l-amino acid oxidase |
| NGF | Nerve growth factor |
| Nuc | Nucleic acid degrading enzymes |
| OP | Other protein |
| PLA2 | Phospholipase A2 |
| PLB | Phospholipase B |
| SP | Serine proteinase |
| SVMP | Snake venom metalloproteinase |
| TBP | Toxin biosynthesis proteins (including aminopeptidases) |
| VEGF | Vascular endothelial growth factor |
References
- Tuniyev, B.S.; Ostrovskikh, S. Two new species of vipers of “kaznakovi” complex (Ophidia, Viperinae) from the Western Caucasus. Russ. J. Herpetol. 2001, 8, 117–126. [Google Scholar]
- Tuniyev, S.B.; Orlov, N.L.; Tuniyev, B.S.; Kidov, F.F. On the taxonomical status of steppe viper from foothills of the south macroslope of the East Caucasus. Russ. J. Herpetol. 2013, 20, 129–146. [Google Scholar]
- Calderón, L.; Lomonte, B.; Gutiérrez, J.M.; Tarkowski, A.; Hanson, L.Å. Biological and biochemical activities of Vipera berus (European viper) venom. Toxicon 1993, 31, 743–753. [Google Scholar] [CrossRef]
- Križaj, I.; Siigur, J.; Samel, M.; Cotič, V.; Gubenšek, F. Isolation, partial characterization and complete amino acid sequence of the toxic phospholipase A2 from the venom of the common viper, Vipera berus berus. Biochim. Biophys. Acta 1993, 1157, 81–85. [Google Scholar] [CrossRef]
- Samel, M.; Vija, H.; Subbi, J.; Siigur, J. Metalloproteinase with factor X-activating and fibrinogenolytic activities from Vipera berus berus venom. Comp. Biochem. Physiol. B. 2003, 135, 575–582. [Google Scholar] [CrossRef]
- Samel, M.; Vija, H.; Rönnholm, G.; Siigur, J.; Kalkkinen, N.; Siigur, E. Isolation and characterization of an apoptotic and platelet aggregation inhibiting l-amino acid oxidase from Vipera berus berus (common viper) venom. Biochim. Biophys. Acta 2006, 1764, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Tsai, I.H.; Wang, Y.M.; Cheng, A.C.; Starkov, V.; Osipov, A.; Nikitin, I.; Makarova, Y.; Ziganshin, R.; Utkin, Y. cDNA cloning, structural, and functional analyses of venom phospholipases A2 and a Kunitz-type protease inhibitor from steppe viper Vipera ursinii renardi. Toxicon 2011, 57, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Ghazaryan, N.A.; Ghulikyan, L.; Kishmiryan, A.; Andreeva, T.V.; Utkin, Y.N.; Tsetlin, V.I.; Lomonte, B.; Ayvazyan, N.M. Phospholipases A2 from Viperidae snakes: Differences in membranotropic activity between enzymatically active toxin and its inactive isoforms. Biochim. Biophys. Acta 2015, 1848, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Vulfius, C.A.; Kasheverov, I.E.; Starkov, V.G.; Osipov, A.V.; Andreeva, T.V.; Filkin, S.Y.; Gorbacheva, E.V.; Astashev, M.E.; Tsetlin, V.I.; Utkin, Y.N. Inhibition of nicotinic acetylcholine receptors, a novel facet in the pleiotropic activities of snake venom phospholipases A2. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Ramazanova, A.S.; Zavada, L.L.; Starkov, V.G.; Kovyazina, I.V.; Subbotina, T.F.; Kostyukhina, E.E.; Dementieva, I.N.; Ovchinnikova, T.V.; Utkin, Y.N. Heterodimeric neurotoxic phospholipases A2—The first proteins from venom of recently established species Vipera nikolskii: Implication of venom composition in viper systematics. Toxicon 2008, 51, 524–537. [Google Scholar] [CrossRef] [PubMed]
- Ramazanova, A.S.; Fil’kin, S.Iu.; Starkov, V.G.; Utkin, Iu.N. Molecular cloning and analysis of cDNA sequences encoding serine proteinase and Kunitz type inhibitor in venom gland of Vipera nikolskii viper. Bioorg. Khim. 2011, 37, 374–385. [Google Scholar] [PubMed]
- Starkov, V.G.; Osipov, A.V.; Utkin, Y.N. Toxicity of venoms from vipers of Pelias group to crickets Gryllus assimilis and its relation to snake entomophagy. Toxicon 2007, 49, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Goçmen, B.; Heiss, P.; Petras, D.; Nalbantsoy, A.; Süssmuth, R.D. Mass spectrometry guided venom profiling and bioactivity screening of the Anatolian Meadow Viper, Vipera anatolica. Toxicon 2015, 107, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.; Ayvazyan, N.; Calvete, J.J. Snake venomics of the Armenian mountain vipers Macrovipera lebetina obtusa and Vipera raddei. J. Proteomics 2008, 71, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, D.; Risch, M.; Kardas, A.; Buck, F.; von Bergen, M.; Betzel, C. Comparative analysis of the venom proteomes of Vipera ammodytes ammodytes and Vipera ammodytes meridionalis. J. Proteome Res. 2008, 7, 866–886. [Google Scholar] [CrossRef] [PubMed]
- Stümpel, N.; Joger, U. Recent advances in phylogeny and taxonomy of near and middle eastern vipers—An update. ZooKeys 2009, 31, 179–191. [Google Scholar] [CrossRef]
- McCleary, R.J.; Kini, R.M. Non-enzymatic proteins from snake venoms: A gold mine of pharmacological tools and drug leads. Toxicon 2013, 62, 56–74. [Google Scholar] [CrossRef] [PubMed]
- Waridel, P.; Frank, A.; Thomas, H.; Surendranath, V.; Sunyaev, S.; Pevzner, P.; Shevchenko, A. Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics 2007, 7, 2318–2329. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.C.; Gorenstein, M.V.; Li, G.Z.; Vissers, J.P.; Geromanos, S.J. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteomics 2006, 5, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, J.; Roschitzki, B.; Panse, C.; Fortes, C.; Barkow-Oesterreicher, S.; Rutishauser, D.; Schlapbach, R. Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J. Proteomics 2010, 73, 1740–1746. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Matic, I.; Hilger, M.; Nagaraj, N.; Selbach, M.; Olsen, J.V.; Mann, M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009, 4, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I.; Aebersold, R. Interpretation of shotgun proteomic data: The protein inference problem. Mol. Cell. Proteomics 2005, 4, 1419–1440. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, T.; Dauga, C.; Ferquel, E.; Choumet, V.; Failloux, A.-B. Molecular phylogeny of Vipera Laurenti, 1768 and the related genera Macrovipera (Reuss, 1927) and Daboia (Gray, 1842), with comments about neurotoxic Vipera aspis aspis populations. Mol. Phylogenet. Evol. 2005, 35, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Kalyabina-Hauf, S.; Schweiger, S.; Joger, U.; Mayer, W.; Orlov, N.; Wink, M. Phylogeny and systematics of adders (Vipera berus complex). Mertensiella 2004, 15, 7–15. [Google Scholar]
- Zinenko, O.; Stumpel, N.; Mazanaeva, L.; Bakiev, A.; Shiryaev, K.; Pavlov, A.; Kotenko, T.; Kukushkin, O.; Chikin, Yu.; Duisebayeva, T.; et al. Mitochondrial phylogeny shows multiple independent ecological transitions and northern dispersion despite of Pleistocene glaciations in meadow and steppe vipers (Vipera ursinii and Vipera renardi). Mol. Phylogenet. Evol. 2015, 84, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Bakiev, A.G.; Garanin, V.I.; Gelashvili, D.B.; Gorelov, R.A.; Doroni, I.V.; Zaytseva, O.V.; Zinenko, F.I.; Klyonina, A.A.; Makarova, T.N.; Malenyov, A.L.; et al. Vipers (Reptilia: Serpentes: Viperidae: Vipera) of the Volga basin. Part. 1; Cassandra: Togliatti, Russia, 2015; p. 234. (In Russian) [Google Scholar]
- Bakiev, А.G.; Garanin, V.I.; Pavlov, А.V.; Shurshina, I.V.; Malenyov, А.L. East steppe viper Vipera renardi (Reptilia, Viperidae) in Volga river basin: Materials on biology, ecology and toxicology. Samar. Luka 2008, 17, 817–845. (In Russian) [Google Scholar]
- Orlov, B.N.; Gelashvili, D.B.; Ibrahimov, A.K. Venomous Animals and Plants of USSR; Vysshaya Shkola: Moscow, Russia, 1990; pp. 111–117. (In Russian) [Google Scholar]
- BelPressa. Available online: http://www.belpressa.ru/news/news/osoba-holodnyh-krovej08494/ (accessed on 14 February 2016). (In Russian)
- Ramazanova, A.S.; Starkov, V.G.; Osipov, A.V.; Ziganshin, R.H.; Filkin, S.Y.; Tsetlin, V.I.; Utkin, Y.N. Cysteine-rich venom proteins from the snakes of Viperinae subfamily—molecular cloning and phylogenetic relationship. Toxicon 2009, 53, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Nilson, G.; Andren, C. The meadow and steppe vipers of Europe and Asia—The Vipera (Acrodiphaga) ursinii complex. Acta Zool. Acad. Sci. Hung. 2001, 47, 87–267. [Google Scholar]
- Munawar, A.; Trusch, M.; Georgieva, D.; Spencer, P.; Frochaux, V.; Harder, S.; Arni, R.K.; Duhalov, D.; Genov, N.; Schluter, H.; et al. Venom peptide analysis of Vipera ammodytes meridionalis (Viperinae) and Bothrops jararacussu (Crotalinae) demonstrates subfamily-specificity of the peptidome in the family Viperidae. Mol. Biosyst. 2011, 7, 3298–3307. [Google Scholar] [CrossRef] [PubMed]
- Zaitseva, О.V.; Маlenyov, А.L.; Bakiev, А.G. Studies of properties of the common viper’s venom in the Volga river basin: Practical value of the results obtained. Samar. Luka 2011, 20, 180–184. (In Russian) [Google Scholar]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, prefractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- NCBI (National Center for Biotechnology Information). Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 17 November 2015).



| Protein No. in SuppData | Protein Name | Taxon | Protein Family 1 | MW, KDa | Seq Cov, % | V. nikolskii | V. kaznakovi | V. orlovi | V. renardi | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prot Abun INT, % | Seq Cov, % | Prot Abun INT, % | Seq Cov, % | Prot Abun INT, % | Seq Cov, % | Prot Abun INT, % | Seq Cov, % | ||||||
| 9 | Natriuretic peptide | Pseudonaja textilis | B-NAP | 14 | 9.8 | 0.007 | 9.8 | 0.029 | 9.8 | 0.012 | 9.8 | - | - |
| 2 | Hemoglobin subunit alpha | Vipera aspis | BP | 15 | 6.4 | 0 | 6.4 | - | - | - | - | 0 | 6.4 |
| 3 | Hemoglobin subunit beta-2 | Naja naja | BP | 16 | 7.5 | 0.016 | 7.5 | - | - | - | - | - | - |
| 4 | Hemoglobin subunit beta | Erythrolamprus miliaris | BP | 15 | 23.3 | 0.018 | 19.2 | - | - | - | - | 0 | 11.6 |
| 33 | Alpha globin | Hydrophis melanocephalus | BP | 16 | 19.7 | 0.011 | 9.2 | 0 | 9.2 | - | - | 0.010 | 19.7 |
| 34 | Alpha globin | Elaphe climacophora | BP | 11 | 25.2 | - | - | - | - | - | - | 0.032 | 25.2 |
| 116 | Murinoglobulin-2 | Ophiophagus hannah | BP | 143 | 2.7 | - | - | - | - | 0.008 | 1.8 | 0.002 | 2.2 |
| 124 | Serum albumin | Protobothrops flavoviridis | BP | 69 | 5.4 | 0.013 | 5.4 | - | - | 0.002 | 5.2 | - | - |
| 158 | Hemoglobin subunit alpha | Hydrophis gracilis | BP | 15 | 15.6 | 0.013 | 15.6 | 0.002 | 15.6 | - | - | 0.017 | 15.6 |
| 165 | Hemoglobin subunit alpha | Crotalus horridus | BP | 15 | 11.3 | 0.012 | 11.3 | 0.003 | 11.3 | - | - | 0.013 | 11.3 |
| 171 | Hemoglobin subunit beta-1 | Boiga irregularis | BP | 16 | 21.8 | 0 | 21.8 | - | - | - | - | 0 | 21.8 |
| 177 | Transferrin | Crotalus adamanteus | BP | 77 | 3.1 | 0.006 | 3.1 | - | 0.004 | 3.1 | 0.009 | 3.1 | |
| 199 | Hemoglobin subunit beta-2 | Thamnophis sirtalis | BP | 16 | 24.5 | 0.021 | 18.4 | - | - | - | - | 0.020 | 12.9 |
| 200 | Hemoglobin subunit beta-1 | Thamnophis sirtalis | BP | 16 | 19.7 | 0.002 | 19.7 | - | - | - | - | - | - |
| 206 | Serum albumin-like | Thamnophis sirtalis | BP | 57 | 8.0 | 0.012 | 8.0 | - | - | - | - | - | - |
| 180 | Cathepsin B-like protein | Crotalus adamanteus | CP | 37 | 5.0 | 0 | 5.0 | - | - | - | - | - | - |
| 7 | Cysteine-rich venom protein | Philodryas patagoniensis | CRISP | 1 | 64.3 | - | - | 0.037 | 64.3 | 0.002 | 64.3 | - | - |
| 18 | Cysteine-rich seceretory protein Dr-CRPK | Daboia russelii | CRISP | 26 | 24.7 | - | - | 0.170 | 24.7 | 0.107 | 21.8 | - | - |
| 19 | Cysteine-rich seceretory protein Dr-CRPB | Daboia russelii | CRISP | 25 | 17.6 | - | - | 0.033 | 17.6 | 0.006 | 15.8 | 0 | 17.6 |
| 89 | Cysteine-rich venom protein | Protobothrops jerdonii | CRISP | 26 | 22.5 | - | - | 0.148 | 22.5 | 0.149 | 22.5 | - | - |
| 90 | Cysteine-rich venom protein | Vipera nikolskii | CRISP | 24 | 83.3 | - | - | 0.112 | 77.4 | 0.050 | 80.1 | 0.116 | 68.3 |
| 91 | Cysteine-rich venom protein | Vipera berus | CRISP | 26 | 77 | 0.345 | 74.1 | 7.916 | 71.5 | 9.924 | 74.1 | 5.125 | 63.2 |
| 145 | Cysteine-rich venom protein triflin | Protobothrops flavoviridis | CRISP | 24 | 23.1 | 0.070 | 23.1 | 2.474 | 23.1 | 2.065 | 23.1 | 3.016 | 23.1 |
| 22 | Snaclec 1 | Sistrurus catenatus edwardsii | CTL | 17 | 6.2 | - | - | 1.404 | 6.2 | 0.168 | 6.2 | - | - |
| 23 | C-type lectin lectoxin-Thr1 | Thrasops jacksonii | CTL | 18 | 7.0 | 0.010 | 6.3 | 0.060 | 7.0 | 0.110 | 7 | 0.069 | 7 |
| 24 | Snaclec A13 | Macrovipera lebetina | CTL | 15 | 33.6 | - | - | 2.042 | 29.0 | 2.555 | 33.6 | - | - |
| 25 | Snaclec A15 | Macrovipera lebetina | CTL | 17 | 46.8 | 0.710 | 46.8 | 0.867 | 46.8 | 0.783 | 46.8 | 0.524 | 46.8 |
| 26 | Snaclec B7 | Macrovipera lebetina | CTL | 15 | 27.2 | - | - | 3.475 | 27.2 | 1.274 | 27.2 | - | - |
| 101 | Snaclec VP12 subunit A | Daboia palaestinae | CTL | 12 | 26.2 | - | - | 0.008 | 26.2 | - | - | - | - |
| 102 | Snaclec VP12 subunit B | Daboia palaestinae | CTL | 15 | 25.6 | - | - | 0.084 | 25.6 | 0.029 | 17.6 | - | - |
| 153 | C-type lectin J | Echis coloratus | CTL | 18 | 14.6 | 0.589 | 14.6 | 0.740 | 14.6 | 0.579 | 14.6 | 0.477 | 14.6 |
| 154 | C-type lectin H | Echis coloratus | CTL | 18 | 10.8 | 0.050 | 10.1 | 0.935 | 10.8 | 0.909 | 10.8 | 0.172 | 10.8 |
| 155 | C-type lectin E | Echis coloratus | CTL | 11 | 12.1 | 0.011 | 7.1 | - | - | 0.008 | 7.1 | 0.045 | 12.1 |
| 156 | C-type lectin B | Echis coloratus | CTL | 13 | 7.1 | - | - | 0.048 | 7.1 | 0.005 | 7.1 | - | - |
| 157 | C-type lectin A | Echis coloratus | CTL | 18 | 10.1 | 0.014 | 10.1 | - | - | - | - | - | - |
| 159 | Snaclec coagulation factor X-activating enzyme light chain 2 | Macrovipera lebetina | CTL | 18 | 6.3 | 0.039 | 6.3 | 0.199 | 6.3 | 0.340 | 6.3 | 0.051 | 6.3 |
| 173 | C-type lectin-like protein 3B | Macrovipera lebetina | CTL | 17 | 42.6 | 2.457 | 42.6 | 2.758 | 42.6 | 2.500 | 42.6 | 1.545 | 42.6 |
| 174 | C-type lectin-like protein 4B | Macrovipera lebetina | CTL | 17 | 14.7 | 0.018 | 12.0 | - | - | 0.017 | 12.0 | 0.353 | 14.7 |
| 175 | Snaclec dabocetin subunit alpha | Daboia siamensis | CTL | 17 | 6.5 | - | - | 0.443 | 6.5 | 0.064 | 6.5 | - | - |
| 181 | Snaclec tokaracetin subunit beta | Protobothrops tokarensis | CTL | 4 | 32.5 | - | - | 0.026 | 32.5 | - | - | - | - |
| 210 | Snaclec anticoagulant protein subunit B | Deinagkistrodon acutus | CTL | 14 | 12.2 | - | - | 0.352 | 12.2 | 0.124 | 12.2 | - | - |
| 14 | Disintegrin VB7A | Vipera berus berus | Dis | 7 | 76.6 | - | - | 0.111 | 23.4 | 0.008 | 23.4 | 12.932 | 76.6 |
| 15 | Disintegrin VB7B | Vipera berus berus | Dis | 6 | 70.3 | - | - | 0.008 | 29.7 | - | - | 0.935 | 70.3 |
| 16 | Disintegrin VLO4 | Macrovipera lebetina obtusa | Dis | 7 | 38.5 | - | - | 0.011 | 24.6 | 0.006 | 24.6 | 0.034 | 38.5 |
| 17 | Disintegrin VA6 | Vipera ammodytes ammodytes | Dis | 7 | 23.4 | - | - | - | - | - | - | 0.133 | 23.4 |
| 191 | Disintegrin lebein-1-alpha | Macrovipera lebetina | Dis | 12 | 30.6 | - | - | 0.389 | 9.0 | 0.578 | 9.0 | 0.006 | 21.6 |
| 86 | Hyaluronidase | Echis ocellatus | Hya | 52 | 13.6 | 0.007 | 13.6 | 0.011 | 7.3 | 0.004 | 2.7 | - | - |
| 176 | Hyaluronidase | Crotalus adamanteus | Hya | 52 | 6.7 | 0.004 | 6.7 | 0.007 | 6.7 | 0.003 | 2.0 | - | - |
| 27 | Inhibitor, chymotrypsin | Vipera ammodytes | Kunitz | 7 | 40.0 | 0.002 | 24.6 | - | - | 0.047 | 29.2 | 0.478 | 29.2 |
| 80 | Protease inhibitor 3 | Walterinnesia aegyptia | Kunitz | 8 | 21.0 | - | - | - | - | 0 | 21.0 | 0.005 | 21.0 |
| 81 | Kunitz-type serine protease inhibitor Vur-KIn | Vipera renardi | Kunitz | 7 | 39.4 | 0.015 | 39.4 | - | - | 0.074 | 39.4 | 0.241 | 39.4 |
| 87 | KP-Sut-1 | Suta fasciata | Kunitz | 13 | 9.4 | 0.072 | 9.4 | - | - | - | - | 0.077 | 9.4 |
| 142 | Kunitz-type serine protease inhibitor ki-VN | Vipera nikolskii | Kunitz | 10 | 34.7 | 0.610 | 34.7 | - | - | - | - | - | - |
| 5 | l-amino-acid oxidase | Macrovipera lebetina | LAAO | 12 | 42.1 | 0.041 | 41.1 | 1.388 | 42.1 | 1.631 | 41.1 | 1.786 | 41.1 |
| 65 | l-amino-acid oxidase | Echis ocellatus | LAAO | 56 | 6.0 | - | - | - | - | 0 | 6.0 | - | - |
| 66 | l-amino-acid oxidase | Vipera ammodytes ammodytes | LAAO | 54 | 13.2 | - | - | 0.009 | 11.2 | 0.012 | 11.2 | 0.007 | 13.0 |
| 75 | l-amino-acid oxidase | Daboia russelii | LAAO | 56 | 11.7 | 0 | 5.8 | 0.316 | 7.7 | 0.045 | 11.7 | 0.049 | 9.9 |
| 92 | Kunitz-type serine protease inhibitor PIVL | Macrovipera lebetina transmediterranea | LAAO | 10 | 8.4 | - | - | - | - | 0 | 8.4 | 0.021 | 8.4 |
| 94 | l-amino-acid oxidase | Gloydius halys | LAAO | 55 | 9.9 | - | - | - | - | 0 | 7.4 | - | - |
| 107 | l-amino acid oxidase | Ovophis okinavensis | LAAO | 58 | 11.0 | 0.001 | 3.9 | 1.893 | 6.8 | 2.106 | 6.8 | 1.378 | 10.9 |
| 144 | l-amino acid oxidase | Protobothrops elegans | LAAO | 57 | 5.7 | - | - | 0 | 5.7 | - | - | - | - |
| 152 | l-amino acid oxidase B variant 1 | Echis coloratus | LAAO | 56 | 17.1 | - | - | 0.425 | 13.1 | 0.213 | 12.9 | 0.216 | 13.9 |
| 164 | l-amino-acid oxidase | Crotalus horridus | LAAO | 58 | 11.2 | 0.001 | 4.5 | 0.042 | 6.6 | 0.005 | 5.8 | 0 | 5.8 |
| 183 | l-amino-acid oxidase | Bothrops moojeni | LAAO | 54 | 12.1 | 0.022 | 6.5 | 0.256 | 7.1 | 0.178 | 9.4 | 0.098 | 9.2 |
| 62 | Venom nerve growth factor 2 | Daboia russelii | NGF | 27 | 14.4 | - | - | 0 | 14.4 | - | - | - | - |
| 63 | Venom nerve growth factor | Vipera ursinii | NGF | 27 | 25.5 | 0.285 | 18.1 | 0.122 | 18.1 | 0.254 | 25.5 | 0.093 | 18.1 |
| 76 | Snake venom 5′-nucleotidase | Gloydius blomhoffii blomhoffii | Nuc | 6 | 27.8 | 0.013 | 27.8 | - | - | - | - | - | - |
| 110 | 5′-nucleotidase | Ovophis okinavensis | Nuc | 55 | 26.6 | 0.091 | 26.6 | 0.012 | 9.1 | 0.018 | 16.5 | 0.057 | 22.4 |
| 125 | Phosphodiesterase | Macrovipera lebetina | Nuc | 96 | 35.4 | 0.243 | 35.4 | 0.103 | 21.4 | 0.088 | 20.7 | 0.038 | 19.5 |
| 126 | 5′-nucleotidase | Macrovipera lebetina | Nuc | 45 | 58.1 | 0.563 | 54.4 | 0.046 | 14.5 | 0.097 | 30.4 | 0.226 | 39.2 |
| 127 | Venom phosphodiesterase 2 | Crotalus adamanteus | Nuc | 91 | 14.0 | 0.007 | 14.0 | - | - | - | - | - | - |
| 132 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3-like | Python bivittatus | Nuc | 93 | 7.5 | 0.004 | 7.5 | 0.002 | 3.5 | 0.003 | 3.5 | - | - |
| 170 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 | Boiga irregularis | Nuc | 100 | 4.5 | - | - | 0.441 | 3.3 | 1.521 | 2.6 | - | - |
| 73 | Proactivator polypeptide-like | Crotalus adamanteus | OP | 58 | 19.7 | 0.036 | 19.7 | 0.011 | 5.2 | 0.024 | 8.7 | 0.006 | 2.5 |
| 104 | ArfGAP with SH3 domain ankyrin repeat and PH domain 3 | Micrurus fulvius | OP | 107 | 2.3 | - | - | 0.027 | 2.3 | 0.033 | 2.3 | - | - |
| 113 | Uncharacterized protein | Ophiophagus hannah | OP | 46 | 3.4 | - | - | 0.040 | 3.4 | 0.015 | 3.4 | - | - |
| 115 | 78 kDa glucose-regulated protein | Ophiophagus hannah | OP | 67 | 6.4 | 0 | 2.8 | 0.004 | 4.4 | 0 | 2.0 | - | - |
| 117 | WD repeat-containing protein 67 | Ophiophagus hannah | OP | 113 | 0.8 | - | - | - | - | - | - | 0.652 | 0.8 |
| 118 | Pituitary adenylate cyclase-activating polypeptide type I receptor | Ophiophagus hannah | OP | 7 | 18.2 | 0.008 | 18.2 | - | - | - | - | - | - |
| 119 | Iron-responsive element-binding protein 2 | Ophiophagus hannah | OP | 90 | 1.8 | - | - | 0.536 | 1.8 | 0.482 | 1.8 | 0.224 | 1.8 |
| 122 | Glutathione peroxidase | Ophiophagus hannah | OP | 29 | 21.6 | 0.078 | 18.9 | 0.047 | 16.7 | 0.095 | 21.6 | 0.081 | 16.7 |
| 128 | PiggyBac transposable element-derived protein 5 | Python bivittatus | OP | 69 | 2.7 | - | - | - | - | - | - | 0.067 | 2.7 |
| 129 | Calmodulin-lysine N-methyltransferase | Python bivittatus | OP | 14 | 12.3 | - | - | - | - | 0 | 12.3 | - | - |
| 130 | Dipeptidase 2 | Python bivittatus | OP | 46 | 7.0 | - | - | - | - | - | - | 0 | 7.0 |
| 131 | Serine/threonine-protein phosphatase 6 regulatory subunit 1 isoform X3 | Python bivittatus | OP | 92 | 1.9 | - | - | 0.199 | 1.9 | 0.117 | 1.8 | 0.279 | 1.8 |
| 133 | E3 ubiquitin-protein ligase MARCH8-like isoform X8 | Python bivittatus | OP | 30 | 7.2 | - | - | - | - | - | - | 0.702 | 7.2 |
| 134 | Nucleolar and coiled-body phosphoprotein 1 isoform X5 | Python bivittatus | OP | 104 | 1.4 | - | - | - | - | 0 | 1.4 | - | - |
| 135 | E3 ubiquitin-protein ligase UBR4 | Python bivittatus | OP | 555 | 0.2 | 0 | 0.2 | - | - | - | - | - | - |
| 136 | Extracellular matrix protein 1 | Python bivittatus | OP | 24 | 8.6 | 0.019 | 8.6 | - | - | 0.004 | 8.6 | - | - |
| 137 | Receptor-type tyrosine-protein phosphatase gamma-like | Python bivittatus | OP | 102 | 1.9 | - | - | 0.026 | 1.9 | 0.018 | 1.9 | 0.018 | 1.9 |
| 138 | Nurim-like | Python bivittatus | OP | 32 | 7.7 | 0.010 | 7.7 | - | - | - | - | - | - |
| 162 | Dickkopf-related protein 3-like | Crotalus horridus | OP | 31 | 5.0 | - | - | 0.005 | 5 | 0.007 | 5.0 | - | - |
| 168 | RNA binding motif protein 6 | Boiga irregularis | OP | 59 | 3.5 | 0.007 | 3.5 | - | - | - | - | - | - |
| 172 | Filamin-B isoform 15 | Boiga irregularis | OP | 282 | 0.9 | - | - | 0.010 | 0.9 | - | - | - | - |
| 197 | Leucine-rich repeats and immunoglobulin-like domains protein 1 | Thamnophis sirtalis | OP | 48 | 1.4 | 0.010 | 1.4 | 0.028 | 1.4 | 0.026 | 1.4 | - | - |
| 201 | Peroxiredoxin-4-like | Thamnophis sirtalis | OP | 31 | 8.7 | 0.002 | 8.7 | - | - | - | - | - | - |
| 202 | Obscurin | Thamnophis sirtalis | OP | 1024 | 0.5 | 0.109 | 0.4 | 0.107 | 0.2 | - | - | - | - |
| 203 | CCR4-NOT transcription complex subunit 3 | Callithrix jacchus | OP | 12 | 15.7 | - | - | 0.008 | 15.7 | 0.046 | 15.7 | - | - |
| 205 | Tyrosine-protein phosphatase non-receptor type 20 | Thamnophis sirtalis | OP | 50 | 4.0 | - | - | 0.019 | 4.0 | 0.020 | 4.0 | - | - |
| 208 | Protein BANP | Thamnophis sirtalis | OP | 40 | 4.4 | - | - | 0.064 | 4.4 | 0.076 | 4.4 | 0.119 | 4.4 |
| 209 | Microtubule-associated serine/threonine-protein kinase 1-like | Thamnophis sirtalis | OP | 94 | 0.7 | - | - | - | - | - | - | 0 | 0.7 |
| 10 | Basic phospholipase A2 chain HDP-1P | Vipera nikolskii | PLA2 | 13 | 86.9 | 20.289 | 86.9 | 5.460 | 9.0 | 0.230 | 9.0 | 0.288 | 27.0 |
| 13 | Basic phospholipase A2 B chain | Vipera aspis zinnikeri | PLA2 | 13 | 80.3 | 0.130 | 80.3 | - | - | - | - | - | - |
| 28 | Phospholipase A2 II | Vipera aspis | PLA2 | 5 | 40.4 | 0.018 | 19.2 | - | - | 0.009 | 21.2 | - | - |
| 29 | Phospholipase A2 III | Vipera aspis | PLA2 | 5 | 66.0 | 0.002 | 66.0 | 6.579 | 66.0 | 5.099 | 66.0 | 0 | 66.0 |
| 30 | Acidic phospholipase A2 PLA-1 | Eristicophis macmahoni | PLA2 | 13 | 22.3 | - | - | - | - | - | - | 0.419 | 22.3 |
| 31 | Acidic phospholipase A2 PLA-2 | Eristicophis macmahoni | PLA2 | 13 | 29.8 | 0 | 28.9 | - | - | 0 | 21.5 | 0.388 | 29.8 |
| 32 | Acidic phospholipase A2 homolog vipoxin A chain | Vipera ammodytes meridionalis | PLA2 | 13 | 94.3 | 34.017 | 94.3 | - | - | - | - | 0.048 | 18.9 |
| 56 | Basic phospholipase A2 3 | Daboia russelii | PLA2 | 13 | 27.3 | - | - | 0.003 | 12.4 | 0.002 | 12.4 | 0.048 | 27.3 |
| 57 | Phospholipase A2 | Agkistrodon piscivorus | PLA2 | 13 | 27.6 | 0 | 16.3 | - | - | - | - | - | - |
| 58 | Phospholipase A2 homolog P-elapitoxin-Aa1a beta chain | Acanthophis antarcticus | PLA2 | 3 | 22.6 | - | - | 0.165 | 22.6 | 0.127 | 22.6 | 0.030 | 22.6 |
| 67 | Acidic phospholipase A2 RV-7 | Daboia siamensis | PLA2 | 13 | 45.1 | 1.078 | 45.1 | - | - | - | - | - | - |
| 78 | Basic phospholipase A2 Pla2Vb | Vipera berus berus | PLA2 | 15 | 46.4 | - | - | 0 | 13.8 | 0.056 | 46.4 | - | - |
| 79 | Acidic phospholipase A2 Vur-PL3 | Vipera renardi | PLA2 | 15 | 69.3 | - | - | 12.956 | 67.2 | 12.705 | 67.2 | 7.113 | 58.4 |
| 82 | Acidic phospholipase A2 PL1 | Vipera renardi | PLA2 | 15 | 75.4 | 4.057 | 70.3 | 5.097 | 59.4 | 4.080 | 52.9 | 10.603 | 75.4 |
| 83 | Acidic phospholipase A2 Vur-PL2B | Vipera renardi | PLA2 | 15 | 72.3 | - | - | 0 | 19.7 | 0.041 | 19.7 | 14.999 | 72.3 |
| 84 | Basic phospholipase A2 homolog Vur-S49 | Vipera renardi | PLA2 | 15 | 63.0 | - | - | 0.009 | 18.8 | 0.055 | 18.8 | 7.676 | 63.0 |
| 85 | Basic phospholipase A2 vurtoxin | Vipera renardi | PLA2 | 15 | 50.0 | - | - | - | - | - | - | 4.220 | 50.0 |
| 95 | Ammodytin I1 | Vipera aspis aspis | PLA2 | 15 | 56.5 | 0.032 | 56.5 | 0.048 | 56.5 | 0.040 | 44.9 | 0.026 | 39.9 |
| 96 | Ammodytin I1 | Vipera ammodytes montandoni | PLA2 | 15 | 75.4 | 1.389 | 75.4 | 1.428 | 75.4 | 0.524 | 63.8 | 1.710 | 59.4 |
| 97 | Ammodytin I2 | Vipera aspis aspis | PLA2 | 15 | 19.7 | - | - | - | - | - | - | 0 | 19.7 |
| 98 | Ammodytin I2 | Vipera berus berus | PLA2 | 15 | 36.5 | - | - | 0.021 | 31.4 | - | - | - | - |
| 99 | Ammodytin I2 | Vipera ursinii | PLA2 | 15 | 45.3 | - | - | - | - | - | - | 0.017 | 45.3 |
| 100 | Ammodytin L | Vipera ammodytes ammodytes | PLA2 | 15 | 14.5 | - | - | - | - | - | - | 0.009 | 14.5 |
| 139 | Basic phospholipase A2 vipoxin B chain | Vipera ammodytes meridionalis | PLA2 | 13 | 80.3 | 0.086 | 80.3 | - | - | - | - | - | - |
| 151 | Phospholipase A2 Group IIE | Echis coloratus | PLA2 | 13 | 5.8 | - | - | - | - | - | - | 0.020 | 5.8 |
| 161 | Basic phospholipase A2 | Azemiops feae | PLA2 | 15 | 7.2 | - | - | - | - | 0.012 | 7.2 | 0.016 | 7.2 |
| 193 | Phospholipase A2-III | Daboia russelii | PLA2 | 13 | 13.1 | 0.019 | 13.1 | - | - | - | - | - | - |
| 195 | Phospholipase A2 ammodytin I1 | Vipera nikolskii | PLA2 | 15 | 75.4 | 4.779 | 75.4 | 4.663 | 75.4 | 4.289 | 63.8 | - | - |
| 196 | Basic phospholipase A2 chain HDP-2P | Vipera nikolskii | PLA2 | 15 | 69.6 | 0.068 | 69.6 | - | - | - | - | 0.006 | 31.2 |
| 109 | Phospholipase b | Ovophis okinavensis | PLB | 64 | 20.8 | - | - | 0.015 | 13.6 | 0.037 | 20.1 | 0.057 | 15.0 |
| 114 | Phospholipase B-like 1 | Ophiophagus hannah | PLB | 58 | 16.8 | 0.022 | 7.0 | 0.085 | 12.0 | 0.104 | 16.2 | 0.088 | 10.2 |
| 169 | Phospholipase B | Boiga irregularis | PLB | 64 | 15.6 | - | - | 0.020 | 11.6 | 0.034 | 11.2 | 0.034 | 9.9 |
| 179 | Phospholipase B | Crotalus adamanteus | PLB | 64 | 26.9 | 0.080 | 16.5 | 0.218 | 15.9 | 0.321 | 21.0 | 0.334 | 12.7 |
| 6 | Unassigned | Calloselasma rhodostoma | SP | 24 | 9.4 | - | - | 0.007 | 5.5 | 0.041 | 8.1 | 0.006 | 8.1 |
| 11 | Snake venom serine protease pallase | Gloydius halys | SP | 26 | 13.1 | 0.002 | 9.3 | 0.005 | 10.6 | 0.002 | 11.9 | 0 | 10.6 |
| 12 | Snake venom serine protease ussurase | Gloydius ussuriensis | SP | 26 | 5.6 | 0.077 | 5.6 | - | - | - | - | - | - |
| 21 | Thrombin-like enzyme KN-BJ 2 | Bothrops jararaca | SP | 27 | 11.3 | 1.017 | 11.3 | 0.329 | 11.3 | 1.944 | 11.3 | 0.050 | 11.3 |
| 35 | Serine protease | Echis coloratus | SP | 28 | 8.5 | - | - | 0.043 | 8.5 | 0.172 | 8.5 | 0.087 | 8.5 |
| 36 | Serine protease | Echis ocellatus | SP | 24 | 6.3 | 0.515 | 6.3 | 1.980 | 3.6 | 2.401 | 6.3 | 0.694 | 6.3 |
| 37 | Serine protease | Echis coloratus | SP | 25 | 7.3 | - | - | 0.079 | 4.7 | 0.047 | 7.3 | - | - |
| 38 | Serine protease | Echis coloratus | SP | 25 | 12.0 | 0.004 | 12.0 | 0 | 9.4 | 0.007 | 12.0 | - | - |
| 39 | Serine protease | Echis coloratus | SP | 26 | 5.9 | 0.110 | 5.9 | 0.048 | 3.4 | 0.015 | 5.9 | 0.005 | 5.9 |
| 40 | Serine protease | Echis carinatus sochureki | SP | 25 | 8.1 | 0.021 | 5.5 | - | - | - | - | 0.036 | 5.5 |
| 64 | Factor V activator RVV-V gamma | Daboia siamensis | SP | 25 | 13.7 | 0.006 | 11.1 | 0.473 | 8.1 | 0.075 | 5.6 | - | - |
| 69 | Serine protease VLSP-3 | Macrovipera lebetina | SP | 28 | 15.5 | 3.526 | 15.5 | 2.548 | 12.0 | 3.479 | 14.3 | 1.303 | 14.3 |
| 70 | Beta-fibrinogenase | Macrovipera lebetina | SP | 28 | 18.3 | 0.017 | 18.3 | 0.008 | 9.3 | 0.078 | 11.7 | 0.224 | 11.7 |
| 71 | Chymotrypsin-like protease VLCTLP | Macrovipera lebetina | SP | 28 | 29.6 | 0.069 | 29.6 | - | - | - | - | - | - |
| 74 | Snake venom serine protease nikobin | Vipera nikolskii | SP | 28 | 43.2 | 12.595 | 43.2 | 2.334 | 34.2 | 8.515 | 32.3 | 1.732 | 28.0 |
| 77 | Snake venom serine protease pallabin | Gloydius halys | SP | 28 | 12.3 | 0.002 | 8.8 | - | - | - | - | 0.010 | 10.0 |
| 103 | Kallikrein-CohID-1 | Crotalus oreganus helleri | SP | 28 | 10.8 | - | - | - | - | - | - | 0 | 10.8 |
| 105 | Serine protease | Protobothrops flavoviridis | SP | 18 | 17.4 | 0 | 17.4 | - | - | - | - | - | - |
| 106 | Serine protease | Ovophis okinavensis | SP | 10 | 23.3 | 0.061 | 20.0 | 0.028 | 23.3 | 0.352 | 20.0 | 0.067 | 23.3 |
| 140 | Factor V activator | Macrovipera lebetina | SP | 28 | 18.9 | 0.049 | 13.9 | 0.776 | 11.6 | 0.088 | 11.2 | 0.005 | 5.0 |
| 141 | Venom serine proteinase-like protein 2 | Macrovipera lebetina | SP | 28 | 30.8 | 1.028 | 30.8 | 0.896 | 26.5 | 1.680 | 25.4 | 1.595 | 26.5 |
| 163 | Serine proteinase 1 | Crotalus horridus | SP | 28 | 13.2 | 1.023 | 10.9 | 0.274 | 4.3 | 2.042 | 6.6 | - | - |
| 187 | Snake venom serine protease HS112 | Bothrops jararaca | SP | 27 | 14.9 | - | - | 0.157 | 12.5 | 0.323 | 14.9 | - | |
| 188 | Snake venom serine protease KN6 | Trimeresurus stejnegeri | SP | 28 | 3.5 | 0.036 | 3.5 | - | - | - | - | - | - |
| 189 | Snake venom serine protease 5 | Trimeresurus stejnegeri | SP | 28 | 11.6 | 0 | 11.6 | - | - | - | - | - | - |
| 190 | Snake venom serine protease catroxase-2 | Crotalus atrox | SP | 27 | 17.4 | 0.352 | 17.4 | 0.162 | 8.5 | 0.531 | 10.9 | 0.012 | 10.9 |
| 198 | Rho GTPase-activating protein 28-like | Thamnophis sirtalis | SP | 24 | 3.2 | - | - | 0.811 | 3.2 | 0.822 | 3.2 | 0.207 | 3.2 |
| 41 | Metalloproteinase | Echis carinatus sochureki | SVMP | 69 | 14.6 | - | - | 1.488 | 8.7 | 1.023 | 11.5 | 0.401 | 14.6 |
| 42 | Metalloproteinase | Echis carinatus sochureki | SVMP | 68 | 6.9 | - | - | - | - | - | - | 0.107 | 6.9 |
| 43 | Metalloproteinase | Echis carinatus sochureki | SVMP | 27 | 6.5 | - | - | 2.987 | 6.5 | 2.973 | 6.5 | 1.419 | 6.5 |
| 44 | Metalloproteinase | Echis coloratus | SVMP | 56 | 6.3 | - | - | - | - | - | - | 0.004 | 6.3 |
| 45 | Metalloproteinase | Echis coloratus | SVMP | 56 | 11.2 | - | - | 0.109 | 9.4 | 0.080 | 9.4 | 0.116 | 9.0 |
| 46 | Metalloproteinase | Echis coloratus | SVMP | 66 | 10.7 | - | - | 0 | 9.2 | 0 | 9.2 | 0 | 10.7 |
| 47 | Metalloproteinase | Echis coloratus | SVMP | 69 | 3.1 | - | - | - | - | - | - | 0.008 | 3.1 |
| 48 | Metalloproteinase | Echis coloratus | SVMP | 69 | 7.4 | - | - | 0.208 | 7.4 | 0.435 | 4.7 | 0.058 | 4.9 |
| 49 | Metalloproteinase | Echis coloratus | SVMP | 68 | 8.8 | - | - | - | - | - | - | 0 | 8.8 |
| 50 | Metalloproteinase | Echis coloratus | SVMP | 68 | 13.8 | - | - | 0 | 8.9 | 0 | 5.2 | 0.488 | 4.9 |
| 51 | Metalloproteinase | Echis coloratus | SVMP | 61 | 5.4 | - | - | 2.479 | 5.4 | 2.296 | 5.4 | 1.071 | 5.4 |
| 52 | Metalloproteinase | Echis coloratus | SVMP | 68 | 6.4 | - | - | 0.019 | 6.4 | 0.012 | 5.6 | 0.021 | 6.4 |
| 53 | Metalloproteinase | Echis pyramidum leakeyi | SVMP | 62 | 8.1 | 0.009 | 2.5 | - | - | - | - | - | - |
| 54 | Metalloproteinase | Echis pyramidum leakeyi | SVMP | 46 | 5.4 | - | - | 0.019 | 5.4 | 0.014 | 5.4 | - | - |
| 55 | Metalloproteinase | Echis carinatus sochureki | SVMP | 54 | 5.6 | - | - | - | - | - | - | 0.010 | 5.6 |
| 59 | Group III snake venom metalloproteinase | Echis ocellatus | SVMP | 62 | 10.7 | 0 | 3.1 | 1.254 | 6.0 | 1.744 | 9.0 | 0.651 | 10.7 |
| 61 | Snake venom metalloproteinase VMP1 | Agkistrodon piscivorus leucostoma | SVMP | 46 | 6.8 | - | - | - | - | - | - | 0.833 | 6.8 |
| 72 | Snake venom metalloproteinase | Crotalus adamanteus | SVMP | 68 | 9.0 | - | - | 0.005 | 3.4 | 0.016 | 4.7 | 0.085 | 7.7 |
| 93 | H3 metalloproteinase 1 | Vipera ammodytes ammodytes | SVMP | 68 | 43.5 | 0.611 | 32.0 | 3.415 | 33.3 | 2.970 | 35.9 | 2.737 | 37.3 |
| 108 | P-III metalloprotease | Ovophis okinavensis | SVMP | 16 | 12.1 | - | - | 2.110 | 12.1 | 1.999 | 12.1 | 0.945 | 12.1 |
| 112 | Metalloproteinase H4-A | Vipera ammodytes ammodytes | SVMP | 68 | 14.2 | 0.022 | 11.4 | 0.342 | 4.2 | 0.006 | 4.2 | - | - |
| 143 | Snake venom metalloproteinase lebetase-4 | Macrovipera lebetina | SVMP | 24 | 19.4 | - | - | - | - | 0.008 | 12.4 | 0.091 | 19.4 |
| 146 | Zinc metalloproteinase-disintegrin-like daborhagin-K | Daboia russelii | SVMP | 69 | 6.2 | - | - | - | - | - | - | 0.112 | 6.2 |
| 147 | Coagulation factor X-activating enzyme heavy chain | Daboia siamensis | SVMP | 69 | 10.0 | 0.003 | 2.9 | 0.473 | 7.3 | 0.088 | 7.1 | 0.010 | 7.3 |
| 160 | Coagulation factor X-activating enzyme heavy chain | Macrovipera lebetina | SVMP | 68 | 11.4 | 0.013 | 5.4 | 1.243 | 11.4 | 0.105 | 6.5 | 0.020 | 5.4 |
| 166 | Metalloproteinase F1 | Vipera ammodytes ammodytes | SVMP | 68 | 25.7 | - | - | - | - | 0.001 | 4.1 | 0.498 | 25.7 |
| 182 | Antihemorrhagic factor cHLP-A | Gloydius brevicaudus | SVMP | 36 | 4.0 | - | - | - | - | - | - | 0 | 4.0 |
| 184 | Zinc metalloproteinase/disintegrin | Macrovipera lebetina | SVMP | 53 | 11.9 | - | - | 0.008 | 4.8 | 0.027 | 8.8 | 0.657 | 7.9 |
| 185 | Zinc metalloproteinase-disintegrin-like VLAIP-B | Macrovipera lebetina | SVMP | 68 | 9.3 | 0 | 4.6 | - | - | - | - | 0.052 | 6.5 |
| 186 | Zinc metalloproteinase-disintegrin-like VLAIP-A | Macrovipera lebetina | SVMP | 68 | 25.2 | 0.005 | 17.7 | 0.138 | 14.9 | 0.110 | 17.5 | 0.048 | 19.0 |
| 192 | Group III snake venom metalloproteinase | Echis ocellatus | SVMP | 69 | 10.7 | - | - | 0 | 7.9 | 0 | 10.7 | - | - |
| 194 | Zinc metalloproteinase-disintegrin-like bothrojarin-2 | Bothrops jararaca | SVMP | 24 | 11.9 | - | - | 0.015 | 11.9 | 0.017 | 11.9 | 0.089 | 11.9 |
| 1 | Renin-like aspartic protease | Echis ocellatus | TBP | 43 | 9.4 | 0.009 | 4.6 | 0.040 | 4.8 | 0.039 | 4.8 | 0.055 | 4.8 |
| 8 | Aminopeptidase A | Gloydius brevicaudus | TBP | 110 | 7.6 | 0 | 2.1 | 0.004 | 2.5 | 0.007 | 2.5 | 0.029 | 7.6 |
| 20 | Aminopeptidase N | Gloydius brevicaudus | TBP | 106 | 1.3 | - | - | - | - | - | - | 0 | 1.3 |
| 68 | Glutaminyl-peptide cyclotransferases | Daboia russelii | TBP | 42 | 37.8 | 0.109 | 37.8 | 0.092 | 29.1 | 0.055 | 32.1 | 0.085 | 37.8 |
| 111 | Glutaminyl-cyclase | Ovophis okinavensis | TBP | 40 | 33.2 | 0.012 | 33.2 | - | - | - | - | - | - |
| 120 | Cathepsin D | Ophiophagus hannah | TBP | 30 | 16.3 | 0.013 | 16.3 | - | - | - | - | - | - |
| 121 | Endoplasmic reticulum aminopeptidase 1 | Ophiophagus hannah | TBP | 91 | 1.6 | 0.003 | 1.6 | - | - | - | - | - | - |
| 123 | Renin | Ophiophagus hannah | TBP | 40 | 5.5 | 0.016 | 2.2 | 0.059 | 5.5 | 0.048 | 5.5 | 0.023 | 5.5 |
| 149 | Renin | Echis coloratus | TBP | 12 | 23.9 | - | - | - | - | 0.013 | 23.9 | - | - |
| 167 | Xaa-Pro aminopeptidase 2 | Boiga irregularis | TBP | 76 | 25.7 | 0.420 | 25.7 | 0.041 | 17.3 | 0.114 | 23.6 | 0.042 | 17.3 |
| 178 | Peptidyl-prolyl cis-trans isomerase | Crotalus adamanteus | TBP | 22 | 12.9 | 0.006 | 12.9 | - | - | - | - | - | - |
| 204 | Dipeptidase 2-like | Thamnophis sirtalis | TBP | 33 | 7.4 | 0.002 | 7.4 | - | - | 0.003 | 7.4 | 0.019 | 7.4 |
| 207 | Xaa-Pro aminopeptidase 2-like | Thamnophis sirtalis | TBP | 27 | 19.4 | 0.057 | 19.4 | 0.011 | 11.3 | 0.016 | 19.4 | - | - |
| 60 | Snake venom vascular endothelial growth factor toxin vammin | Vipera ammodytes ammodytes | VEGF | 16 | 49.7 | 5.315 | 31.0 | 4.239 | 44.1 | 5.109 | 36.6 | 2.446 | 32.4 |
| 88 | Snake venom vascular endothelial growth factor toxin HF | Vipera aspis aspis | VEGF | 12 | 65.5 | 0.083 | 65.5 | 0.002 | 58.2 | 0.002 | 48.2 | - | - |
| 148 | Vascular endothelial growth factor A | Echis coloratus | VEGF | 22 | 34.4 | 0.017 | 34.4 | - | - | 0.004 | 17.2 | - | - |
| 150 | Vascular endothelial growth factor F | Echis coloratus | VEGF | 16 | 28.5 | - | - | 0.390 | 28.5 | 0.640 | 28.5 | 0.030 | 18.8 |
| Protein Family 3 | # of Identified Proteins | Protein Abundance 1 LFQ/INT, % (# of Identified Proteins 2) | |||
|---|---|---|---|---|---|
| V. nikolskii | V. kaznakovi | V. orlovi | V. renardi | ||
| PLA2 | (29) | 64.68/65.96 (14) | 41.03/36.43 (11) | 24.21/27.27 (14) | 44.05/47.64 (18) |
| SVMP | (32) | 0.66/0.66 (8) | 16.15/16.31 (19) | 14.77/13.92 (21) | 11.98/10.53 (28) |
| CTL | (18) | 4.01/3.9 (9) | 12.48/13.44 (15) | 11.2/9.46 (15) | 3.46/3.24 (8) |
| SP | (27) | 19.34/20.51 (20) | 10.79/10.96 (18) | 23.97/22.61 (19) | 7.87/6.03 (15) |
| CRISP | (7) | 0.66/0.41 (2) | 9.72/10.89 (7) | 12.2/12.3 (7) | 7.98/8.26 (4) |
| LAAO | (11) | 0.08/0.07 (5) | 3.99/4.33 (8) | 4.59/4.19 (10) | 4.21/3.56 (8) |
| VEGF | (4) | 7.57/5.42 (3) | 3.96/4.63 (2) | 4.2/5.76 (4) | 2.92/2.48 (2) |
| Dis | (5) | 0/0 (0) | 0.53/0.52 (4) | 0.56/0.59 (3) | 13.43/14.04 (5) |
| OP | (28) | 0.17/0.28 (11) | 0.49/1.13 (13) | 0.95/0.96 (16) | 1.8/2.15 (8) |
| PLB | (4) | 0.12/0.1 (2) | 0.32/0.34 (3) | 0.52/0.5 (3) | 0.54/0.51 (4) |
| Nuc | (7) | 0.88/0.92 (6) | 0.21/0.6 (4) | 2.12/1.73 (5) | 0.47/0.32 (3) |
| TBP | (13) | 0.68/0.65 (11) | 0.17/0.25 (5) | 0.3/0.3 (8) | 0.33/0.25 (7) |
| NGF | (2) | 0.33/0.28 (1) | 0.14/0.12 (2) | 0.25/0.25 (1) | 0.12/0.09 (1) |
| Hya | (2) | 0/0.01 (2) | 0.01/0.02 (2) | 0/0.01 (2) | 0/0 (0) |
| BP | (14) | 0.15/0.12 (12) | 0/0.01 (2) | 0.01/0.01 (3) | 0.06/0.1 (9) |
| B-NAP | (1) | 0.01/0.01 (0) | 0/0.03 (1) | 0/0.01 (1) | 0/0 (0) |
| Kunitz | (5) | 0.66/0.7 (4) | 0/0 (0) | 0.15/0.12 (3) | 0.79/0.8 (4) |
| CP | (1) | 0/0 (1) | 0/0 (0) | 0/0 (0) | 0/0 (0) |
| total | (210) | (111) | (116) | (135) | (124) |
| Family | Number of Proteins | Number of Peptides |
|---|---|---|
| CTL | 4 | 5 |
| Dis | 3 | 7 |
| Kunitz | 2 | 2 |
| LAAO | 2 | 3 |
| NAP | 6 | 14 |
| PLA2 | 1 | 1 |
| SP | 2 | 2 |
| SVMP | 11 | 22 |
| VEGF | 1 | 1 |
| Snake Venom Protein Family | V. kaznakovi Venom | V. renardi Venom | V. orlovi Venom | V. nikolskii Venom | V. anatolica Venom 1 | V. raddei Venom 2 | V. a. ammodytes Venom 3 | V. a. meridionalis Venom 3,4 |
|---|---|---|---|---|---|---|---|---|
| PLA2 | + | + | + | + | + | + | + | + 3 |
| SP | + | + | + | + | + | + | + | + 3 |
| Dis | + | + | + | − | + | + | + | + 3 |
| CRISP | + | + | + | + | + | + | + | − |
| Kunitz | − | + | + | + | + | + | − | + 4 |
| LAAO | + | + | + | + | − | + | + | + 3 |
| SVMP | + | + | + | + | + | + | + | + 3 |
| NGF | + | + | + | + | − | + | + | + 3 |
| CTL | + | + | + | + | + | + | − | − |
| PLB | + | + | + | + | − | − | − | − |
| VEGF | + | + | + | + | − | + | + | + 3 |
| Nuc | + | + | + | + | − | − | − | − |
| B-NAP | + | + | + | + | − | + | − | + 4 |
| Hya | + | − | + | + | − | − | − | − |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kovalchuk, S.I.; Ziganshin, R.H.; Starkov, V.G.; Tsetlin, V.I.; Utkin, Y.N. Quantitative Proteomic Analysis of Venoms from Russian Vipers of Pelias Group: Phospholipases A2 are the Main Venom Components. Toxins 2016, 8, 105. https://doi.org/10.3390/toxins8040105
Kovalchuk SI, Ziganshin RH, Starkov VG, Tsetlin VI, Utkin YN. Quantitative Proteomic Analysis of Venoms from Russian Vipers of Pelias Group: Phospholipases A2 are the Main Venom Components. Toxins. 2016; 8(4):105. https://doi.org/10.3390/toxins8040105
Chicago/Turabian StyleKovalchuk, Sergey I., Rustam H. Ziganshin, Vladislav G. Starkov, Victor I. Tsetlin, and Yuri N. Utkin. 2016. "Quantitative Proteomic Analysis of Venoms from Russian Vipers of Pelias Group: Phospholipases A2 are the Main Venom Components" Toxins 8, no. 4: 105. https://doi.org/10.3390/toxins8040105
APA StyleKovalchuk, S. I., Ziganshin, R. H., Starkov, V. G., Tsetlin, V. I., & Utkin, Y. N. (2016). Quantitative Proteomic Analysis of Venoms from Russian Vipers of Pelias Group: Phospholipases A2 are the Main Venom Components. Toxins, 8(4), 105. https://doi.org/10.3390/toxins8040105