Tentacle Transcriptome and Venom Proteome of the Pacific Sea Nettle, Chrysaora fuscescens (Cnidaria: Scyphozoa)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Construction of a Protein Database from the C. fuscescens Tentacle Transcriptome
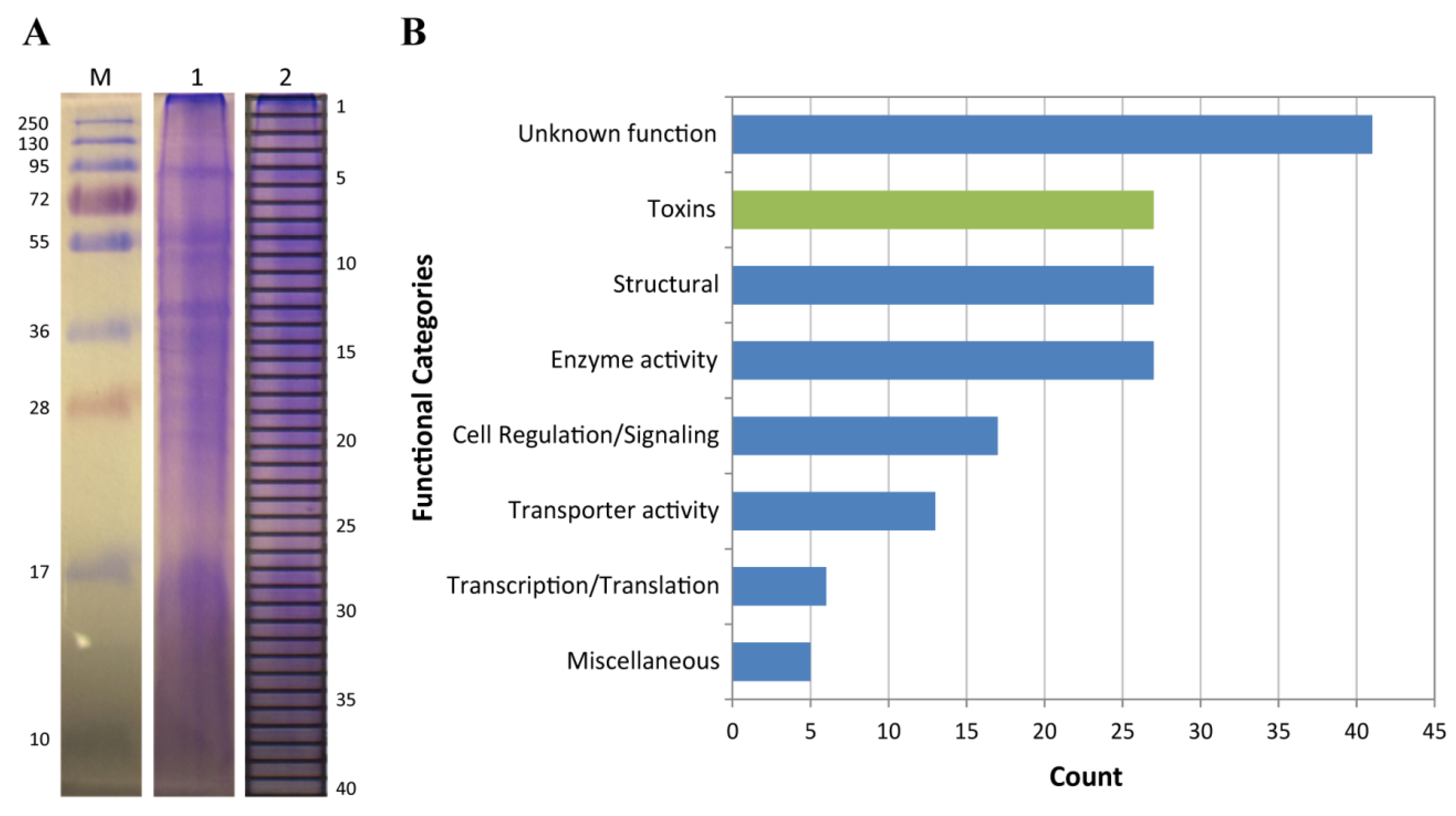
2.2. Proteomic Analysis of C. fuscescens Venom
2.2.1. Proteases
2.2.2. Pore-Forming Toxins
2.2.3. Venom Allergens
2.2.4. Other Potential Toxin Proteins Identified in the C. fuscescens Venom Proteome
2.3. Putative Venom Proteins with ShKT Domains
2.4. Putative Venom Proteins Identified Exclusively in the C. fuscescens Transcriptome
2.5. Comparison of C. fuscescens Transcriptome and Proteome With Other Cnidarians
3. Conclusions
4. Materials and Methods
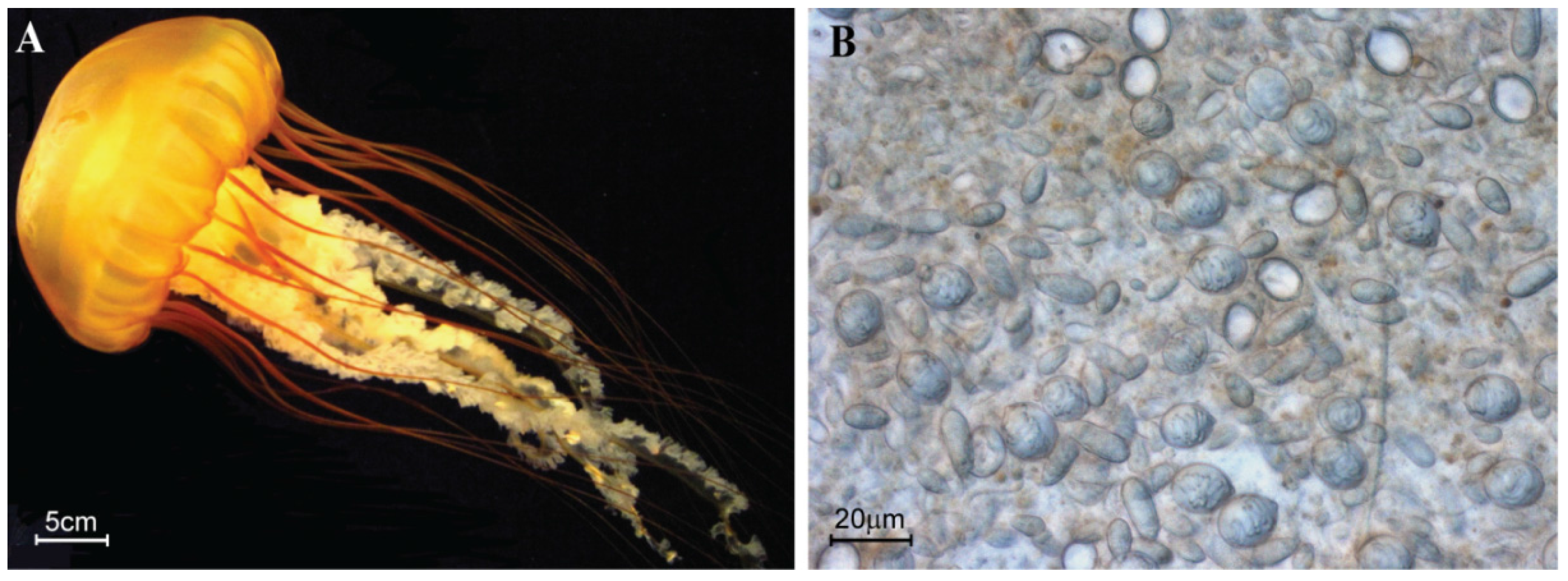
4.1. Jellyfish Collection
4.2. cDNA Library Construction and Illumina Sequencing
4.3. De Novo Transcriptome Assembly
4.4. Functional Annotation of Assembled Transcriptome
4.5. Venom Sample Preparation for Proteomic Analysis
4.6. SDS-PAGE and In-Gel Digestion
4.7. Tandem Mass Spectrometry
4.8. Spectral Searches and Bioinformatics Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kayal, E.; Roure, B.; Philippe, H.; Collins, A.G.; Lavrov, D.V. Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evol. Biol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.G. Phylogeny of Medusozoa and the evolution of cnidarian life cycles. J. Evol. Biol. 2002, 15, 418–432. [Google Scholar] [CrossRef]
- Jouiaei, M.; Yanagihara, A.A.; Madio, B.; Nevalainen, T.J.; Alewood, P.; Fry, B.G. Ancient venom systems: A review on Cnidaria toxins. Toxins 2015, 7, 2251–2271. [Google Scholar] [CrossRef] [PubMed]
- Badré, S. Bioactive toxins from stinging jellyfish. Toxicon 2014, 91, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Frazão, B.; Vasconcelos, V.; Antunes, A. Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: An overview. Mar. Drugs 2012, 10, 1812–1851. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, D.L.; Burnell, J.N. Biochemical and molecular characterisation of cubozoan protein toxins. Toxicon 2009, 54, 1162–1173. [Google Scholar] [CrossRef] [PubMed]
- Newman-Martin, G. Manual of envenomation and poisoning: Australian fauna and flora; Defence Publishing Service: Canberra, Australia, 2007. [Google Scholar]
- Cegolon, L.; Heymann, W.C.; Lange, J.H.; Mastrangelo, G. Jellyfish stings and their management: A review. Mar. Drugs 2013, 11, 523–550. [Google Scholar] [CrossRef] [PubMed]
- Kimball, A.B.; Arambula, K.Z.; Stauffer, A.R.; Levy, V.; Davis, V.W.; Liu, M.; Rehmus, W.E.; Lotan, A.; Auerbach, P.S. Efficacy of a jellyfish sting inhibitor in preventing jellyfish stings in normal volunteers. Wilderness Environ. Med. 2004, 15, 102–108. [Google Scholar] [CrossRef]
- Radwan, F.F.Y.; Gershwin, L.A.; Burnett, J.W. Toxinological studies on the nematocyst venom of Chrysaora achlyos. Toxicon 2000, 38, 1581–1591. [Google Scholar] [CrossRef]
- Vega, M.A.; Ogalde, J.P. First results on qualitative characteristics and biological activity of nematocyst extracts from Chrysaora plocamia (Cnidaria, Scyphozoa). Lat. Am. J. Aquat. Res. 2008, 36, 83–86. [Google Scholar] [CrossRef]
- Neeman, I.; Calton, G.J.; Burnett, J.W. Cytotoxicity and dermonecrosis of sea nettle (Chrysaora quinquecirrha) venom. Toxicon 1980, 18, 55–63. [Google Scholar] [CrossRef]
- Burnett, J.W.; Goldner, R. Effects of Chrysaora quinquecirrha (sea nettle) toxin on the rat cardiovascular system. Proc. Soc. Exp. Biol. Med. 1969, 132, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Kleinhaus, A.L.; Cranefield, P.F.; Burnett, J.W. The effects on canine cardiac Purkinje fibers of Chrysaora quinquecirrha (sea nettle) toxin. Toxicon 1973, 11, 341–349. [Google Scholar] [CrossRef]
- Shryock, J.C.; Bianchi, C.P. Sea nettle (Chrysaora quinquecirrha) nematocyst venom: mechanism of action on muscle. Toxicon 1983, 21, 81–95. [Google Scholar] [CrossRef]
- Burnett, J.W.; Calton, G.J. A comparison of the toxicology of the nematocyst venom from sea nettle fishing and mesenteric tentacles. Toxicon 1976, 14, 109–115. [Google Scholar] [CrossRef]
- Warnick, J.E.; Weinreich, D.; Burnett, J.W. Sea nettle (Chrysaora quinquecirrha) toxin on electrogenic and chemosensitive properties of nerve and muscle. Toxicon 1981, 19, 361–371. [Google Scholar] [CrossRef]
- Houck, H.E.; Lipsky, M.M.; Marzella, L.; Burnett, J.V. Toxicity of sea nettle (Chrysaora quinquecirrha) fishing tentacle nematocyst venom in cultured rat hepatocytes. Toxicon 1996, 34, 771–778. [Google Scholar] [CrossRef]
- Cao, C.J.; Eldefrawi, M.E.; Eldefrawi, A.T.; Burnett, J.W.; Mioduszewski, R.J.; Menking, D.E.; Valdes, J.J. Toxicity of sea nettle toxin to human hepatocytes and the protective effects of phosphorylating and alkylating agents. Toxicon 1998, 36, 269–281. [Google Scholar] [CrossRef]
- Weston, A.J.; Chung, R.; Dunlap, W.C.; Morandini, A.C.; Marques, A.C.; Moura-da-Silva, A.M.; Ward, M.; Padilla, G.; Ferreira da Silva, L.F.; Andreakis, N.; et al. Proteomic characterisation of toxins isolated from nematocysts of the South Atlantic jellyfish Olindias sambaquiensis. Toxicon 2013, 71, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Turk, T.; Kem, W.R. The phylum Cnidaria and investigations of its toxins and venoms until 1990. Toxicon 2009, 54, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Escoubas, P.; King, G.F. Venomics as a drug discovery platform. Expert Rev. Proteomics 2009, 6, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, H.; Xue, W.; Yue, Y.; Liu, S.; Xing, R.; Li, P. Jellyfish venomics and venom gland transcriptomics analysis of Stomolophus meleagris to reveal the toxins associated with sting. J. Proteomics 2014, 106, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Rachamim, T.; Morgenstern, D.; Aharonovich, D.; Brekhman, V.; Lotan, T.; Sher, D. The dynamically evolving nematocyst content of an anthozoan, a scyphozoan, and a hydrozoan. Mol. Biol. Evol. 2015, 32, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Praher, D.; Schlesinger, A.; Ayalon, A.; Tal, Y.; Technau, U. Analysis of soluble protein contents from the nematocysts of a model sea anemone sheds light on venom evolution. Mar.Biotechnol. 2013, 15, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, D.L.; Jia, X.; Potriquet, J.; Kumar, D.; Dash, D.; Kvaskoff, D.; Mulvenna, J. Transcriptome and venom proteome of the box jellyfish Chironex fleckeri. BMC Genomics 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.C.; Ryan, J.F.; Watson, J.A.; Webb, J.; Mullikin, J.C.; Rokhsar, D.S. StellaBase: The Nematostella vectensis genomics database. Nucleic Acids Res. 2006, 34, 495–496. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.A.; Kirkness, E.F.; Simakov, O.; Hampson, S.E.; Mitros, T.; Weinmaier, T.; Rattei, T.; Balasubramanian, P.G.; Borman, J.; Busam, D.; et al. The dynamic genome of Hydra. Nature 2010, 464, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Shinzato, C.; Shoguchi, E.; Kawashima, T.; Hamada, M.; Hisata, K.; Tanaka, M.; Fujie, M.; Fujiwara, M.; Koyanagi, R.; Ikuta, T.; et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 2011, 476, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Riesgo, A.; Andrade, S.C.; Sharma, P.P.; Novo, M.; Perez-Porro, A.R.; Vahtera, V.; Gonzalez, V.L.; Kawauchi, G.Y.; Giribet, G. Comparative description of ten transcriptomes of newly sequenced invertebrates and efficiency estimation of genomic sampling in non-model taxa. Front. Zool. 2012, 9. [Google Scholar] [CrossRef] [PubMed]
- Margres, M.J.; McGivern, J.J.; Wray, K.P.; Seavy, M.; Calvin, K.; Rokyta, D.R. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). J. Proteomics 2014, 96, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 2008. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhou, Y.; Liu, D.; Wang, Q.; Ruan, Z.; He, Q.; Zhang, L. Global transcriptome analysis of the tentacle of the jellyfish Cyanea capillata using deep sequencing and expressed sequence tags: Insight into the toxin- and degenerative disease-related transcripts. PloS one 2015, 10, e0142680. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, P.G.; Beckmann, A.; Warnken, U.; Schnolzer, M.; Schuler, A.; Bornberg-Bauer, E.; Holstein, T.W.; Ozbek, S. Proteome of Hydra nematocyst. J. Biol. Chem. 2012, 287, 9672–9681. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, D.L.; Aziz, A.; Loukas, A.; Potriquet, J.; Seymour, J.; Mulvenna, J. Venom proteome of the box jellyfish Chironex fleckeri. PloS one 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Fujimura, Y.; Titani, K. Snake venom proteases affecting hemostasis and thrombosis. Biochim. Biophys. Acta 2000, 1477, 146–156. [Google Scholar] [CrossRef]
- King, G.F.; Hardy, M.C. Spider-venom peptides: structure, pharmacology, and potential for control of insect pests. Annu. Rev. Entomol. 2013, 58, 475–496. [Google Scholar] [CrossRef] [PubMed]
- Jouiaei, M.; Casewell, N.R.; Yanagihara, A.A.; Nouwens, A.; Cribb, B.W.; Whitehead, D.; Jackson, T.N.; Ali, S.A.; Wagstaff, S.C.; Koludarov, I.; et al. Firing the sting: chemically induced discharge of cnidae reveals novel proteins and peptides from box jellyfish (Chironex fleckeri) venom. Toxins 2015, 7, 936–950. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.H.; Woo, T.H.; Kim, C.B.; Park, J.H.; Kim, H.; Lee, S.; Lee, S.H. Differential gene expression profiles in the venom gland/sac of Orancistrocerus drewseni (Hymenoptera: Eumenidae). Arch. Insect Biochem. Physiol. 2009, 71, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Safavi-Hemami, H.; Moller, C.; Mari, F.; Purcell, A.W. High molecular weight components of the injected venom of fish-hunting cone snails target the vascular system. J. Proteomics 2013, 91, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Z.; Cun, S.J.; Xie, X.J.; Lin, J.H.; Wei, J.W.; Yang, W.L.; Mou, C.Y.; Yu, C.L.; Ye, L.T.; Lu, Y.; et al. EST analysis of gene expression in the tentacle of Cyanea capillata. FEBS Lett. 2003, 538, 183–191. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, D.L.; Konstantakopoulos, N.; McInerney, B.V.; Mulvenna, J.; Seymour, J.E.; Isbister, G.K.; Hodgson, W.C. Chironex fleckeri (box jellyfish) venom proteins: expansion of a cnidarian toxin family that elicits variable cytolytic and cardiovascular effects. J. Biol. Chem. 2014, 289, 4798–4812. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, D.; Burnell, J. Identification, cloning and sequencing of two major venom proteins from the box jellyfish, Chironex fleckeri. Toxicon 2007, 50, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, D.; Burnell, J. Partial purification of cytolytic venom proteins from the box jellyfish, Chironex fleckeri. Toxicon 2008, 51, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Takuwa, K.; Nakao, M.; Ito, E.; Miyake, M.; Noda, M.; Nakajima, T. Novel proteinaceous toxins from the box jellyfish (sea wasp) Carybdea rastoni. Biochem. Biophys. Res. Commun. 2000, 275, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Takuwa, K.; Nakao, M.; Sakamoto, B.; Crow, G.L.; Nakajima, T. Isolation and characterization of a novel protein toxin from the Hawaiian box jellyfish (sea wasp) Carybdea alata. Biochem. Biophys. Res. Commun. 2000, 275, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Takuwa-Kuroda, K.; Nakao, M.; Oshiro, N.; Iwanaga, S.; Nakajima, T. A novel protein toxin from the deadly box jellyfish (sea wasp, habu-kurage) Chiropsalmus quadrigatus. Biosci. Biotechnol. Biochem. 2002, 66, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Yanagihara, A.A.; Shohet, R.V. Cubozoan venom-induced cardiovascular collapse is caused by hyperkalemia and prevented by zinc gluconate in mice. PloS one 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Ponce, D.; Brinkman, D.L.; Luna-Ramírez, K.; Wright, C.E.; Dorantes-Aranda, J.J. Comparative study of the toxic effects of Chrysaora quinquecirrha (Cnidaria: Scyphozoa) and Chironex fleckeri (Cnidaria: Cubozoa) venoms using cell-based assays. Toxicon 2015, 106, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Lassen, S.; Helmholz, H.; Ruhnau, C.; Prange, A. A novel proteinaceous cytotoxin from the northern Scyphozoa Cyanea capillata (L.) with structural homology to cubozoan haemolysins. Toxicon 2011, 57, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Gacesa, R.; Chung, R.; Dunn, S.; Weston, A.; Jaimes-Becerra, A.; Marques, A.; Morandini, A.; Hranueli, D.; Starcevic, A.; Ward, M.; et al. Gene duplications are extensive and contribute significantly to the toxic proteome of nematocysts isolated from Acropora digitifera (Cnidaria: Anthozoa: Scleractinia). BMC Genomics 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Sanchez, J.; Kouskoura, T.; Crickmore, N. N-terminal activation is an essential early step in the mechanism of action of the Bacillus thuringiensis Cry1Ac insecticidal toxin. J. Biol. Chem. 2002, 277, 23985–23987. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Soria, G. Molecular Characterization of Carukia barnesi and Malo kingi, Cnidaria; Cubozoa; Carybdeidae. Ph.D. Thesis, James Cook University, Townsville City, Australia, 5 November 2009. [Google Scholar]
- Toshino, S.; Miyake, H.; Shibata, H. Meteorona kishinouyei, a new family, genus and species (Cnidaria, Cubozoa, Chirodropida) from Japanese waters. ZooKeys 2015, 503, 1–21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lewis, C.; Bentlage, B. Clarifying the identity of the Japanese Habu-kurage, Chironex yamaguchii, sp. nov. (Cnidaria: Cubozoa: Chirodropida). Zootaxa 2009, 2030, 59–65. [Google Scholar]
- Gershwin, L.A. Two new species of jellyfishes (Cnidaria: Cubozoa: Carybdeida) from tropical western australia, presumed to cause Irukandji syndrome. Zootaxa 2005, 1–30. [Google Scholar]
- Yamazaki, Y.; Morita, T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon 2004, 44, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Morita, T. C-type lectin-related proteins from snake venoms. Curr. Drug Targets Cardiovasc. Haematol. Disord. 2004, 4, 357–373. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, R.B.; Chaim, O.M.; Mangili, O.C.; Gremski, W.; Dietrich, C.P.; Nader, H.B.; Veiga, S.S. Hyaluronidases in Loxosceles intermedia (Brown spider) venom are endo-beta-N-acetyl-d-hexosaminidases hydrolases. Toxicon 2007, 49, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Schweitz, H.; Heurteaux, C.; Bois, P.; Moinier, D.; Romey, G.; Lazdunski, M. Calcicludine, a venom peptide of the Kunitz-type protease inhibitor family, is a potent blocker of high-threshold Ca2+ channels with a high affinity for L-type channels in cerebellar granule neurons. Proc. Natl. Acad. Sci. 1994, 91, 878–882. [Google Scholar] [CrossRef] [PubMed]
- Morjen, M.; Honore, S.; Bazaa, A.; Abdelkafi-Koubaa, Z.; Ellafi, A.; Mabrouk, K.; Kovacic, H.; El Ayeb, M.; Marrakchi, N.; Luis, J. PIVL, a snake venom Kunitz-type serine protease inhibitor, inhibits in vitro and in vivo angiogenesis. Microvasc. Res. 2014, 95, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Peigneur, S.; van Der Haegen, A.; Möller, C.; Waelkens, E.; Diego-García, E.; Marí, F.; Naudé, R.; Tytgat, J. Unraveling the peptidome of the South African cone snails Conus pictus and Conus natalis. Peptides 2013, 41, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Isaeva, M.P.; Chausova, V.E.; Zelepuga, E.A.; Guzev, K.V.; Tabakmakher, V.M.; Monastyrnaya, M.M.; Kozlovskaya, E.P. A new multigene superfamily of Kunitz-type protease inhibitors from sea anemone Heteractis crispa. Peptides 2012, 34, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Dy, C.Y.; Buczek, P.; Imperial, J.S.; Bulaj, G.; Horvath, M.P. Structure of conkunitzin-S1, a neurotoxin and Kunitz-fold disulfide variant from cone snail. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Dai, H.; Qiu, S.; Li, T.; He, Y.; Ma, Y.; Chen, Z.; Wu, Y.; Li, W.; Cao, Z. SdPI, the first functionally characterized Kunitz-type trypsin inhibitor from scorpion venom. PloS one 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Tews, I.; Perrakis, A.; Oppenheim, A.; Dauter, Z.; Wilson, K.S.; Vorgias, C.E. Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nature Struct. Biol. 1996, 3, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Purcell, J.E. Predation on zooplankton by large jellyfish, Aurelia labiata, Cyanea capillata and Aequorea aequorea, in Prince William Sound, Alaska. Mar. Ecol. Prog. Ser. 2003, 246, 137–152. [Google Scholar] [CrossRef]
- Castañeda, O.; Harvey, A.L. Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon 2009, 54, 1119–1124. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Gurevitz, M. When positive selection of neurotoxin genes is missing. The riddle of the sea anemone Nematostella vectensis. FEBS J. 2006, 273, 3886–3892. [Google Scholar] [CrossRef] [PubMed]
- Putnam, N.H.; Srivastava, M.; Hellsten, U.; Dirks, B.; Chapman, J.; Salamov, A.; Terry, A.; Shapiro, H.; Lindquist, E.; Kapitonov, V.V.; et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Lapébie, P.; Ruggiero, A.; Barreau, C.; Chevalier, S.; Chang, P.; Dru, P.; Houliston, E.; Momose, T. Differential responses to Wnt and PCP disruption predict expression and developmental function of conserved and novel genes in a cnidarian. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Gröger, H.; Schmid, V.; Spring, J. A toxin homology domain in an astacin-like metalloproteinase of the jellyfish Podocoryne carnea with a dual role in digestion and development. Dev. Genes Evol. 1998, 208, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, T.V.; Balandin, S.V.; Aleshina, G.M.; Tagaev, A.A.; Leonova, Y.F.; Krasnodembsky, E.D.; Men’shenin, A.V.; Kokryakov, V.N. Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem. Biophys. Res. Commun. 2006, 348, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Li, R.F.; Yu, H.; Xing, R.; Liu, S.; Qing, Y.; Li, K.; Li, B.; Meng, X.; Cui, J.; Li, P. Application of nano LC-MS/MS to the shotgun proteomic analysis of the nematocyst proteins from jellyfish Stomolophus meleagris. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2012, 899, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Sher, D.; Knebel, A.; Bsor, T.; Nesher, N.; Tal, T.; Morgenstern, D.; Cohen, E.; Fishman, Y.; Zlotkin, E. Toxic polypeptides of the hydra—A bioinformatic approach to cnidarian allomones. Toxicon 2005, 45, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Da Silveira, R.B.; Wille, A.C.; Chaim, O.M.; Appel, M.H.; Silva, D.T.; Franco, C.R.; Toma, L.; Mangili, O.C.; Gremski, W.; Dietrich, C.P.; et al. Identification, cloning, expression and functional characterization of an astacin-like metalloprotease toxin from Loxosceles intermedia (brown spider) venom. Biochem. J. 2007, 406, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Lotan, A.; Fishman, L.; Zlotkin, E. Toxin compartmentation and delivery in the cnidaria: The nematocyst’s tubule as a multiheaded poisonous arrow. J. Exp. Zool. 1996, 275, 444–451. [Google Scholar] [CrossRef]
- Nevalainen, T.J.; Peuravuori, H.J.; Quinn, R.J.; Llewellyn, L.E.; Benzie, J.A.H.; Fenner, P.J.; Winkel, K.D. Phospholipase A2 in Cnidaria. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2004, 139, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Yu, H.; Xing, R.; Liu, S.; Wang, L.; Cai, S.; Li, P. Partial characterization of the hemolytic activity of the nematocyst venom from the jellyfish Cyanea nozakii Kishinouye. Toxicol. In Vitro 2010, 24, 1750–1756. [Google Scholar] [CrossRef] [PubMed]
- Gusmani, L.; Avian, M.; Galil, B.; Patriarca, P.; Rottini, G. Biologically active polypeptides in the venom of the jellyfish Rhopilema nomadica. Toxicon 1997, 35, 637–648. [Google Scholar] [CrossRef]
- Helmholz, H.; Ruhnau, C.; Schutt, C.; Prange, A. Comparative study on the cell toxicity and enzymatic activity of two northern scyphozoan species Cyanea capillata (L.) and Cyanea lamarckii (Peron & Leslieur). Toxicon 2007, 50, 53–64. [Google Scholar] [PubMed]
- Radwan, F.F.; Burnett, J.W.; Bloom, D.A.; Coliano, T.; Eldefrawi, M.E.; Erderly, H.; Aurelian, L.; Torres, M.; Heimer-de la Cotera, E.P. A comparison of the toxinological characteristics of two Cassiopea and Aurelia species. Toxicon 2001, 39, 245–257. [Google Scholar] [CrossRef]
- Radwan, F.F.; Roman, L.G.; Baksi, K.; Burnett, J.W. Toxicity and mAChRs binding activity of Cassiopea xamachana venom from Puerto Rican coasts. Toxicon 2005, 45, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.H.; Yu, H.H.; Li, C.P.; Xing, R.G.; Liu, S.; Wang, L.; Cai, S.B.; Li, P.C. Isolation and characterization of lethal proteins in nematocyst venom of the jellyfish Cyanea nozakii Kishinouye. Toxicon 2010, 55, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Harrison, R.A.; Wuster, W.; Wagstaff, S.C. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genomics 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Corrêa-Netto, C.; Junqueira-de-Azevedo, I.L.M.; Silva, D.A.; Ho, P.L.; Leitão-de-Araújo, M.; Alves, M.L.M.; Sanz, L.; Foguel, D.; Zingali, R.B.; Calvete, J.J. Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J. Proteomics 2011, 74, 1795–1809. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, K.; Midorikawa, S.; Ishida, M.; Nagashima, Y.; Nagai, H. Plancitoxins, lethal factors from the crown-of-thorns starfish Acanthaster planci, are deoxyribonucleases II. Toxicon 2004, 44, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, K.; Yamamoto, S.; Yamanaka, H.; Kikuchi, T. Purification and characterization of a lethal factor in venom from the crown-of-thorns starfish (Acanthaster planci). Toxicon 1988, 26, 1077–1083. [Google Scholar] [CrossRef]
- Shiomi, K.; Yamamoto, S.; Yamanaka, H.; Kikuchi, T.; Konno, K. Liver damage by the crown-of-thorns starfish (Acanthaster planci) lethal factor. Toxicon 1990, 28, 469–475. [Google Scholar] [CrossRef]
- Watanabe, A.; Nagai, H.; Nagashima, Y.; Shiomi, K. Structural characterization of plancitoxin I, a deoxyribonuclease II-like lethal factor from the crown-of-thorns starfish Acanthaster planci, by expression in Chinese hamster ovary cells. Fisheries Sci. 2009, 75, 225–231. [Google Scholar] [CrossRef]
- Nevalainen, T.J. Phospholipases A2 in the genome of the sea anemone Nematostella vectensis. Comp. Biochem. Physiol. Part D Genomics Proteomics 2008, 3, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Weinberger, H.; Sullivan, J.C.; Reitzel, A.M.; Finnerty, J.R.; Gurevitz, M. Concerted evolution of sea anemone neurotoxin genes is revealed through analysis of the Nematostella vectensis genome. Mol. Biol. Evol. 2008, 25, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Crossley, S.M.G.; George, A.L.; Keller, C.J. A method for eradicating amphipod parasites (Hyperiidae) from host jellyfish, Chrysaora fuscescens (Brandt, 1835), in a closed recirculating system. J. Zoo Wildlife Med. 2009, 40, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 March 2016).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic. Available online: http://www.usadellab.org/cms/?page=trimmomatic (accessed on 24 March 2016).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. Basic local alignment search tool. J. Mol. Evol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Howson, R.; Neupane, R.; Shengqiang, S. Metazome. Available online: http://www.metazome.net/ (accessed on 24 March 2016).
- Jungo, F. Animal toxin annotation project. Available online: http://www.uniprot.org/program/Toxins (accessed on 24 March 2016).
- Mitchell, A.; Chang, H.Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology (GO) project. Available online: http://geneontology.org/ (accessed on 24 March 2016).
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Stoffel, W. TMbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 1993, 374, 166–170. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Iseli, C.; Jongeneel, C.V.; Bucher, P. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999, 138, 138–148. [Google Scholar]
- Bloom, D.A.; Burnett, J.W.; Alderslade, P. Partial purification of box jellyfish (Chironex fleckeri) nematocyst venom isolated at the beachside. Toxicon 1998, 36, 1075–1085. [Google Scholar] [CrossRef]
- Marchini, B.; De Nuccio, L.; Mazzei, M.; Mariottini, G.L. A fast centrifuge method for nematocyst isolation from Pelagia noctiluca Forskal (Cnidaria: Scyphozoa). Riv. Biol. 2004, 97, 505–515. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Mendoza, L.; Shteynberg, D.; Farrah, T.; Lam, H.; Tasman, N.; Sun, Z.; Nilsson, E.; Pratt, B.; Prazen, B.; et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics 2010, 10, 1150–1159. [Google Scholar] [CrossRef] [PubMed]

| Assembly | Count |
|---|---|
| Raw reads (paired-end) | 26,991,925 |
| After cleaning | 17,319,746 |
| Contigs | 30,317 |
| Average length ± SD | 628.70 ± 840.07 |
| Length (min and max) | 201 to 31,945 |
| GC content | 40.42% |
| Raw reads mapped to contigs | 97.69% |
| CDS | Count |
| Containing a coding region | 23,534 (78%) |
| Transcripts with significant BLAST hit (1 × 10−5) | 16,925 (72%) |
| With homologues in databases: | |
| GenBank non-redundant Cnidarian protein sequences | 15,987 (53%) |
| H. vulgaris | 14,261 (47%) |
| SwissProt | 13,375 (44%) |
| N. vectensis | 12,144 (40%) |
| Uniprot animal toxin and venom | 549 (2%) |
| Sequence analysis | Count |
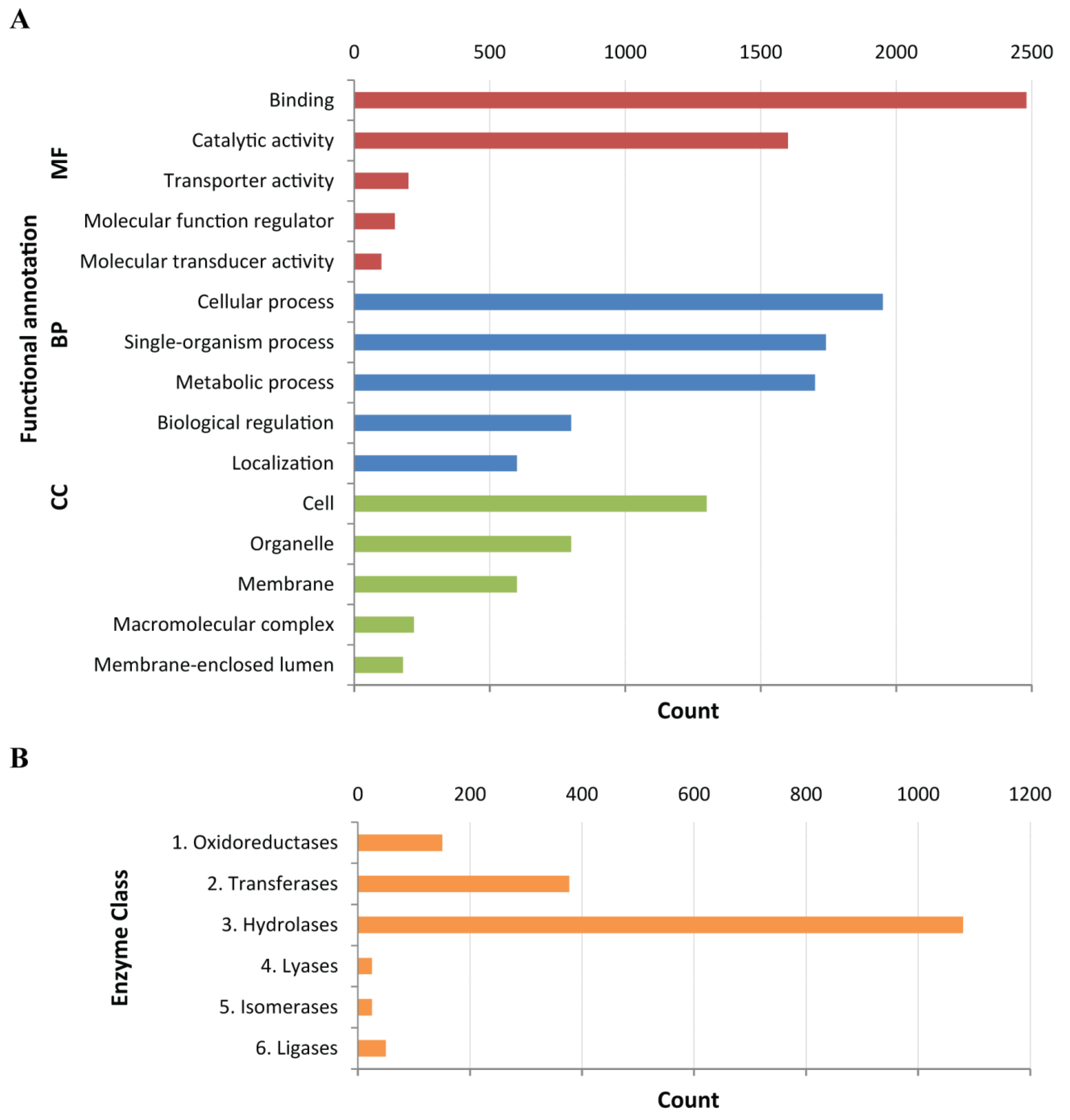
| Returning GO term | 11,586 (49%) |
| GO terms returned: | |
| Molecular function | 8265 (35%) |
| Biological process | 4768 (20%) |
| Cellular component | 2173 (9%) |
| Predicted proteins with signal sequences * | 1012 (4%) |
| Predicted proteins with two or more transmembrane helices | 641 (2%) |
| Transcript | Category | Unique Peptides (n) | Coverage (%) | InterProScan Protein Feature | Signal Peptide (Yes/No) | Transmembrane Domain (Yes/No) | BLAST Analysis | |
|---|---|---|---|---|---|---|---|---|
| Protein Identity | Species of Closest Homology | |||||||
| comp13691_c0_seq2 | Metalloprotease | 18 | 76.6 | Peptidase M1, alanine aminopeptidase | n | n | Aminopeptidase N-like | H. vulgaris |
| comp12218_c0_seq1 | Metalloprotease | 16 | 76.2 | Peptidase M13 | n | y (27–49) | Endothelin-converting enzyme 1-like | H. vulgaris |
| comp13767_c0_seq1 | Metalloprotease | 10 | 80.4 | Peptidase M13 | y (1–29) | y (12–29) | Endothelin-converting enzyme 2-like | H. vulgaris |
| comp11996_c0_seq1 | Metalloprotease | 8 | 62.2 | Peptidase M1, alanine aminopeptidase | n | n | Endoplasmic reticulum aminopeptidase 1-like | H. vulgaris |
| comp11571_c0_seq2 | Metalloprotease | 8 | 75.4 | Peptidase M13 | n | n | Endothelin-converting enzyme 1-like | H. vulgaris |
| comp10942_c1_seq1 | Metalloprotease | 7 | 71.0 | Peptidase M13 | n | y (31–53) | Endothelin-converting enzyme 1-like | H. vulgaris |
| comp12208_c0_seq1 | Metalloprotease | 6 | 61.0 | Peptidase M14, carboxypeptidase A | y (1–20) | y (292–309) | Carboxypeptidase D-like | H. vulgaris |
| comp9530_c0_seq2 | Metalloprotease | 6 | 87.3 | Peptidase M13 | n | n | Endothelin-converting enzyme 1-like | N. vectensis |
| comp14393_c0_seq1 | Metalloprotease | 5 | 68.2 | Peptidase M2, peptidyl-dipeptidase A | n | n | Angiotensin-converting enzyme-like isoform | H. vulgaris |
| comp14137_c0_seq1 | Metalloprotease | 2 | 42.1 | Peptidase M2, peptidyl-dipeptidase A | n | n | Angiotensin-converting enzyme | H. vulgaris |
| comp14070_c0_seq1 | Metalloprotease | 3 | 67.3 | Peptidase M18 | n | y (493–510) | Aspartyl aminopeptidase-like | Lepisosteus oculatus |
| comp13494_c0_seq1 | Aspartyl protease | 2 | 39.6 | Aspartic peptidase | y (1–17) | y (161–193) | Cathepsin D | Pteria penguin |
| comp12883_c0_seq1 | Aspartyl protease | 2 | 48.9 | Aspartic peptidase | y (1–18) | y (2–24) | Aspartyl protease | Placozoa sp. H4 |
| comp13655_c0_seq2 | Serine protease | 2 | 67.8 | Peptidase S8/S53 | n | y (156–178, 323–339, 748–771) | PC3-like endoprotease variant B isoform X1 | H. vulgaris |
| comp13207_c0_seq1 | Pore-forming toxin | 6 | 75.4 | Delta-endotoxin, N-terminal | y (1–19) | y (104–125) | Toxin TX2 | A. aurita |
| comp12925_c0_seq1 | Pore-forming toxin | 8 | 76.0 | Delta endotoxin, N-terminal | n | y (15–37) | Uncharacterized protein LOC105843890 | H. vulgaris |
| comp13855_c0_seq5 | Venom allergen | 37 | 83.0 | Cysteine-rich secretory protein, allergen V5/Tpx-1-related | n | n | Cell wall protein PRY3-like | H. vulgaris |
| comp13672_c0_seq1 | Venom allergen | 18 | 81.4 | Cysteine-rich secretory protein, allergen V5/Tpx-1-related | n | n | Cell wall protein PRY3-like | H. vulgaris |
| comp13791_c0_seq1 | Venom allergen | 15 | 75.2 | Cysteine-rich secretory protein, allergen V5/Tpx-1-related | n | n | Cell wall protein PRY3-like | H. vulgaris |
| comp13791_c0_seq3 | Venom allergen | 9 | 70.6 | Cysteine-rich secretory protein, allergen V5/Tpx-1-related | n | n | Cell wall protein PRY3-like | H. vulgaris |
| comp13342_c1_seq2 | Venom allergen | 2 | 76.1 | Cysteine-rich secretory protein, allergen V5/Tpx-1-related | n | n | Cell wall protein PRY3-like | H. vulgaris |
| comp12264_c0_seq1 | Venom allergen | 2 | 81.7 | Cysteine-rich secretory protein, allergen V5/Tpx-1-related | y (1–20) | y (7–29) | PRY2-like protein | Pyronema omphalodes CBS 100304 |
| comp13629_c0_seq1 | C-type lectin | 194 | 85.0 | C-type lectin | y (1–20) | y (344–362) | Golgi-associated plant pathogenesis-related protein 1 | H. vulgaris |
| comp13792_c0_seq2 | C-type lectin | 44 | 70.0 | C-type lectin | n | y (96–116) | Golgi-associated plant pathogenesis-related protein 1 | H. vulgaris |
| comp13880_c0_seq1 | C-type lectin | 8 | 56.5 | C-type lectin | y (1–21) | y (348–368) | Golgi-associated plant pathogenesis-related protein 1 | H. vulgaris |
| comp13219_c0_seq1 | Glycoside hydrolase | 18 | 59.4 | Beta-hexosaminidase | y (1–20) | y (416–438) | beta-hexosaminidase subunit alpha-like isoform X1 | H. vulgaris |
| comp7130_c0_seq1 | Enzyme inhibitor | 4 | 64.0 | Peptidase S8/S53 domain | y (1–19) | y (484–507) | Tripeptidyl-peptidase 1-like | H. vulgaris |
| Toxin ID | Jellyfish Species | BLAST | UniProt Accession No. | Reference | |
|---|---|---|---|---|---|
| Identity (%) | E-Value | ||||
| TX2 * | A. aurita | 48 | 2 × 10−124 | I3VAS2 | UniProt |
| TX1 * | A. aurita | 37 | 2 × 10−96 | I3VAS1 | UniProt |
| CfTX-A | C. fleckeri | 25 | 1 × 10−27 | T1PRE3 | [45] |
| CrTX-A | Carybdea rastoni 1 | 25 | 3 × 10−26 | Q9GV72 | [48] |
| CfTX-1 | C. fleckeri | 24 | 9 × 10−26 | A7L035 | [46] |
| CfTX-2 | C. fleckeri | 24 | 3 × 10−27 | A7L036 | [46] |
| CqTX-A | Chiropsalmus quadrigatus 2 | 24 | 9 × 10−25 | P58762 | [50] |
| CaTX-A | Carybdea alata 3 | 24 | 1 × 10−23 | Q9GNN8 | [49] |
| CfTX-Bt | C. fleckeri | 24 | 2 × 10−16 | W0K4S7 | [45] |
| CfTX-B | C. fleckeri | 23 | 1 × 10−26 | T1PQV6 | [45] |
| MkTX-A1 * | M. kingi | 20 | 6 × 10−10 | D2DRC0 | [56] |
| MkTX-A2 * | M. kingi | 20 | 1 × 10−9 | D2DRC1 | [56] |
| Toxin Family | Class Scyphozoa | Class Cubozoa | Class Hydrozoa | Class Anthozoa | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyanea capillata | Stomolophus meleagris | Aurelia aurita | Chrysaora fuscescens | Chironex fleckeri | Malo kingi | Carukia barnesi | Olindias sambaquiensis | Hydra vulgaris | Anemonia viridis | Nematostella vectensis | Acropora digitifera | ||
| Proteinases | (E) Cysteine protease; (T) astacin, zinc and matrix metallo-proteinases, serine proteases; ECE-1 | (T) Zinc metallo-proteinases | (T) Zinc metallo-proteinases; ECE-1 | (T, P) Serine peptidases, zinc metallo-proteinases, ECE-1 and -2 | (T,P) Serine peptidases; astacin and zinc metallo-proteinases; (P) ECE-2 and 2-like | (E) Serine peptidases; carboxy-peptidases; metalloproteases | (E) Serine peptidases | (P) Serine peptidases; zinc metallo-proteinases | (P) Serine peptidases; zinc metallo-proteinase; (T, P) ECE-1 | (T) ECE-1 | (T) Astacin-like metallo-proteinase | (T, P) Serine peptidases; astacin and other metallo-proteinases | |
| Lipases | (E, T) PLA2; (T) PLD; LALs | (T) PLA2 and PLB1 | (T, P) PLA2 | (T) LALs; PLA2 and PLB2 | (T) LALs; PLA2 | - | - | (P) PLA2 | (E, T, P) PLA2 | - | (T) | (T, P) LALs; endothelial lipase; PLB; PLA1; PLA2 | |
| Deoxyribonu-cleases | (T) Plancitoxin-like | - | - | (T) Plancitoxin-like | - | - | - | - | (E) Plancitoxin-like | - | - | - | |
| Cytolysins | (E) Hemolysin C | (T) Hemolysins (homologues to ryncolin, veficolin, hemolysin hlyIII) | - | - | - | - | - | (P) AvTX-60A and PsTX-like | (E) Actinoporins and hydralysins | (T) Actinoporin-like | - | (T, P) Bandaporin; actinoporin; urticinatoxin | |
| Pore-forming (cnidarian toxin family) | (T) CfTX-like; (P) CcTX-1 | - | (T,P) 1 AaTX-1 and -2; CaTX-like | (T, P) CfusTX-1 | (T, P) CfTXs | (E) MkTXs (CfTX-like) | - | - | (T, P) CaTX-like | (T) CaTX-like | - | (T, P) CfTX-1-like | |
| Pore-forming (MAC-PF) | - | - | (T, P) | - | - | - | - | - | (T, P) | (T, P) | - | - | |
| CRISPs | - | - | * | (T, P) allergen V5/Tpx-1-related | (T) allergen V5/Tpx-1-related | - | - | - | (E) venom allergen 5 | * | (T) venom allergen 5 /PR-1-like | - | |
| C-type lectins | - | (T) | (T) | (T, P) | (T, P) | - | - | - | (T, P) | - | (T) | (T, P) | |
| Protease inhibitors | (T) Kazal-type; Kunitz-type | (T) Kunitz-type | (T) Kunitz-type | (T, P) Kunitz-type | (T, P) Cysteine protease inhibitors; Kunitz-type; Kazal-type | (E) Kazal-type | - | - | (T, P) Cysteine protease inhibitors; Kunitz-type | (P) Kunitz-type | - | (T, P) Kunitz-type | |
| Proteins with ShKT domains | - | (T) 1–4 domains + other protein domains | (T) 1–3 domains + other protein domains | (T) 1–4 domains + other protein domains; (P) CRISP-like protein and C-type lectin | (T, P) + astacin domains | (E) | (E) | - | (E, T, P) + metalloprotease-like protein domains | (T, P) | (T) | - | |
| Neurotoxins (modulators of nicotinic receptors or presynaptic nerve endings) | - | (T) Botulinum neurotoxins; α-latrocrusto-toxin-Lt1a | - | - | - | - | - | (P) κ-4-Bungarotoxin; α-latrocrusto-toxin-Lt1a | - | - | - | - | |
| Neurotoxins (K+ channel blockers) | - | - | - | - | - | - | - | - | (E) Kalicludine-like | (T, P) ShK toxins, BDS-like | - | - | |
| Neurotoxins (Na+ channel blockers) | - | - | - | - | - | - | - | - | (T, P) Av1, Av2 and Av3 | (T) Nv1 | (T, P) Av1-like | ||
| Associated References | [35,43,53] | [23] | [24] | Present work | [26,37,40] | [56] | [56] | [20] | [24,36,79] | [5,24] | [25,72,73,95,96] | [54] | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponce, D.; Brinkman, D.L.; Potriquet, J.; Mulvenna, J. Tentacle Transcriptome and Venom Proteome of the Pacific Sea Nettle, Chrysaora fuscescens (Cnidaria: Scyphozoa). Toxins 2016, 8, 102. https://doi.org/10.3390/toxins8040102
Ponce D, Brinkman DL, Potriquet J, Mulvenna J. Tentacle Transcriptome and Venom Proteome of the Pacific Sea Nettle, Chrysaora fuscescens (Cnidaria: Scyphozoa). Toxins. 2016; 8(4):102. https://doi.org/10.3390/toxins8040102
Chicago/Turabian StylePonce, Dalia, Diane L. Brinkman, Jeremy Potriquet, and Jason Mulvenna. 2016. "Tentacle Transcriptome and Venom Proteome of the Pacific Sea Nettle, Chrysaora fuscescens (Cnidaria: Scyphozoa)" Toxins 8, no. 4: 102. https://doi.org/10.3390/toxins8040102
APA StylePonce, D., Brinkman, D. L., Potriquet, J., & Mulvenna, J. (2016). Tentacle Transcriptome and Venom Proteome of the Pacific Sea Nettle, Chrysaora fuscescens (Cnidaria: Scyphozoa). Toxins, 8(4), 102. https://doi.org/10.3390/toxins8040102







