Selection and Characterization of a Novel DNA Aptamer for Label-Free Fluorescence Biosensing of Ochratoxin A
Abstract
:1. Introduction
2. Results and Discussion
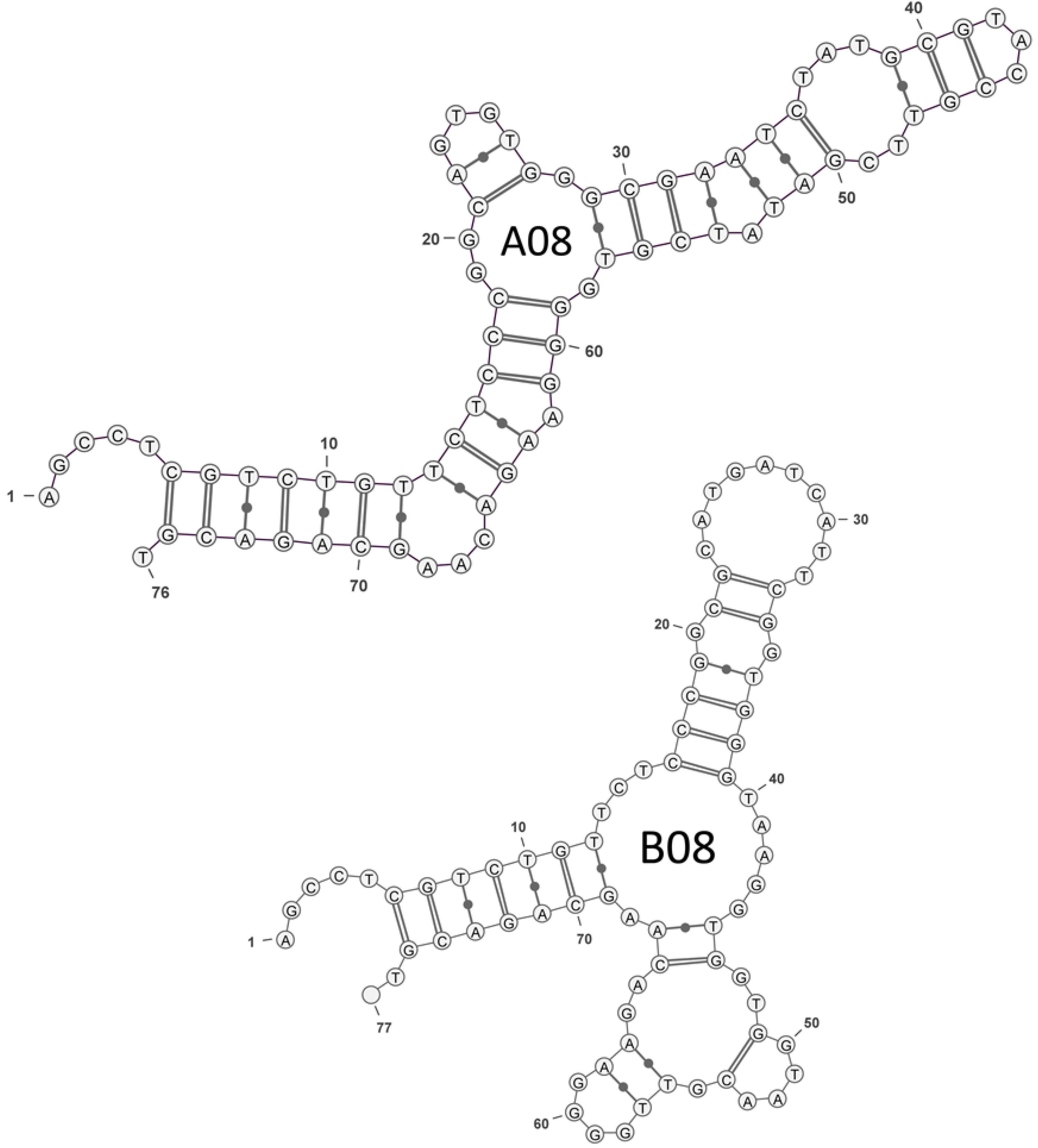
2.1. Selection of OTA Aptamers
| Aptamer | 5'-AGCCTCGTCTGTTCTCCC-N40-GGGAAGACAAGCAGACGT-3' |
|---|---|
| A08 | GGCAGTGTGGGCGAATCTATGCGTACCGTTCGATATCGTG |
| B08 | GGCGCATGATCATTCGGTGGGTAAGGTGGTGGTAACGTTG |
2.2. Measuring Aptamer Affinity with a Magnetic Beads-Based Isocratic Elution Assay
| Aptamer | KD (nM) |
|---|---|
| 1.12.2 | 370 ± 250 |
| A08 | 290 ± 150 |
| B08 | 110 ± 50 |
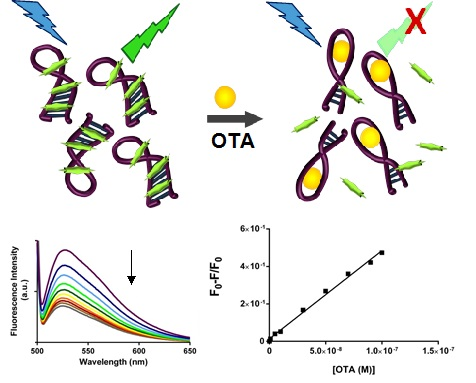
2.3. SYBR Green I Assay
2.3.1. Principle of Assay
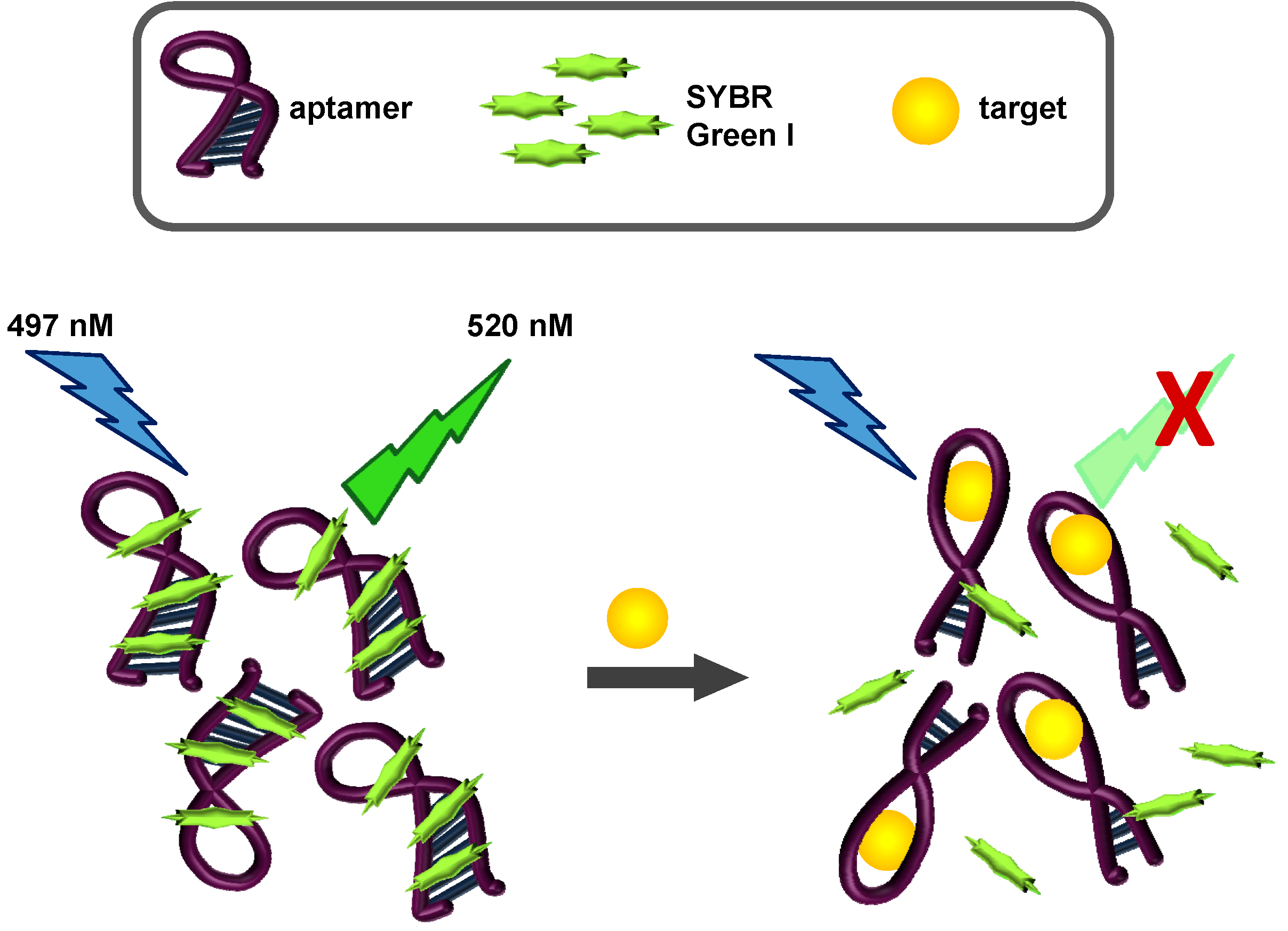
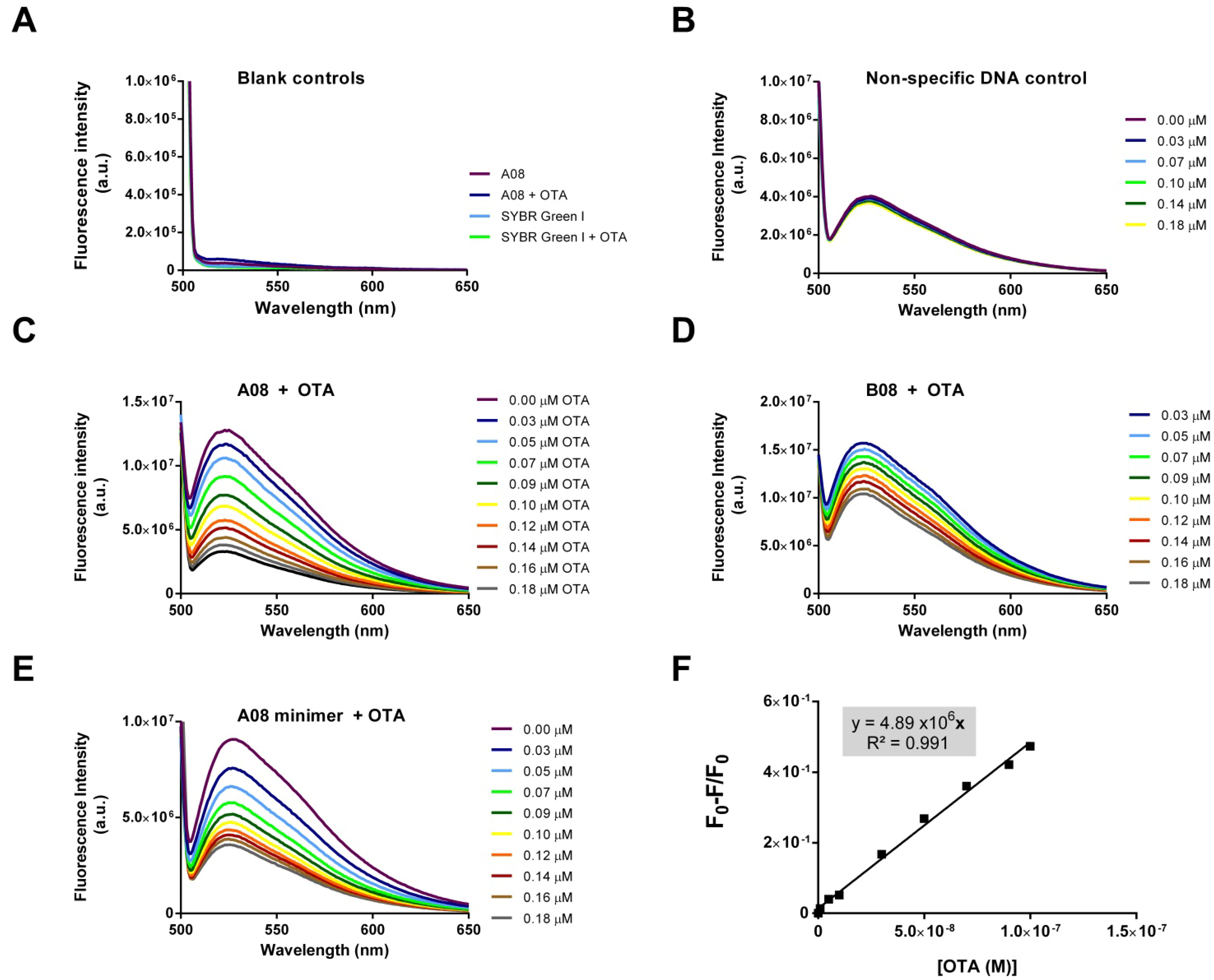
2.3.2. Biosensing of OTA

3. Experimental Section
3.1. Materials
3.2. OTA SELEX Experiments
3.2.1. Preparation of OTA Resin
3.2.2. SELEX Library
3.2.3. In Vitro Selection
3.3. Oligonucleotides
3.4. Characterization of OTA Aptamers
3.4.1. Aptamer Synthesis
| Aptamer | Sequence 5' to 3' |
|---|---|
| A08 | AGCCTCGTCTGTTCTCCCGGCAGTGTGGGCGAATCTATGCGTACCGTTCGATATCGTGGGGAAGACAAGCAGACGT |
| B08 | AGCCTCGTCTGTTCTCCCGGCGCATGATCATTCGGTGGGTAAGGTGGTGGTAACGTTGGGGAAGACAAGCAGACGT |
| A08 minimer | GGCAGTGTGGGCGAATCTATGCGTACCGTTCGATATCGTG |
| 1.12.2 | GATCGGGTGTGGGTGGCGTAAAGGGAGCATCGGACA |
| Non-specific control | AGCACGTTGGTAGGTCGGTTTGGGTTTCGTGC |
3.4.2. Magnetic Beads Binding Assay
3.4.2.1. Conjugating OTA to Magnetic Beads
3.4.2.2. Determining Dissociation Constants with a Magnetic Beads-Based Isocratic Elution Method
3.5. Biosensing of OTA with the SG Assay
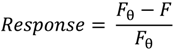
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van Der Merwe, K.J.; Steyn, P.S.; Fourie, L.; Scott, D.B.; Theron, J.J. Ochratoxin A, A toxic metabolite produced by aspergillus ochraceus wilh. Nature 1965, 205, 1112–1113. [Google Scholar]
- Duarte, S.C.; Lino, C.M.; Pena, A. Mycotoxin food and feed regulation and the specific case of ochratoxin A: A review of the worldwide status. Food Addit. Contam. Part AChem. Anal. Control Expo. Risk Assess. 2010, 27, 1440–1450. [Google Scholar] [CrossRef]
- Tittlemier, S.A.; Varga, E.; Scott, P.M.; Krska, R. Sampling of cereals and cereal-based foods for the determination of ochratoxin A: An overview. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2011, 28, 775–785. [Google Scholar] [CrossRef]
- Mayura, K.; Parker, R.; Berndt, W.O.; Phillips, T.D. Ochratoxin A-induced teratogenesis in rats: Partial protection by phenylalanine. Appl. Environ. Microbiol. 1984, 48, 1186–1188. [Google Scholar]
- Creppy, E.E.; Chiarappa, P.; Baudrimont, I.; Borracci, P.; Moukha, S.; Carratù, M.R. Synergistic effects of fumonisin B1 and ochratoxin A: Are In vitro cytotoxicity data predictive of In vivo acute toxicity? Toxicology 2004, 201, 115–123. [Google Scholar] [CrossRef]
- Pitout, M.J. The effect of ochratoxin A on glycogen storage in the rat liver. Toxicol. Appl. Pharmacol. 1968, 13, 299–306. [Google Scholar] [CrossRef]
- Creppy, E.E.; Stormer, F.C.; Roschenthaler, R.; Dirheimer, G. Effects of two metabolites of ochratoxin A, (4R)-4-hydroxyochratoxin A and ochratoxin alpha, on immune response in mice. Infect. Immun. 1983, 39, 1015–1018. [Google Scholar]
- Pfohl-Leszkowicz, A.; Manderville, R.A. Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 2007, 51, 61–99. [Google Scholar] [CrossRef]
- Creppy, E.E.; Lugnier, A.A.J.; Fasiolo, F.; Heller, K.; Röschenthaler, R.; Dirheimer, G. In vitro inhibition of yeast phenylalanyl-tRNA synthetase by ochratoxin A. Chem.-Biol. Interact. 1979, 24, 257–261. [Google Scholar] [CrossRef]
- European Union. Commission regulation (Ec) No. 123/2005 Of 26 January 2005 amending regulation (Ec) No. 466/2001 as regards ochratoxin A. Off. J. Eur. Union 2005, L25, 3–5. [Google Scholar]
- Entwisle, A.C.; Williams, A.C.; Mann, P.J.; Russell, J.; Slack, P.T.; Gilbert, J. Combined phenyl silane and immunoaffinity column cleanup with liquid chromatography for determination of ochratoxin A in roasted coffee: Collaborative study. J. AOAC Int. 2001, 84, 444–450. [Google Scholar]
- Visconti, A.; de Girolamo, A. Fitness for purpose—Ochratoxin A analytical developments. Food Addit. Contam. 2005, 22 (Suppl. 1), 37–44. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Cho, E.J.; Lee, J.W.; Ellington, A.D. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2009, 2, 241–264. [Google Scholar] [CrossRef]
- Mckeague, M.; Giamberardino, A.; DeRosa, M.C. Advances in aptamer-based biosensors for food safety. In Environmental Biosensors; Somerset, V., Ed.; Intech: Rijeka, Croatia, 2011; pp. 17–42. [Google Scholar]
- Mckeague, M.; DeRosa, M.C. Challenges and opportunities for small molecule aptamer development. J. Nucleic Acids 2012, 2012, 748913. [Google Scholar]
- Stead, S.L.; Ashwin, H.; Johnston, B.; Dallas, A.; Kazakov, S.A.; Tarbin, J.A.; Sharman, M.; Kay, J.; Keely, B.J. An RNA-aptamer-based assay for the detection and analysis of malachite green and leucomalachite green residues in fish tissue. Anal. Chem. 2010, 82, 2652–2660. [Google Scholar] [CrossRef]
- Cruz-Aguado, J.A.; Penner, G. Determination of ochratoxin A with a DNA aptamer. J. Agric. Food Chem. 2008, 56, 10456–10461. [Google Scholar] [CrossRef]
- Sheng, L.; Ren, J.; Miao, Y.; Wang, J.; Wang, E. Pvp-coated graphene oxide for selective determination of ochratoxin A via quenching fluorescence of free aptamer. Biosens. Bioelectron. 2011, 26, 3494–3499. [Google Scholar] [CrossRef]
- Huizenga, D.E.; Szostak, J.W. A DNA aptamer that binds adenosine and Atp. Biochemistry 1995, 34, 656–665. [Google Scholar] [CrossRef]
- Barthelmebs, L.; Jonca, J.; Hayat, A.; Prieto-Simon, B.; Marty, J.-L. Enzyme-linked aptamer assays (Elaas), based on a competition format for a rapid and sensitive detection of ochratoxin A in wine. Food Control 2011, 22, 737–743. [Google Scholar] [CrossRef]
- Silverman, S.K. Artificial functional nucleic acids: Aptamers, ribozymes, and deoxyribozymes identified by in vitro selection. Funct. Nucleic Acids Anal. Appl. 2009, 1, 47–108. [Google Scholar] [CrossRef]
- Rhouati, A.; Yang, C.; Hayat, A.; Marty, J.-L. Aptamers: A promosing tool for ochratoxin A detection in food analysis. Toxins 2013, 5, 1988–2008. [Google Scholar] [CrossRef]
- De Girolamo, A.; Mckeague, M.; Miller, J.D.; DeRosa, M.C.; Visconti, A. Determination of ochratoxin A in wheat after clean-up through a DNA aptamer-based solid phase extraction column. Food Chem. 2011, 127, 1378–1384. [Google Scholar] [CrossRef]
- Wang, L.; Chen, W.; Ma, W.; Liu, L.; Ma, W.; Zhao, Y.; Zhu, Y.; Xu, L.; Kuang, H.; Xu, C. Fluorescent strip sensor for rapid determination of toxins. Chem. Commun. 2011, 47, 1574–1576. [Google Scholar]
- Guo, Z.; Ren, J.; Wang, J.; Wang, E. Single-walled carbon nanotubes based quenching of free fam-aptamer for selective determination of ochratoxin A. Talanta 2011, 85, 2517–2521. [Google Scholar] [CrossRef]
- Chen, J.; Fang, Z.; Liu, J.; Zeng, L. A simple and rapid biosensor for ochratoxin A based on a structure-switching signaling aptamer. Food Control 2012, 25, 555–560. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Yang, G.; Chen, J.; Wang, S. A signal-on fluorescent aptasensor based on Tb3+ and structure-switching aptamer for label-free detection of ochratoxin A in wheat. Biosens. Bioelectron. 2013, 41, 704–709. [Google Scholar] [CrossRef]
- Zhao, Q.; Geng, X.; Wang, H. Fluorescent sensing ochratoxin A with single fluorophore-labeled aptamer. Anal. Bioanal. Chem. 2013, 405, 6281–6286. [Google Scholar] [CrossRef]
- Yang, C.; Lates, V.; Prieto-Simon, B.; Marty, J.L.; Yang, X. Aptamer-dnazyme hairpins for biosensing of ochratoxin A. Biosens. Bioelectron. 2012, 32, 208–212. [Google Scholar] [CrossRef]
- Yang, C.; Lates, V.; Prieto-Simon, B.; Marty, J.L.; Yang, X. Rapid high-throughput analysis of ochratoxin A by the self-assembly of dnazyme-aptamer conjugates in wine. Talanta 2013, 116, 520–526. [Google Scholar] [CrossRef]
- Wang, Z.; Duan, N.; Hun, X.; Wu, S. Electrochemiluminescent aptamer biosensor for the determination of ochratoxin A at a gold-nanoparticles-modified gold electrode using N-(aminobutyl)-N-ethylisoluminol as a luminescent label. Anal. Bioanal. Chem. 2010, 398, 2125–2132. [Google Scholar] [CrossRef]
- Kuang, H.; Chen, W.; Xu, D.; Xu, L.; Zhu, Y.; Liu, L.; Chu, H.; Peng, C.; Xu, C.; Zhu, S. Fabricated aptamer-based electrochemical “signal-off” sensor of ochratoxin A. Biosens. Bioelectron. 2010, 26, 710–716. [Google Scholar] [CrossRef]
- Prabhakar, N.; Matharu, Z.; Malhotra, B.D. Polyaniline langmuir–blodgett film based aptasensor for ochratoxin A detection. Biosens. Bioelectron. 2011, 26, 4006–4011. [Google Scholar] [CrossRef]
- Bonel, L.; Vidal, J.C.; Duato, P.; Castillo, J.R. An electrochemical competitive biosensor for ochratoxin A based on a DNA biotinylated aptamer. Biosens. Bioelectron. 2011, 26, 3254–3259. [Google Scholar] [CrossRef]
- Rhouati, A.; Hayat, A.; Hernandez, D.B.; Meraihi, Z.; Munoz, R.; Marty, J.-L. Development of an automated flow-based electrochemical aptasensor for on-line detection of ochratoxin A. Sens. Actuators B 2013, 176, 1160–1166. [Google Scholar] [CrossRef]
- Wu, J.; Chu, H.; Mei, Z.; Deng, Y.; Xue, F.; Zheng, L.; Chen, W. Ultrasensitive one-step rapid detection of ochratoxin A by the folding-based electrochemical aptasensor. Anal. Chim. Acta 2012, 753, 27–31. [Google Scholar] [CrossRef]
- Castillo, G.; Lamberti, I.; Mosiello, L.; Hianik, T. Impedimetric DNA aptasensor for sensitive detection of ochratoxin A in food. Electroanalysis 2012, 24, 512–520. [Google Scholar] [CrossRef]
- Tong, P.; Zhang, L.; Xu, J.J.; Chen, H.Y. Simply amplified electrochemical aptasensor of ochratoxin A based on exonuclease-catalyzed target recycling. Biosens. Bioelectron. 2011, 29, 97–101. [Google Scholar] [CrossRef]
- Tong, P.; Zhao, W.W.; Zhang, L.; Xu, J.J.; Chen, H.Y. Double-probe signal enhancing strategy for toxin aptasensing based on rolling circle amplification. Biosens. Bioelectron. 2012, 33, 146–151. [Google Scholar] [CrossRef]
- Hayat, A.; Sassolas, A.; Marty, J.L.; Radi, A.E. Highly sensitive ochratoxin A impedimetric aptasensor based on the immobilization of azido-aptamer onto electrografted binary film via click chemistry. Talanta 2013, 103, 14–19. [Google Scholar] [CrossRef]
- Zhu, Z.; Yang, C.; Zhou, X.; Qin, J. Label-free aptamer-based sensors for l-argininamide by using nucleic acid minor groove binding dyes. Chem. Commun. 2011, 47, 3192–3194. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, X.; Xie, Y.; Zhao, R.; Tan, C.; Jiang, Y. Label-free fluorescent assays based on aptamer-target recognition. Analyst 2012, 137, 2309–2312. [Google Scholar] [CrossRef]
- Babendure, J.R.; Adams, S.R.; Tsien, R.Y. Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc. 2003, 125, 14716–14717. [Google Scholar] [CrossRef]
- Jin, Y.; Bai, J.; Li, H. Label-free protein recognition using aptamer-based fluorescence assay. Analyst 2010, 135, 1731–1735. [Google Scholar] [CrossRef]
- Huang, C.C.; Chang, H.T. Aptamer-based fluorescence sensor for rapid detection of potassium ions in urine. Chem. Commun. 2008, 1461–1463. [Google Scholar] [CrossRef]
- Cruz-Toledo, J.; Mckeague, M.; Zhang, X.; Giamberardino, A.; Mcconnell, E.; Francis, T.; Derosa, M.C.; Dumontier, M. Aptamer base: A collaborative knowledge base to describe aptamers and selex experiments. Database: J. Biol. Databases Curation 2012, 2012, Bas006. [Google Scholar]
- Tok, J.B.; Fischer, N.O. Single microbead selex for efficient ssdna aptamer generation against botulinum neurotoxin. Chem. Commun. 2008, 1883–1885. [Google Scholar]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef]
- Mathews, D.H.; Disney, M.D.; Childs, J.L.; Schroeder, S.J.; Zuker, M.; Turner, D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7287–7292. [Google Scholar] [CrossRef]
- Luo, X.; Mckeague, M.; Pitre, S.; Dumontier, M.; Green, J.; Golshani, A.; DeRosa, M.C.; Dehne, F. Computational approaches toward the design of pools for the in vitro selection of complex aptamers. RNA (N. Y.) 2010, 16, 2252–2262. [Google Scholar] [CrossRef]
- Kikin, O.; D’antonio, L.; Bagga, P.S. Qgrs mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef]
- Mergny, J.L.; Phan, A.T.; Lacroix, L. Following G-quartet formation by Uv-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef]
- Darty, K.; Denise, A.; Ponty, Y. Varna: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef]
- Mckeague, M.; Bradley, C.R.; de Girolamo, A.; Visconti, A.; Miller, J.D.; DeRosa, M.C. Screening and initial binding assessment of fumonisin B1 aptamers. Int. J. Mol. Sci. 2010, 11, 4864–4881. [Google Scholar] [CrossRef]
- Mckeague, M.; Foster, A.; Miguel, Y.; Giamberardino, A.; Verdin, C.; Chan, J.Y.S.; DeRosa, M.C. Development of a DNA aptamer for direct and selective homocysteine detection in human serum. RSC Adv. 2013, 3, 24415–24422. [Google Scholar]
- Chang, A.L.; Mckeague, M.; Liang, J.C.; Smolke, C.D. Kinetic and equilibrium binding characterization of aptamers to small molecules using a label-free, sensitive, and scalable platform. Anal. Chem. 2014, 86, 3273–3278. [Google Scholar]
- Carothers, J.M.; Goler, J.A.; Kapoor, Y.; Lara, L.; Keasling, J.D. Selecting RNA aptamers for synthetic biology: Investigating magnesium dependence and predicting binding affinity. Nucleic Acids Res. 2010, 38, 2736–2747. [Google Scholar] [CrossRef]
- Zipper, H.; Brunner, H.; Bernhagen, J.; Vitzthum, F. Investigations on DNA intercalation and surface binding by sybr green I, its structure determination and methodological implications. Nucleic Acids Res. 2004, 32, E103. [Google Scholar] [CrossRef]
- Schmidt, D.M.; Ernst, J.D. A fluorometric assay for the quantification of RNA in solution with nanogram sensitivity. Anal. Biochem. 1995, 232, 144–146. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Herrmann, M.G.; Gundry, C.N.; Elenitoba-Johnson, K.S. Real-time multiplex pcr assays. Methods 2001, 25, 430–442. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Ririe, K.M.; Andrew, R.V.; David, D.A.; Gundry, R.A.; Balis, U.J. The lightcycler: A microvolume multisample fluorimeter with rapid temperature control. Biotechniques 1997, 22, 176–181. [Google Scholar]
- Suzuki, T.; Fujikura, K.; Higashiyama, T.; Takata, K. DNA staining for fluorescence and laser confocal microscopy. J. Histochem. Cytochem.: Off. J. Histochem. Soc. 1997, 45, 49–53. [Google Scholar] [CrossRef]
- Sarpong, K.; Datta, B. Nucleic-acid-binding chromophores as efficient indicators of aptamer-target interactions. J. Nucleic Acids 2012, 2012, 247–280. [Google Scholar]
- Kong, L.; Xu, J.; Xu, Y.; Xiang, Y.; Yuan, R.; Chai, Y. A universal and label-free aptasensor for fluorescent detection of ATP and thrombin based on SYBR green I dye. Biosens. Bioelectron. 2013, 42, 193–197. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, J.; Xiang, Y.; Yuan, R.; Chai, Y. Target-induced structure switching of hairpin aptamers for label-free and sensitive fluorescent detection of ATP via exonuclease-catalyzed target recycling amplification. Biosens. Bioelectron. 2014, 51, 293–296. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev./Aust. Assoc. Clin. Biochem. 2008, 29 (Suppl. 1), S49–S52. [Google Scholar]
- Carothers, J.M.; Oestreich, S.C.; Szostak, J.W. Aptamers selected for higher-affinity binding are not more specific for the target ligand. J. Am. Chem. Soc. 2006, 128, 7929–7937. [Google Scholar] [CrossRef]
- Murphy, M.B.; Fuller, S.T.; Richardson, P.M.; Doyle, S.A. An improved method for the in vitro evolution of aptamers and applications in protein detection and purification. Nucleic Acids Res. 2003, 31, E110. [Google Scholar] [CrossRef]
- Rosenthal, A.; Coutelle, O.; Craxton, M. Large-scale production of DNA sequencing templates by microtitre format Pcr. Nucleic Acids Res. 1993, 21, 173–174. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
McKeague, M.; Velu, R.; Hill, K.; Bardóczy, V.; Mészáros, T.; DeRosa, M.C. Selection and Characterization of a Novel DNA Aptamer for Label-Free Fluorescence Biosensing of Ochratoxin A. Toxins 2014, 6, 2435-2452. https://doi.org/10.3390/toxins6082435
McKeague M, Velu R, Hill K, Bardóczy V, Mészáros T, DeRosa MC. Selection and Characterization of a Novel DNA Aptamer for Label-Free Fluorescence Biosensing of Ochratoxin A. Toxins. 2014; 6(8):2435-2452. https://doi.org/10.3390/toxins6082435
Chicago/Turabian StyleMcKeague, Maureen, Ranganathan Velu, Kayla Hill, Viola Bardóczy, Tamás Mészáros, and Maria C. DeRosa. 2014. "Selection and Characterization of a Novel DNA Aptamer for Label-Free Fluorescence Biosensing of Ochratoxin A" Toxins 6, no. 8: 2435-2452. https://doi.org/10.3390/toxins6082435
APA StyleMcKeague, M., Velu, R., Hill, K., Bardóczy, V., Mészáros, T., & DeRosa, M. C. (2014). Selection and Characterization of a Novel DNA Aptamer for Label-Free Fluorescence Biosensing of Ochratoxin A. Toxins, 6(8), 2435-2452. https://doi.org/10.3390/toxins6082435