Deletion and Gene Expression Analyses Define the Paxilline Biosynthetic Gene Cluster in Penicillium paxilli
Abstract
:1. Introduction

2. Results and Discussion
| Gene/ ORF | Exon # | Predicted product size (aa) | Predicted function | Top BlastP hit | ||||
|---|---|---|---|---|---|---|---|---|
| Description | InterProScan | Protein | Organism | E-value (%ID) | Accession/Ref | |||
| PP101 | 2 | 402 | FAD-dependent oxidoreductase | IPR002938 IPR003042 | Pc16g05940 | P. chrysogenum | 0.0 (78) | CAP93264 |
| PP102 | 3 | 508 | β-1,3- glucanosyltransferase | IPR004886 IPR012946 IPR013781 IPR013781 | A. oryzae | 0.0 (66) | BAE66482 | |
| PP103 | 3 | 103 | Unknown | IPR018809 | P. digitatum | 3 × 10−47 (74) | EKV06610 | |
| PP104 | 2 | 477 | Zn(II)2Cys6 transcription factor | IPR001138 | N. fischeri | 3 × 10−81 (34) | EAW19936 | |
| PP105 | 7 | 684 | Zn(II)2Cys6 transcription factor | IPR001138 IPR007219 | A. oryzae | 0.0 (51) | XP_003189175 | |
| PP106 | 1 | 345 | Unknown | IPR011042 | Pc16g00180 | P. chrysogenum | 5 × 10−164 (66) | CAP92688 |
| PP107 | 4 | 385 | NADH oxidoreductase | IPR001327 IPR013027 IPR023753 | Pc16g00160 | P. chrysogenum | 0.0 (65) | CAP92686 |
| PP108 | 1 | 543 | Capsule associated protein | IPR006598 | CAP1 | Metarhizium anisopliae | 0.0 (55) | EFY96463 |
| PP109 | 1 | 175 | Acetyltransferase | IPR000182 IPR016181 | M. anisopliae | 3 × 10−80 (68) | EFY95041 | |
| PP110 | 3 | 818 | Unknown | No hits | A. niger | 0.0 (44) | EHA28514 | |
| PP111 | 3 | 478 | Transporter (MFS) | IPR005828 IPR016196 IPR020846 | A. oryzae | 0.0 (75) | BAE63453 | |
| PP112 | 1 | 291 | DUF829-Conserved protein family of unknown function | IPR008547 | Pc13g04190 | P. chrysogenum | 5 × 10−158 (71) | CAP91488 |
| paxG (PP113) | 4 | 371 | Geranylgeranyl diphosphate synthase | IPR000092 IPR008949 IPR017446 | Pc20g01860 | P. chrysogenum | 1 × 10-162 (64) | CAP85515 |
| paxA (PP114) | 2 | 356 | Integral membrane protein | No hits | AtmA | A. flavus | 4 × 10−462 (33) | CAP53940/ [14] |
| paxM (PP115) | 3 | 477 | FAD-dependent monooxygenase | IPR002938 IPR003042 | Pc20g01850 | P. chrysogenum | 0.0 (60) | CAP85514 |
| paxB (PP116) | 2 | 243 | Integral membrane protein | No hits | AtmB | A. flavus | 1 × 10−103 (62) | CAP53939/ [14] |
| paxC (PP117) | 3 | 317 | Prenyl transferase | IPR000092 IPR008949 IPR017446 | Pc20g01840 | P. chrysogenum | 3 × 10−162 (69) | CAP85513 |
| paxP (PP118) | 6 | 515 | Cytochrome P450 monooxygenase | IPR001128 IPR002403 | A. oryzae | 0.0 (64) | EIT78616 | |
| paxQ (PP119) | 10 | 512 | Cytochrome P450 monooxygenase | IPR001128 IPR002401 IPR017972 | AtmQ | A. flavus | 0.0 (60) | CAP53938/ [14] |
| paxD (PP120) | 2 | 438 | Indole dimethylallyl transferase | IPR012148 IPR017795 | AtmD | A. flavus | 2 × 10−74 (35) 5 × 10−74 (35) | EED52847 CAP53937/ [13,14] |
| PP121 | 4 | 418 | FAD-binding oxidoreductase | IPR006094 IPR016166 IPR016167 IPR016169 | W97_07461 | Coniosporium apollinis | 9 × 10−88 (34) | EON68203 |
| PP122 | 3 | 306 | Arabinase/Xylanase | IPR006710 IPR023296 | Pc12g01330 | P. chrysogenum | 2 × 10−42 (38) | CAP79760 |
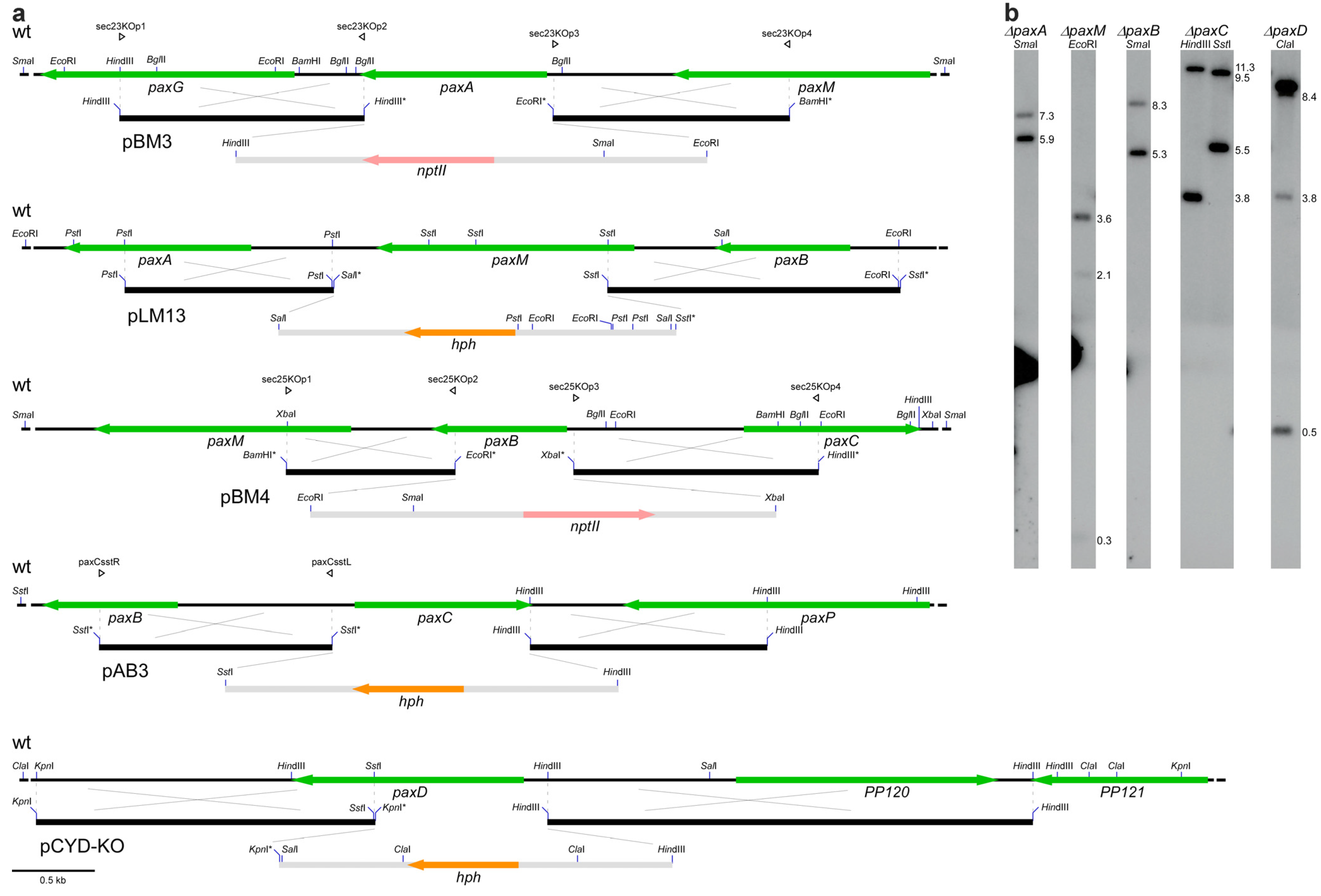
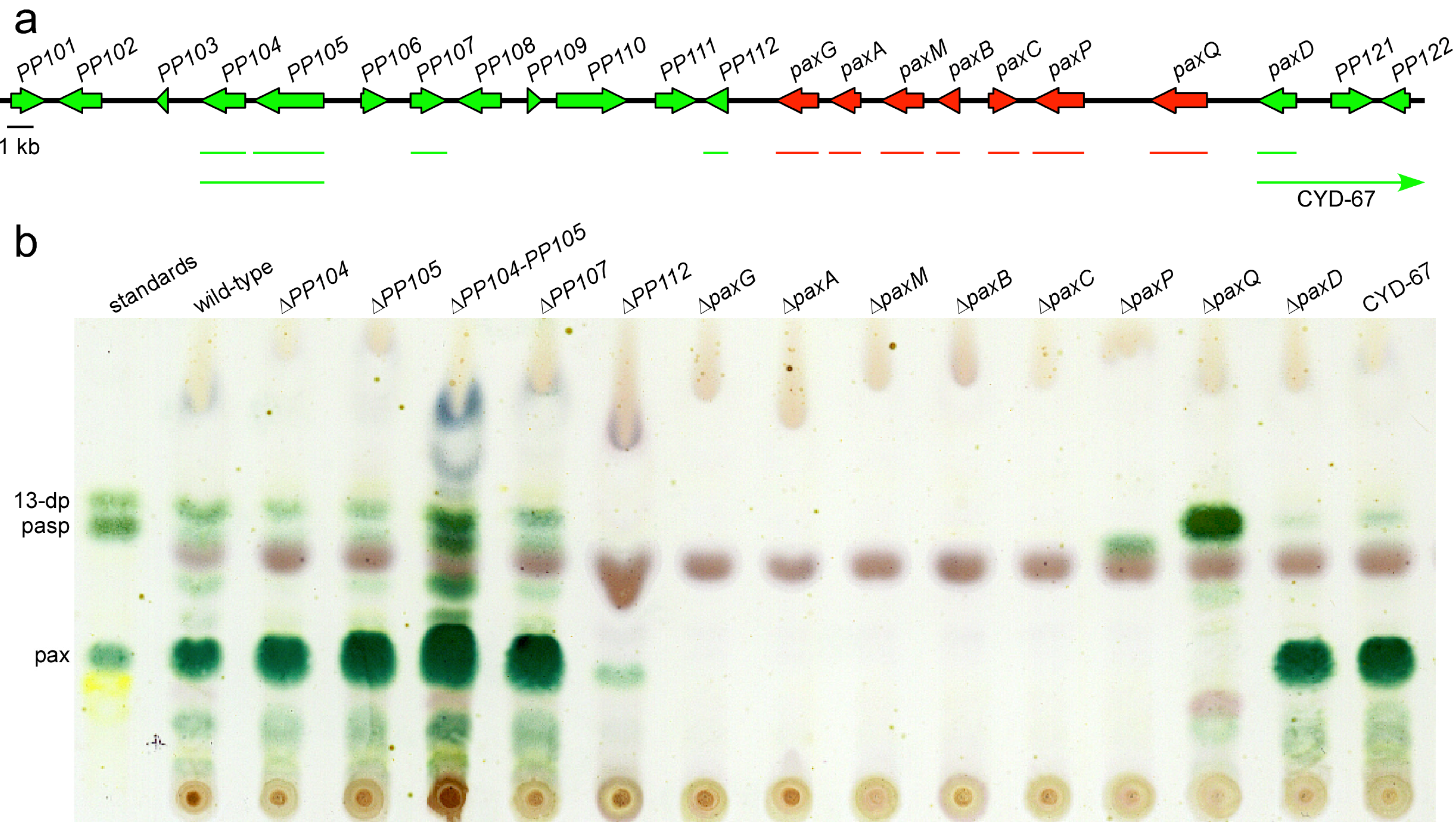
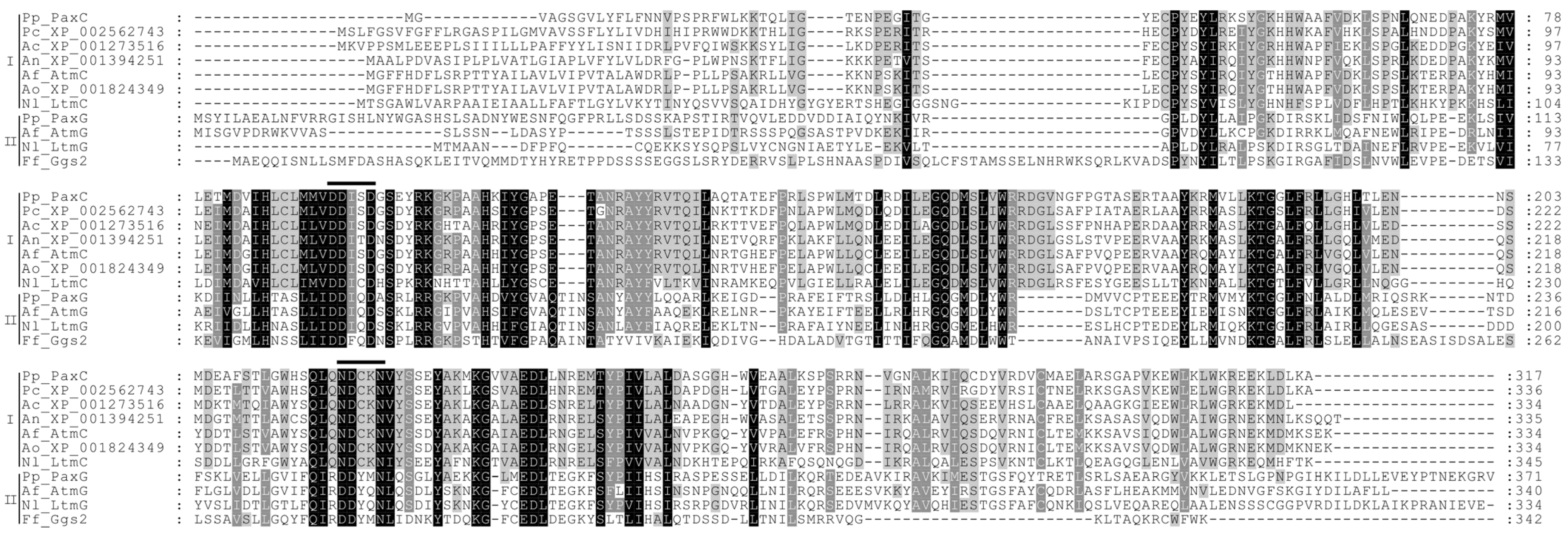


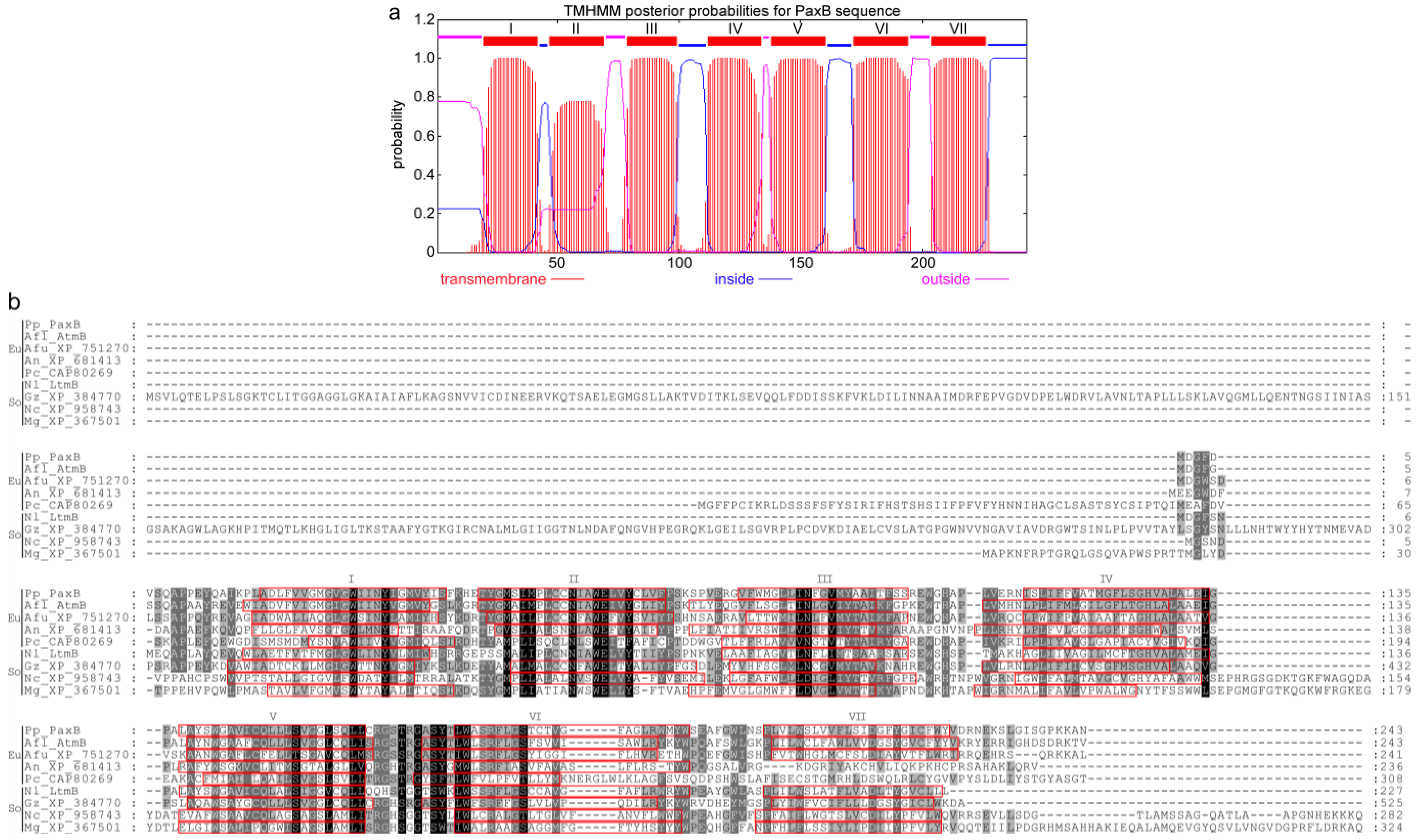
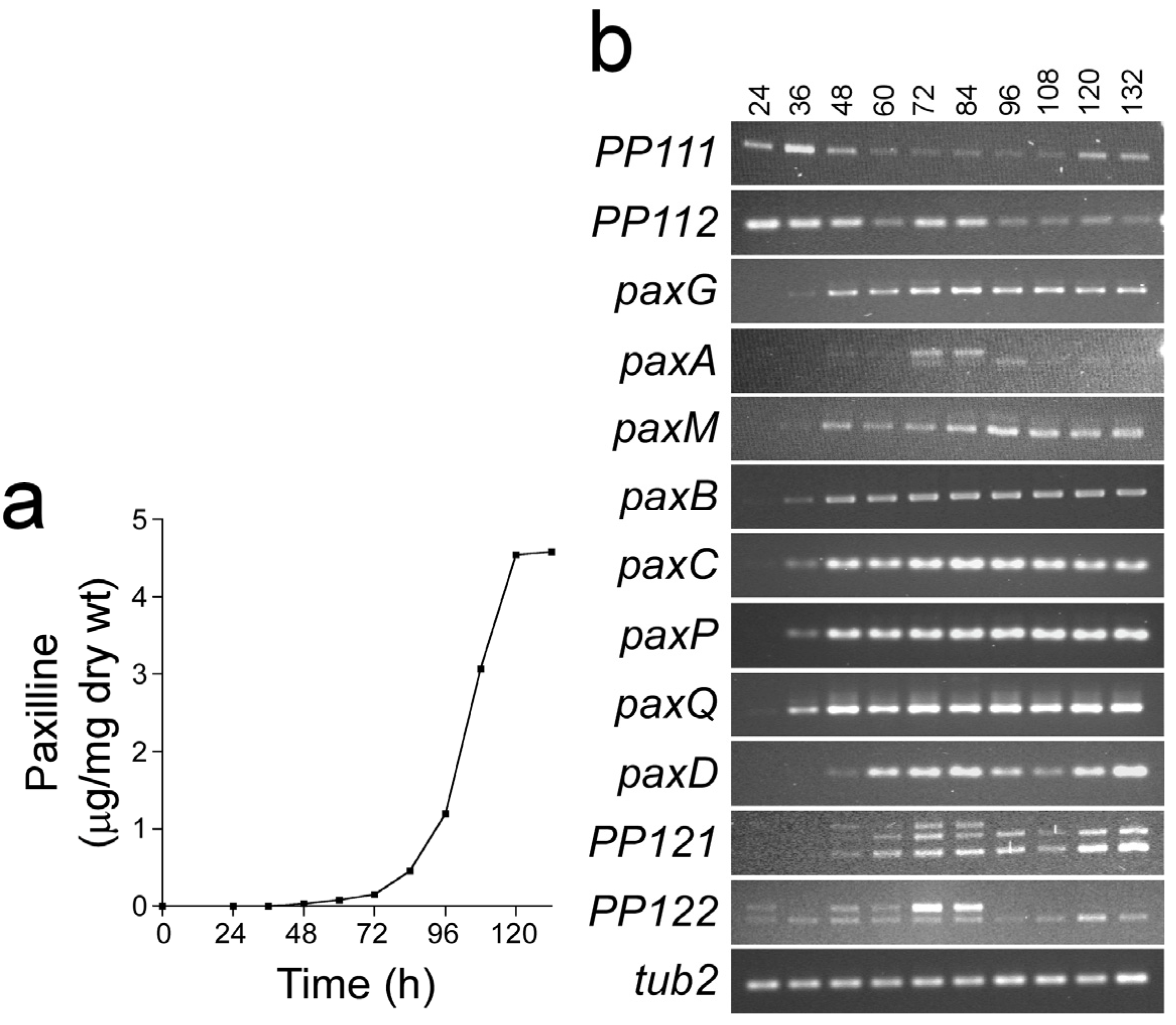

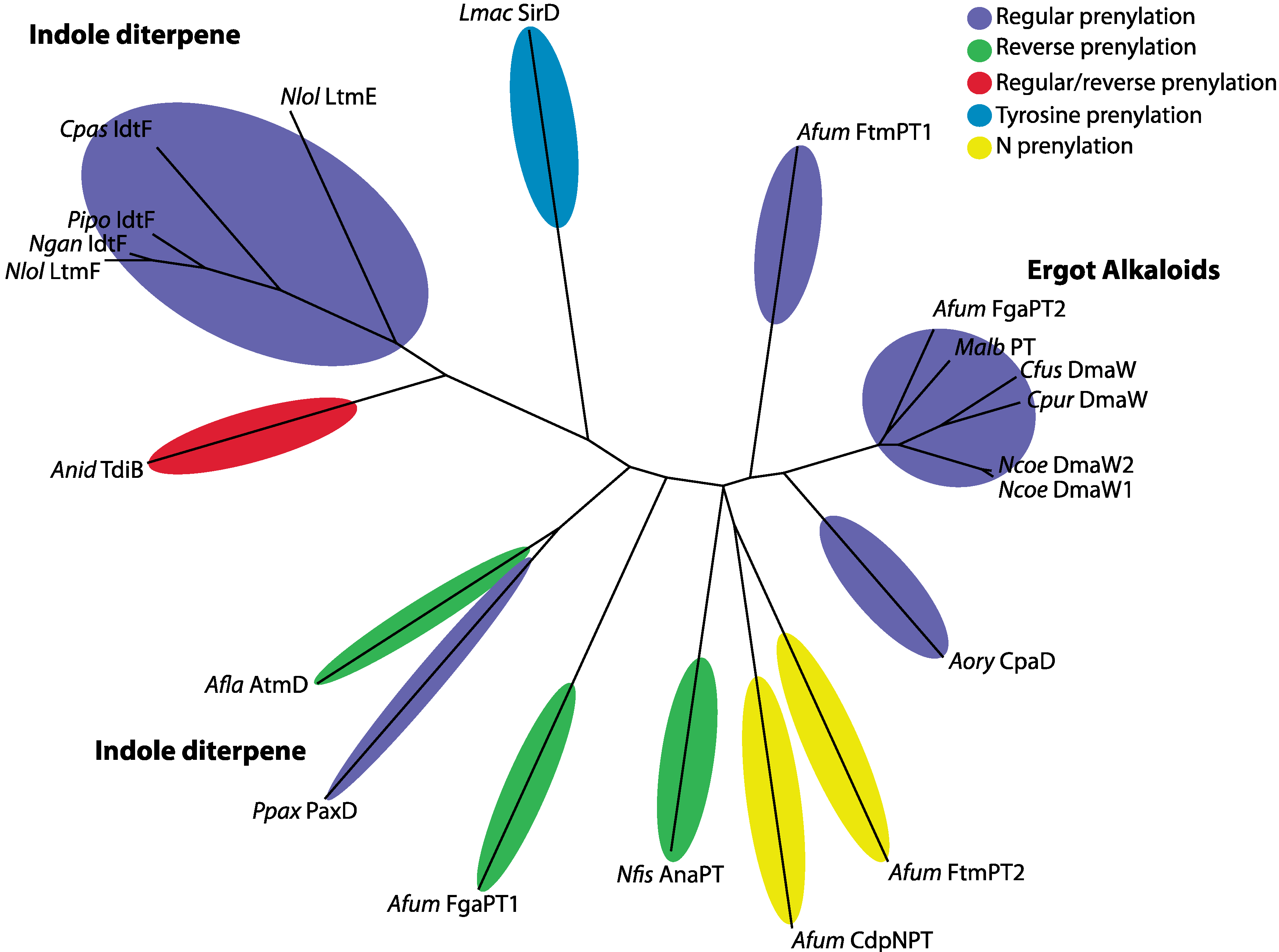
3. Experimental Section
3.1. Bacterial Strains and Plasmids
| Biological material | Targeted gene | Strain | Relevant characteristics | Reference |
|---|---|---|---|---|
| Fungal strains | ||||
| Penicillium paxilli | ||||
| PN2013 | Wild-type; paxilline positive | [7] | ||
| PN2262 | PP104 | LMS-218 | PN2013/∆PP104::P trpC-hph; HygR; paxilline positive | This study |
| PN2254 | PP105 | LMR-36 | PN2013/∆PP105::P trpC-hph; HygR; paxilline positive | This study |
| PN2263 | PP104-PP105 | LMRS-173 | PN2013/∆PP104, ∆PP105::PtrpC-hph; HygR; paxilline positive | This study |
| PN2434 | PP107 | ABH-17 | PN2013/∆PP107::P trpC-nptII-TtrpC; GenR; paxilline positive | This study |
| PN2456 | PP112 | BMU-13 | PN2013/∆PP112::P trpC-nptII-TtrpC; GenR; paxilline positive | This study |
| PN2255 | paxG | LMG-23 | PN2013/∆ paxG::PtrpC-hph; HygR; paxilline negative | [8] |
| PN2457 | paxA | sec23-22 | PN2013/∆ paxA::PtrpC-nptII-TtrpC; GenR; paxilline negative | This study |
| PN2257 | paxM | LMM-100 | PN2013/∆ paxM::PglcA-hph-trpC; HygR; paxilline negative | This study |
| PN2458 | paxB | sec25-2 | PN2013/∆ paxB::PtrpC-nptII-trpC; GenR; paxilline negative | This study |
| PN2290 | paxC | ABC-83 | PN2013/∆ paxC::PtrpC-hph; HygR; paxilline negative | This study |
| PN2258 | paxP | LMP-1 | PN2013/∆ paxP::PtrpC-hph; HygR; paspaline positive | [9] |
| PN2259 | paxQ | LMQ-226 | PN2013/∆ paxQ::PtrpC-hph; HygR; 13-desoxypaxilline positive | [9] |
| PN2260 | paxD | CYD-162 | PN2013/∆ paxD::PtrpC-hph; HygR; paxilline positive | This study |
| PN2261 | paxD-PP122+ | CYD-67 | PN2013/∆ paxD::PtrpC-hph; HygR; paxilline positive; extended deletion | This study |
| Plasmids | ||||
| pLM14 (pLMS-KO) | PP104 | pUChph hygromycin resistance | This study | |
| pLM15 (pLMR-KO) | PP105 | pUChph hygromycin resistance | This study | |
| pLM16 (pLMRS-KO) | PP104-PP105 | pUChph hygromycin resistance | This study | |
| pAB5 (pABH-KO) | PP107 | pII99 geneticin resistance | This study | |
| pBM2 (ppaxU-KO) | PP112 | PN1942 | pII99 geneticin resistance | This study |
| pBM3 (psec23-KO) | paxA | PN1944 | pII99 geneticin resistance | This study |
| pLM13 (pLMM-KO) | paxM | PN1659 | pCWHyg1 hygromycin resistance | This study |
| pBM4 (psec25-KO) | paxB | PN1946 | pII99 geneticin resistance | This study |
| pAB3 (ppaxC-KO) | paxC | pUChph hygromycin resistance | This study | |
| pCY1 (pCYD-KO) | paxD | PN1642 | pCWHyg1 hygromycin resistance | This study |
3.2. Fungal Strains and Growth Conditions
3.3. Molecular Biology
3.4. Preparation of Deletion Constructs
3.5. Penicillium paxilli Transformation and Screening
3.6. Indole-Diterpene Analysis
3.7. Bioinformatic Analyses
4. Conclusions
Acknowledgments
Conflict of Interest
Appendix
| Species | Isolate | Prenylation Type | Position | Protein | Compound | Reference |
|---|---|---|---|---|---|---|
| Aspergillus flavus | NRRL 6541 | reverse | C4 | AtmD | aflatrem (IDT) | [14] |
| Aspergillus fumigatus | Af293 | regular | C2 | FtmPT1 | Brevianamide F | [51] |
| Aspergillus fumigatus | Af293 | reverse | C2 | FgaPT1 | fumigaclavine C | [35] |
| Aspergillus fumigatus | Af293 | regular | C4 | FgaPT2 | fumigaclavine C | [35] |
| Aspergillus fumigatus | Af293 | N-PT-reverse | N1 | CdpNPT | cyclic dipeptide N-prenylated | [52,53] |
| Aspergillus fumigatus | Af293 | N-PT-regular | N1 | FtmPT2 | fumitremorgin B | [54,55] |
| Aspergillus nidulans | Un-specified | regular and reverse | Reg = quinone; Rev = C2 | TdiB | terrequinone A | [56] |
| Aspergillus oryzae | RIB 40 | regular | C4 | CpaD | cyclopiazonic acid | [33,36] |
| Claviceps fusiformis | ATCC26245 | regular | C4 | DmaW | ergot alkaloid | [32] |
| Claviceps paspali | RRC-1481 | regular | C5 | IdtF | paspalitrem A (IDT) | [25] |
| Claviceps purpurea | P1 | regular | C4 | DmaW | ergot alkaloid | [30] |
| Leptosphaeria maculans | ICBN 18 | Tyrosine-regular | O | SirD | sirodesmin | [57] |
| Malbranchea aurantiaca | RRC1813 | regular | C4 | MaPT | ergot alkaloid | [58] |
| Neosartorya fischeri | NRRL 181 | reverse | C3 | AnaPT | acetylaszonalenin | [59] |
| Neotyphodium coenophialum | ATCC90664 | regular | C4 | DmaW | ergot alkaloid | [60] |
| Neotyphodium gansuense | e7080 | regular | IdtF | [25] | ||
| Neotyphodium lolii | Lp19 | regular | ggpp moiety | LtmF | lolitrem B (IDT) | [61] |
| Neotyphodium lolii | Lp19 | regular | C4, C5 | LtmE | lolitrem B (IDT) | [61] |
| Penicillium paxilli | ATCC26601 | regular (di) | C5, C6 | PaxD | Indole-diterpene (IDT) | [8] |
| Periglandula ipomoeae | IasaF13 | regular | ggpp moiety | IdtF | terpendole K (IDT) | [25] |
| Gene | Primer 1 | Sequence (5'–3') a | Primer 2 | Sequence (5'–3') | Size (kb) | Application |
|---|---|---|---|---|---|---|
| PP104 | paxU1SstIL | CTTGTTGGCgaGCTcCATATGAC | pax66 | CGCGATGGCGTACTGTAGAC | 1.03 | KO construct |
| PP104 | paxRU1HindIIIL | TTTAGTAGAAGCTTGGCC | paxRU1HindIIIR | TCGTTGAAGcTTGCAGTA | 1.16 | KO construct |
| PP105 | paxRBamL | ATTGACGgATCCCGATTATC | paxRBamR | GGATccGAGATGGGTGTATAC | 1.1 | KO construct |
| PP105 | KORH | GGGGTATAaGcTTAACATAGAGCAG | KORHS | GTTACATGCTTCCATTTAAAGTTGGGAGCTGTC | 1.1 | KO construct |
| PP104 & PP105 | paxRU1BamR | CAACGTTGTGGATCCATTCGG | paxRU1BamL | CCCTATCGGGATGCAATTTTCAAAC | 1.1 | KO construct |
| PP107 | pax251Hind | TGCAAGCTTCCGCTATAG | pax186 | AGTCAACACCAAGACAGG | 1.27 | KO construct |
| PP107 | pax175 | TCGACGACTTCGACCAGA | pax183 | ATGTCATCTTCCGCAATC | 1.5 | KO construct |
| PP112 | paxUKOp3 | ACGTTGCTAGTcTAGaTGGAAGC | paxUKOp4 | AGTTCGTaAGCTTGATGTGTTG | 1.5 | KO construct |
| PP112 | paxUKOp1 | GTGATGGATcCCAAAATTCATTGG | paxUKOp2 | GCAAGAaTTCAAATGCCTGGAAG | 1.6 | KO construct |
| paxA | sec23KOp3 | TGGCCGAaTTCCGAGAATAGAGT | sec23KOp4 | AAGAAATACGTGgATCCTGACAG | 1.4 | KO construct |
| paxA | sec23KOp2 | GCTAAAGcTtAACAACTGGACCA | sec23KOp1 | CTCGACAAGcTTAGAAAAGTCAC | 1.5 | KO construct |
| paxB | sec25KOp3/ | CGTTGAATAGCTcTAGATTGAAGG | sec25KOp4 | TGAGCCAAgcTTTGTGTAACTCG | 1.0 | KO construct |
| paxB | sec25KOp1/ | GTGATgGATCCCAAAATTCATTGG | sec25KOp2 | GCAAGaATTCAAATGCCTGGAAG | 1.5 | KO construct |
| paxC | paxCSstR | GTTGAGCTCAATCCACCAACGC | paxCSstL | GAGTGAGCTCTGCTTGGTAGGC | 1.4 | KO construct |
| PP112 | paxU RT-F | TCGTCCTATCTCGCACCTTTC | paxU RT-R | AGAGTCTGTTCGGTTCGATGG | 474 | RT-PCR |
| paxG | paxG RT-F | ACACTGCATCCCTTCTTATCG | paxG RT-R | TATCGAGAAGCTCGGAGCTCT | 528 | RT-PCR |
| paxA | paxA RT-F | CAACCTTTCAGGGTGAGATTC | paxA RT-R | CAGATGAGCAAGCCAAGGCAA | 489 | RT-PCR |
| paxM | paxM RT-F | TCATCGATCAAAGGTTCGGTT | paxM RT-R | AACTCGACCGTAAGCTTGGAA | 301 | RT-PCR |
| paxB | paxB RT-F | GAACTGGTCTACTGTCTGGTC | paxB RT-R | ACGGTCGACGTACCAGAAACA | 504 | RT-PCR |
| paxC | paxC RT-F | ATGATGGTCGACGATATCTCC | paxC RT-R | CAATTGCGAATGCCAGCCAAG | 385 | RT-PCR |
| paxP | paxP RT-F | CCACCTACAAGACCAATGTCA | paxP RT-R | AAGCGAATTGATCATCGCATG | 417 | RT-PCR |
| paxQ | paxQ RT-F | CAGCCTTACAGAGAGATTCGT | paxQ RT-R | GATGTGCGACAACTCTTGCAC | 562 | RT-PCR |
| paxD | paxD RT-F | CAGTCTGGAGCTTATGCCATC | paxD RT-R | CGTCCTTGACGAATGCCTTGA | 456 | RT-PCR |
| PP121 | paxO RT-F | GTGGCTGCTACTAAGCTGGTA | paxO RT-R | CACAGGAAGAAGCGATCTGGT | 456 | RT-PCR |
| PP122 | Pax248 | AGTTCGACAGCGCTTGGGAGA | Pax249 | CAGTGGCTCCTTAACTCTCGT | 592 | RT-PCR |
| tubA | Tub2 RT-F | ACACTCCTGATCTCCAAGATC | Tub2 RT-R | GATGTCGTACAGAGCCTCGTT | 258 | RT-PCR |
References
- Steyn, P.S.; Vleggaar, R. Tremorgenic mycotoxins. Prog. Chem. Org. Nat. Prod. 1985, 48, 1–80. [Google Scholar] [CrossRef]
- Byrne, K.M.; Smith, S.K.; Ondeyka, J.G. Biosynthesis of nodulisporic acid A: Precursor studies. J. Am. Chem. Soc. 2002, 124, 7055–7060. [Google Scholar] [CrossRef]
- De Jesus, A.E.; Gorst-Allman, C.P.; Steyn, P.S.; van Heerden, F.R.; Vleggar, R.; Wessels, P.L.; Hull, W.E. Tremorgenic mycotoxins from Penicillium crustosum. Biosynthesis of Penitrem A. J. Chem. Soc. Perkin Trans. 1983, 1863–1868. [Google Scholar]
- Saikia, S.; Parker, E.J.; Koulman, A.; Scott, B. Four gene products are required for the fungal synthesis of the indole diterpene paspaline. FEBS Lett. 2006, 580, 1625–1630. [Google Scholar] [CrossRef]
- Parker, E.J.; Scott, D.B. Indole-diterpene Biosynthesis in Ascomycetous Fungi. In Handbook of Industrial Mycology; An, Z., Ed.; Marcel Dekker: New York, NY, USA, 2004; Chapter 14; pp. 405–426. [Google Scholar]
- Young, C.; Itoh, Y.; Johnson, R.; Garthwaite, I.; Miles, C.O.; Munday-Finch, S.C.; Scott, B. Paxilline-negative mutants of Penicillium paxilli generated by heterologous and homologous plasmid integration. Curr. Genet. 1998, 33, 368–377. [Google Scholar] [CrossRef]
- Itoh, Y.; Johnson, R.; Scott, B. Integrative transformation of the mycotoxin-producing fungus, Penicillium paxilli. Curr. Genet. 1994, 25, 508–513. [Google Scholar] [CrossRef]
- Young, C.; McMillan, L.; Telfer, E.; Scott, B. Molecular cloning and genetic analysis of an indole-diterpene gene cluster from Penicillium paxilli. Mol. Microbiol. 2001, 39, 754–764. [Google Scholar] [CrossRef]
- McMillan, L.K.; Carr, R.L.; Young, C.A.; Astin, J.W.; Lowe, R.G.T.; Parker, E.J.; Jameson, G.B.; Finch, S.C.; Miles, C.O.; McManus, O.B.; et al. Molecular analysis of two cytochrome P450 monooxygenase genes required for paxilline biosynthesis in Penicillium paxilli, and effects of paxilline intermediates on mammalian maxi-K ion channels. Mol. Gen. Genomics 2003, 270, 9–23. [Google Scholar] [CrossRef]
- Saikia, S.; Parker, E.J.; Koulman, A.; Scott, B. Defining paxilline biosynthesis in Penicillium paxilli: Functional characterization of two cytochrome P450 monooxygenases. J. Biol. Chem. 2007, 282, 16829–16837. [Google Scholar] [CrossRef]
- Saikia, S.; Scott, B. Functional analysis and subcellular localization of two geranylgeranyl diphosphate synthases from Penicillium paxilli. Mol. Genet. Genomics 2009, 282, 257–271. [Google Scholar] [CrossRef]
- Tagami, K.; Liu, C.; Minami, A.; Noike, M.; Isaka, T.; Fueki, S.; Shichijo, Y.; Toshima, H.; Gomi, K.; Dairi, T.; et al. Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae. J. Am. Chem. Soc. 2013, 135, 1260–1263. [Google Scholar] [CrossRef]
- Liu, C.; Noike, M.; Minami, A.; Oikawa, H.; Dairi, T. Functional analysis of a prenyltransferase gene (paxD) in the paxilline biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 2013. [Google Scholar] [CrossRef]
- Nicholson, M.J.; Koulman, A.; Monahan, B.J.; Pritchard, B.L.; Payne, G.A.; Scott, B. Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 2009, 75, 7469–7481. [Google Scholar] [CrossRef]
- Chen, A.; Kroon, P.A.; Poulter, C.D. Isoprenyl diphosphate synthases: Protein sequence comparisons, a phylogenetic tree, and predictions of secondary structure. Protein Sci. 1994, 3, 600–607. [Google Scholar] [CrossRef]
- Wierenga, R.K.; Terpstra, P.; Hol, W.G.J. Predictions of the occurrence of the ADP-binding bab-fold in proteins using an amino acid sequence fingerprint. J. Mol. Biol. 1986, 187, 101–107. [Google Scholar] [CrossRef]
- Vallon, O. New sequence motifs in flavoproteins: Evidence for common ancestry and tools to predict structure. Proteins Struct. Funct. Genet. 2000, 38, 95–114. [Google Scholar] [CrossRef]
- Eggink, G.; Engel, H.; Vriend, G.; Terpstra, P.; Witholt, B. Rubredoxin reductase of Pseudomonas oleovorans: Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J. Mol. Biol. 1990, 212, 135–142. [Google Scholar] [CrossRef]
- You, I.-S.; Ghosal, D.; Gunsalus, I.C. Nucleotide sequence analysis of the Pseudomonas putida PpG7 salicylate hydroxylase gene (nahG) and its 3'-flanking region. Biochemistry 1991, 30, 1635–1641. [Google Scholar] [CrossRef]
- Marin, E.; Nussaume, L.; Quesada, A.; Gonneau, M.; Sotta, B.; Hugueney, P.; Frey, A.; Marion-Poll, A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996, 15, 2331–2342. [Google Scholar]
- Nowrousian, M. A novel polyketide biosynthesis gene cluster is involved in fruiting body morphogenesis in the filamentous fungi Sordaria macrospora and Neurospora crassa. Curr. Genet. 2009, 55, 185–198. [Google Scholar] [CrossRef]
- Hicks, K.A.; O’Leary, S.E.; Begley, T.P.; Ealick, S.E. Structural and mechanistic studies of HpxO, a novel flavin adenine dinucleotide-dependent urate oxidase from Klebsiella pneumoniae. Biochemistry 2013, 52, 477–487. [Google Scholar] [CrossRef]
- Lindqvist, Y.; Koskiniemi, H.; Jansson, A.; Sandalova, T.; Schnell, R.; Liu, Z.; Mantsala, P.; Niemi, J.; Schneider, G. Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis. J. Mol. Biol. 2009, 393, 966–977. [Google Scholar] [CrossRef]
- Treiber, N.; Schulz, G.E. Structure of 2,6-dihydroxypyridine 3-hydroxylase from a nicotine-degrading pathway. J. Mol. Biol. 2008, 379, 94–104. [Google Scholar] [CrossRef]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013, 9, e1003323. [Google Scholar] [CrossRef]
- Saikia, S.; Takemoto, D.; Tapper, B.A.; Lane, G.A.; Fraser, K.; Scott, B. Functional analysis of an indole-diterpene gene cluster for lolitrem B biosynthesis in the grass endosymbiont Epichloë festucae. FEBS Lett. 2012, 586, 2563–2569. [Google Scholar] [CrossRef]
- Buchan, D.W.; Ward, S.M.; Lobley, A.E.; Nugent, T.C.; Bryson, K.; Jones, D.T. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010, 38, W563–W568. [Google Scholar] [CrossRef]
- Saikia, S.; Nicholson, M.J.; Young, C.; Parker, E.J.; Scott, B. The genetic basis for indole-diterpene chemical diversity in filamentous fungi. Mycol. Res. 2008, 112, 184–199. [Google Scholar] [CrossRef]
- Steffan, N.; Grundmann, A.; Yin, W.B.; Kremer, A.; Li, S.M. Indole prenyltransferases from fungi: A new enzyme group with high potential for the production of prenylated indole derivatives. Curr. Med. Chem. 2009, 16, 218–231. [Google Scholar] [CrossRef]
- Tudzynski, P.; Hölter, K.; Correia, T.; Arntz, C.; Grammel, N.; Keller, U. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol. Gen. Genet. 1999, 261, 133–141. [Google Scholar] [CrossRef]
- Schneider, P.; Weber, M.; Hoffmeister, D. The Aspergillus nidulans enzyme TdiB catalyzes prenyltransfer to the precursor of bioactive asterriquinones. Fungal Genet. Biol. 2008, 45, 302–309. [Google Scholar] [CrossRef]
- Tsai, H.-F.; Wang, H.; Gebler, J.C.; Poulter, C.D.; Schardl, C.L. The Claviceps purpurea gene encoding dimethylallyltryptophan synthase, the committed step for ergot alkaloid biosynthesis. Biochem. Biophys. Res. Commun. 1995, 216, 119–125. [Google Scholar] [CrossRef]
- Liu, X.; Walsh, C.T. Characterization of cyclo-acetoacetyl-L-tryptophan dimethylallyltransferase in cyclopiazonic acid biosynthesis: Substrate promiscuity and site directed mutagenesis studies. Biochemistry 2009, 48, 11032–11044. [Google Scholar] [CrossRef]
- Metzger, U.; Schall, C.; Zocher, G.; Unsold, I.; Stec, E.; Li, S.M.; Heide, L.; Stehle, T. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 14309–14314. [Google Scholar]
- Unsöld, I.A.; Li, S.M. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: Gene expression, purification, and characterization of fumigaclavine C synthase FGAPT1. ChemBioChem 2006, 7, 158–164. [Google Scholar] [CrossRef]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef]
- Liu, M.; Panaccione, D.G.; Schardl, C.L. Phylogenetic analyses reveal monophyletic origin of the ergot alkaloid gene dmaW in fungi. Evol. Bioinform. 2009, 5, 1–17. [Google Scholar]
- Bullock, W.O.; Fernandez, J.M.; Short, J.M. XL1-Blue: A high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechniques 1987, 5, 376–378. [Google Scholar]
- Vieira, J.; Messing, J. Production of single-stranded plasmid DNA. Meth. Enzymol. 1987, 153, 3–11. [Google Scholar] [CrossRef]
- Namiki, F.; Matsunaga, M.; Okuda, M.; Inoue, I.; Nishi, K.; Fujita, Y.; Tsuge, T. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant-Microbe Interact. 2001, 14, 580–584. [Google Scholar] [CrossRef]
- Yoder, O.C. Cochliobolus heterostrophus, cause of southern corn leaf blight. Adv. Plant Pathol. 1988, 6, 93–112. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; Apweiler, R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Lobley, A.; Sadowski, M.I.; Jones, D.T. pGenTHREADER and pDomTHREADER: New methods for improved protein fold recognition and superfamily discrimination. Bioinformatics 2009, 25, 1761–1767. [Google Scholar] [CrossRef]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Grundmann, A.; Li, S.M. Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology 2005, 151, 2199–2207. [Google Scholar] [CrossRef]
- Ruan, H.L.; Yin, W.B.; Wu, J.Z.; Li, S.M. Reinvestigation of a cyclic dipeptide N-prenyltransferase reveals rearrangement of N-1 prenylated indole derivatives. ChemBioChem 2008, 9, 1044–1047. [Google Scholar] [CrossRef]
- Yin, W.B.; Ruan, H.L.; Westrich, L.; Grundmann, A.; Li, S.M. CdpNPT, an N-prenyltransferase from Aspergillus fumigatus: Overproduction, purification and biochemical characterisation. ChemBioChem 2007, 8, 1154–61. [Google Scholar] [CrossRef]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef]
- Grundmann, A.; Kuznetsova, T.; Afiyatullov, S.; Li, S.M. FtmPT2, an N-prenyltransferase from Aspergillus fumigatus, catalyses the last step in the biosynthesis of fumitremorgin B. ChemBioChem 2008, 9, 2059–2063. [Google Scholar] [CrossRef]
- Balibar, C.J.; Howard-Jones, A.R.; Walsh, C.T. Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nature Chem. Biol. 2007, 3, 584–592. [Google Scholar] [CrossRef]
- Gardiner, D.M.; Cozijnsen, A.J.; Wilson, L.M.; Pedras, M.S.; Howlett, B.J. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol. Microbiol. 2004, 53, 1307–1318. [Google Scholar] [CrossRef]
- Ding, Y.; Williams, R.M.; Sherman, D.H. Molecular analysis of a 4-dimethylallyltryptophan synthase from Malbranchea aurantiaca. J. Biol. Chem. 2008, 283, 16068–16076. [Google Scholar] [CrossRef]
- Yin, W.B.; Grundmann, A.; Cheng, J.; Li, S.M. Acetylaszonalenin biosynthesis in Neosartorya fischeri. Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J. Biol. Chem. 2009, 284, 100–109. [Google Scholar]
- Wang, J.; Machado, C.; Panaccione, D.G.; Tsai, H.-F.; Schardl, C.L. The determinant step in ergot alkaloid biosynthesis by an endophyte of perennial ryegrass. Fungal Genet. Biol. 2004, 41, 189–198. [Google Scholar] [CrossRef]
- Young, C.A.; Felitti, S.; Shields, K.; Spangenberg, G.; Johnson, R.D.; Bryan, G.T.; Saikia, S.; Scott, B. A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fungal Genet. Biol. 2006, 43, 679–693. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Scott, B.; Young, C.A.; Saikia, S.; McMillan, L.K.; Monahan, B.J.; Koulman, A.; Astin, J.; Eaton, C.J.; Bryant, A.; Wrenn, R.E.; et al. Deletion and Gene Expression Analyses Define the Paxilline Biosynthetic Gene Cluster in Penicillium paxilli. Toxins 2013, 5, 1422-1446. https://doi.org/10.3390/toxins5081422
Scott B, Young CA, Saikia S, McMillan LK, Monahan BJ, Koulman A, Astin J, Eaton CJ, Bryant A, Wrenn RE, et al. Deletion and Gene Expression Analyses Define the Paxilline Biosynthetic Gene Cluster in Penicillium paxilli. Toxins. 2013; 5(8):1422-1446. https://doi.org/10.3390/toxins5081422
Chicago/Turabian StyleScott, Barry, Carolyn A. Young, Sanjay Saikia, Lisa K. McMillan, Brendon J. Monahan, Albert Koulman, Jonathan Astin, Carla J. Eaton, Andrea Bryant, Ruth E. Wrenn, and et al. 2013. "Deletion and Gene Expression Analyses Define the Paxilline Biosynthetic Gene Cluster in Penicillium paxilli" Toxins 5, no. 8: 1422-1446. https://doi.org/10.3390/toxins5081422
APA StyleScott, B., Young, C. A., Saikia, S., McMillan, L. K., Monahan, B. J., Koulman, A., Astin, J., Eaton, C. J., Bryant, A., Wrenn, R. E., Finch, S. C., Tapper, B. A., Parker, E. J., & Jameson, G. B. (2013). Deletion and Gene Expression Analyses Define the Paxilline Biosynthetic Gene Cluster in Penicillium paxilli. Toxins, 5(8), 1422-1446. https://doi.org/10.3390/toxins5081422





