Current Understanding on Aflatoxin Biosynthesis and Future Perspective in Reducing Aflatoxin Contamination
Abstract
:1. Introduction on Aspergillus flavus, Aflatoxins, and the Importance for the Economy
2. Genetics and Molecular Biology of Aflatoxin Biosynthesis
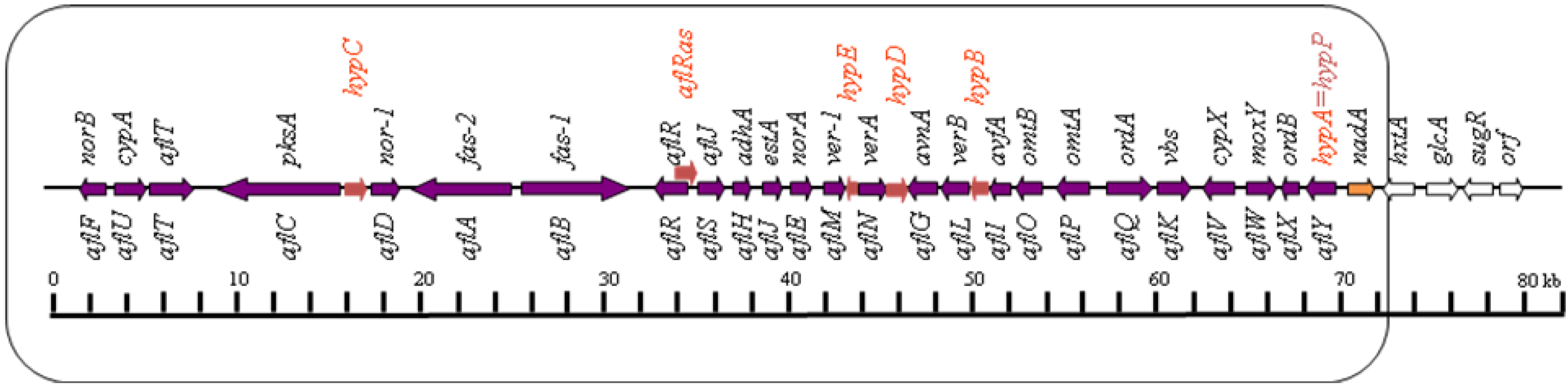
2.1. Conversion of Acetate to Norsolorinic Acid (NOR)
2.2. Conversion of Norsolorinic Acid (NOR) to Averantin (AVN)

2.3. Conversion of Averantin (AVN) to 5'-Hydroxyaverantin (HAVN)
2.4. Conversion of 5'-Hydroxyaverantin (HAVN) to Oxoaverantin (OAVN), and Averufin (AVF)
2.5. Conversion of Averufin (AVF) to Versiconal Hemiacetal Acetate (VHA)
2.6. Conversion of Versiconal Hemiacetal Acetate (VHA) to Versiconal (VHOH, Also Abbreviated as VAL)
2.7. Conversion of Versiconal (VHOH) to Versicolorin B (VER B)
2.8. Conversion of Versicolorin B (VER B) to Versicolorin A (VER A)
2.9. Conversion of Versicolorin A (VER A) to Demethylsterigmatocystin (DMST) and Versicolorin B (VER B) to Demethyldihydrosterigmatocystin (DMDHST)
2.10. Conversion of Demethylsterigmatocystin (DMST) to Sterigmatocystin (ST) and Dihydrodemethylsterigmatocystin (DHDMST) to Dihydrosterigmatocystin (DHST)
2.11. Conversion of Sterigmatocystin (ST) to O-Methylsterigmatocystin (OMST) and Dihydrosterigmatocystin (DHST) to Dihydro-O-methylsterigmatocystin (DHOMST)
2.12. Conversion of O-Methylsterigmatocystin (OMST) to Aflatoxin B1 (AFB1) and Aflatoxin G1 (AFG1) and Dihydro-O-methylsterigmatocystin (DHOMST) to Aflatoxin B2 (AFB2) and Aflatoxin G2 (AFG2)
2.13. Aflatoxins M1 and M2
3. Genetic Regulation of Aflatoxin Biosynthesis
3.1. Genetic Control by aflR Gene, Encoding the Pathway-Specific Transcription Factor, AflR
3.2. Genetic Control by aflS (aflJ) Gene, Encoding a Putative Transcriptional Co-activator, AflS
3.3. Genetic Control on Secondary Metabolism by laeA Gene, Encoding a Global Regulator, LaeA
3.4. Genetic Control on Fungal Development and Mycotoxin Formation by veA Gene, Encoding a Regulator, VeA
4. Factors that Affect Aflatoxin Biosynthesis
4.1. Carbon
4.2. Nitrogen
4.3. Temperature
4.4. Water Activity
4.5. Culture pH
4.6. Developmental Stage
4.7. Oxidative Stress
4.8. Plant Metabolites
5. Future Perspective in Control of Aflatoxin Contamination
5.1. Detection and Screening
5.2. Preharvest Control
5.3. Postharvest Control
5.4. Biological Control
5.5. Breeding for Commercial Crops that Are Resistant to Fungal Growth and/or Aflatoxin Formation
6. Conclusions
References
- Brakhage, A.A.; Schuemann, J.; Bergmann, S.; Scherlach, K.; Schroeckh, V.; Hertweck, C. Activation of fungal silent gene clusters: A new avenue to drug discovery. Prog. Drug Res. 2008, 66, 3–12. [Google Scholar]
- Hoffmeister, D.; Keller, N.P. Natural products of filamentous fungi: Enzymes, genes, and their regulatio. Nat. Prod. Rep. 2007, 24, 393–416. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—From biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Sweeney, M.J.; Dobson, A.D. Mycotoxin production by Aspergillus, Fusarium and Penicillium species. Int. J. Food Microbiol. 1998, 43, 141–158. [Google Scholar] [CrossRef]
- Klich, M.A. Soil fungi of some low-altitude desert cotton fields and ability of their extracts to inhibit Aspergillus flavus. Mycopathologia 1998, 142, 97–100. [Google Scholar] [CrossRef]
- Bennett, J.W.; Leong, P.M.; Kruger, S.J.; Keyes, D. Sclerotial and low aflatoxigenic morphological variants from haploid and diploid Aspergillus parasiticus. Experientia 1986, 42, 848–851. [Google Scholar] [CrossRef]
- Cotty, P. Aflatoxin and sclerotial production by Aspergillus flavus: Influence of pH. Phytopathol 1988, 78, 1250–1253. [Google Scholar] [CrossRef]
- Chang, P.K.; Bennett, J.W.; Cotty, P.J. Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia 2002, 153, 41–48. [Google Scholar] [CrossRef]
- St Leger, R.J.; Screen, S.E.; Shams-Pirzadeh, B. Lack of host specialization in Aspergillus flavus. Appl. Environ. Microbiol. 2000, 66, 320–324. [Google Scholar] [CrossRef]
- Richard, J.L.; Payne, G.A. Mycotoxins: Risks in Plant, Animal and Human Systems; Council for Agricultural Science and Technology (CAST): Ames, IA, USA, 2003. [Google Scholar]
- Robens, J.F.; Cardwell, K. The costs of mycotoxin management to the USA: Management of aflatoxins in the United States. J. Toxicol. 2003, 22, 139–152. [Google Scholar]
- Robens, J.F.; Cardwell, K. The Cost of Mycotoxin Management in the United States. In Aflatoxin and Food Safety; Abbas, H.K., Ed.; CRC Press: Boca Raton, FL, USA, 2005; pp. 1–12. [Google Scholar]
- Allcroft, R.; Carnaghan, R.B.A.; Sargeant, K.; O’Kelly, J. A toxic factor in Brazilian groundnut meal. Vet. Rec. 1961, 73, 428–429. [Google Scholar]
- Lancaster, M.D.; Jenkins, F.P.; Philip, J.M. Toxicity associated with certain samples of ground nuts. Nature 1961, 192, 1095–1096. [Google Scholar]
- Van Egmond, H.P. Current situation on regulations for mycotoxins. Overview of tolerances and status of standard methods of sampling and analysis. J. Food Addit. Contam. 1989, 6, 139–188. [Google Scholar] [CrossRef]
- Goto, T.; Wicklow, D.T.; Ito, Y. Aflatoxin and cyclopiazonic acid production by a sclerotium-producing Aspergillus tamarii strain. Appl. Environ. Microbiol. 1996, 62, 4036–4038. [Google Scholar]
- Eaton, D.; Gallagher, E. Mechanisms of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1994, 34, 135–172. [Google Scholar] [CrossRef]
- Hsieh, D.P.H. Potential Human Health Hazards of Mycotoxins. In Mycotoxins and Phycotoxins; Natori, S., Hashimoto, H., Ueno, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 1989; pp. 69–80. [Google Scholar]
- Ngindu, A.; Johnson, B.K.; Kenya, P.R.; Ngira, J.A.; Ocheng, D.M.; Nandwa, H.; Omondi, T.N.; Jansen, A.J.; Ngare, W.; Kaviti, J.N.; et al. Outbreak of acute hepatitis caused by aflatoxin poisoning in Kenya. Lancet 1982, 1, 1346–1348. [Google Scholar]
- Lewis, L.; Onsongo, M.; Njapau, H.; Schurz-Rogers, H.; Luber, G.; Kieszak, S.; Nyamongo, J.; Backer, L.; Dahiye, A.M.; Misore, A.; et al. Aflatoxin contamination of commercial maize products during an outbreak of acute aflatoxicosis in eastern and central Kenya. Environ. Health Perspect. 2005, 113, 1763–1767. [Google Scholar] [CrossRef]
- Baertschi, S.W.; Raney, K.D.; Shimada, T.; Harris, T.M.; Guengerich, F.P. Comparison rates of enzymatic oxidation of aflatoxin B1, aflatoxin G1, and sterigmatocystin and activities of the epoxides in forming guanyl-N adducts and inducing different genetic responses. Chem. Res. Toxicol. 1989, 2, 114–122. [Google Scholar] [CrossRef]
- Bressac, B.; Kew, M.; Wands, J.; Ozturk, M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 1991, 350, 429–431. [Google Scholar]
- Coursaget, P.; Depril, N.; Chabaud, M.; Nandi, R.; Mayelo, V.; LeCann, P.; Yvonnet, B. High prevalence of mutations at codon 249 of the p53 gene in hyptocellular carcinomas from Senegal. Br. J. Cancer 1993, 67, 1395–1397. [Google Scholar] [CrossRef]
- Hsu, I.C.; Metcalf, R.A.; Sun, T.; Welsh, J.A.; Wang, N.J.; Harris, C.C. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature 1991, 350, 427–428. [Google Scholar]
- Ozturk, M. p53 mutation in hepatocellular carcinoma after aflatoxin exposure. Lancet 1991, 338, 1356–1359. [Google Scholar]
- Busby, W.F., Jr.; Wogan, G.N. Aflatoxins. In Mycotoxins and N-Nitrosocompounds: Environmental Risks; Shank, R.C., Ed.; CRC Press: Boca Raton, FL, USA, 1981; Volume 2, pp. 3–45. [Google Scholar]
- Groopman, J.D.; Wogan, G.N.; Roebuck, B.D.; Kensler, T.W. Molecular biomarkers for aflatoxins and their application to human cancer prevention. Cancer Res. 1994, 54, 190–191. [Google Scholar]
- Raisuddin, S.; Singh, K.P.; Zaidi, S.I.; Paul, B.N.; Ray, P.K. Immunosuppressive effects of aflatoxin in growing rats. Mycopathologia 1993, 124, 189–194. [Google Scholar] [CrossRef]
- Kelly, J.D.; Eaton, D.L.; Guengerich, F.P.; Coulombe, R.A., Jr. Aflatoxin B1 activation in human lung. Toxicol. Appl. Pharmacol. 1997, 144, 88–95. [Google Scholar] [CrossRef]
- Wild, C.P.; Shrestha, S.M.; Anwar, W.A.; Montesano, R. Field studies of aflatoxin exposure, metabolism and induction of genetic alterations in relation to HBV infection and hepatocellular carcinoma in the Gambia and Thailand. Toxicol. Lett. 1992, 64-65, 455–461. [Google Scholar] [CrossRef]
- Peers, F.; Bosch, X.; Kaldor, J.; Linsell, A.; Pluijmen, M. Aflatoxin exposure, hepatitis B virus infection and liver cancer in Swaziland. Int. J.Cancer 1987, 39, 545–553. [Google Scholar] [CrossRef]
- McGlynn, K.A.; Hunter, K.; LeVoyer, T.; Roush, J.; Wise, P.; Michielli, R.A.; Shen, F.M.; Evans, A.A.; London, W.T.; Buetow, K.H. Susceptibility to aflatoxin B1-related primary hepatocellular carcinoma in mice and humans. Cancer Res. 2003, 63, 4594–4601. [Google Scholar]
- Arsura, M.; Cavin, L.G. Nuclear factor-kappaB and liver carcinogenesis. Cancer Lett. 2005, 229, 157–169. [Google Scholar] [CrossRef]
- Chen, C.J.; Wang, L.Y.; Lu, S.N.; Wu, M.H.; You, S.L.; Zhang, Y.J.; Wang, L.W.; Santella, R.M. Elevated aflatoxin exposure and increased risk of hepatocellular carcinoma. Hepatology 1996, 24, 38–42. [Google Scholar]
- Chen, C.J.; Yu, M.W.; Liaw, Y.F.; Wang, L.W.; Chiamprasert, S.; Matin, F.; Hirvonen, A.; Bell, D.A.; Santella, R.M. Chronic hepatitis B carriers with null genotypes of glutathione S transferase MI and TI polymorphisms who are exposed to aflatoxin are at increased risk of hepatocellular carcinoma. Am. J. Human Genet. 1996, 59, 128–134. [Google Scholar]
- Williams, J.H.; Phillips, T.D.; Jolly, P.E.; Stiles, J.K.; Jolly, C.M.; Aggarwal, D. Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 2004, 80, 1106–1122. [Google Scholar]
- Krishnamachari, K.A.; Bhat, R.V.; Nagarajan, V.; Tilak, T.B. Hepatitis due to aflatoxicosis: An outbreak of hepatitis in parts of western India. Lancet 1975, 1, 1061–1063. [Google Scholar]
- Fung, F.; Clark, R.F. Health effects of mycotoxins: A toxicological overview. J. Toxicol. Clin. Toxicol. 2004, 42, 217–234. [Google Scholar] [CrossRef]
- Wogan, G.N. Aflatoxins as risk factors for hepatocellular carcinoma in humans. Cancer Res. 1992, 52, 2114–2118. [Google Scholar]
- Wogan, G.N. Impacts of chemicals on liver cancer risk. Semin. Cancer Biol. 2000, 10, 201–210. [Google Scholar] [CrossRef]
- The US Food and Drug Administration. Toxic pet food may have killed dozens of dogs. Available online: http://www.msnbc.msn.com/id/10771943/ns/health-pet_health/t/toxic-pet-food-may-have-killed-dozens-dogs/ (accessed on 10 May 2006).
- Van Egmond, H.P.; Schothorst, R.C.; Jonker, M.A. Regulations relating to mycotoxins in food: Perspectives in a global and European context. Anal. Bioanal. Chem. 2007, 389, 147–157. [Google Scholar] [CrossRef]
- Van Egmond, H.P.; Jonker, M.A. Worldwide Regulations on Aflatoxins; Abbas, H.K., Ed.; CRC Press: Boca Raton, FL, USA, 2005; pp. 77–93. [Google Scholar]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef]
- Bennett, J.W. Mycotoxins, mycotoxicoses, mycotoxicology and mycopathologia. Mycopathologia 1987, 100, 3–5. [Google Scholar] [CrossRef]
- Bennett, J.W.; Lee, L.S. Mycotoxins—Their biosynthesis in fungi: Aflatoxins and other bisfuranoids. J. Food Protection 1979, 42, 805–809. [Google Scholar]
- Bhatnagar, D.; Brown, R.; Ehrlich, K.; Cleveland, T.E. Mycotoxins Contaminating Cereal Grain Crops: Their Occurrence and Toxicity. In Applied Mycology and Biotechnology; Khachatourians, G.G., Arora, D.K., Eds.; Elsevier: New York, NY, USA, 2002; Volume 2, pp. 171–196. [Google Scholar]
- Cleveland, T.E.; Bhatnagar, D. Molecular Strategies for Reducing Aflatoxin Levels in Crops Before Harvest. In Molecular Approaches to Improving Food Quality and Safety; Bhatnagar, D., Cleveland, T.E., Eds.; Van Nostrand Reinhold: New York, NY, USA, 1992; pp. 205–228. [Google Scholar]
- Eaton, D.L.; Groopman, J.D. The Toxicology of Aflatoxins: Human Health, Veterinary, and Agricultural Significance; Academic Press: San Diego, CA, USA, 1994. [Google Scholar]
- Hall, A.J.; Wild, C.P. Epidemiology of Aflatoxin-Related Disease. In The Toxicology of Aflatoxins: Human Health, Veterinary, and Agricultural Significance; Eaton, D.L., Groopman, J.D., Eds.; Academic Press: San Diego, CA, USA, 1994; pp. 233–258. [Google Scholar]
- Jelinek, C.F.; Pohland, A.E.; Wood, G.E. Worldwide occurrence of mycotoxins in food and feeds—An update. J. Assoc. Off. Anal. Chem. 1989, 72, 223–230. [Google Scholar]
- Papa, K.E. Linkage groups in Aspergillus flavus. Mycologia 1976, 68, 159–165. [Google Scholar] [CrossRef]
- Papa, K.E. Genetics of Aspergillus flavus: Complementation and mapping of aflatoxin mutants. Genet. Res. 1979, 34, 1–9. [Google Scholar] [CrossRef]
- Papa, K.E. Genetics of Aspergillus flavus: Linkage of aflatoxin mutants. Can. J. Microbiol. 1984, 30, 68–73. [Google Scholar] [CrossRef]
- Bennett, J.W. Microbiological aspects of the aflatoxin problem. Am. Ass. Feed Microscop. Off. Proc. 1970, 18, 118–131. [Google Scholar]
- Bennett, J.W.; Goldblatt, L.A. The isolation of mutants of Aspergillus flavus and A. parasiticus with altered aflatoxin producing ability. Sabouraudia 1973, 11, 235–241. [Google Scholar] [CrossRef]
- Bennett, J.W.; Lee, L.S.; Cucullu, A.F. Effect of dichlorvos on aflatoxin and versicolorin A production in Aspergillus parasiticus. Bot. Gaz. 1976, 137, 318–324. [Google Scholar]
- Bennett, J.W. Aflatoxins and anthraquinones from diploids of Aspergillus parasiticus. J. Gen. Microbiol. 1979, 113, 127–136. [Google Scholar] [CrossRef]
- Bennett, J.W.; Kronberg, F.G.; Goodman, L.A.; Seltman, M.A. Isolation of an anthraquinoe-accumulating mutant of Aspergillus parasiticus and partial characterization by dry column chromatography. Mycologia 1983, 75, 202–208. [Google Scholar]
- Bennett, J.W.; Lee, L.S.; Vinnett, C.H. The correlation of aflatoxin and norsolorinic acid production. J. Am. Oil Chem. Soc. 1971, 48, 368–370. [Google Scholar] [CrossRef]
- Bennett, J.W.; Kronberg, F.; Gougis, G. Pigmented isolates from anthraquinone-producing mutants of Aspergillus parasiticus. Am. Soc. Microbiol. 1976, 76, 6. [Google Scholar]
- Bennett, J.W.; Chang, P.K.; Bhatnagar, D. One gene to whole pathway: The role of norsolorinic acid in aflatoxin research. Adv. Appl. Microbiol. 1997, 45, 1–15. [Google Scholar] [CrossRef]
- Hsieh, D.P.; Lin, M.T.; Yao, R.C.; Singh, R. Biosynthesis of aflatoxin. Conversion of norsolorinic acid and other hypothetical intermediates into aflatoxin B1. J. Agric. Food Chem. 1976, 24, 1170–1174. [Google Scholar] [CrossRef]
- Dutton, M.F. Enzymes and aflatoxin biosynthesis. Microbiol. Rev. 1988, 52, 274–295. [Google Scholar]
- Hsieh, D.P.; Mateles, R.I. The relative contribution of acetate and glucose to aflatoxin biosynthesis. Biochem. Biophys. Acta 1970, 208, 482–486. [Google Scholar] [CrossRef]
- Hsieh, D.P.; Lin, M.T.; Yao, R.C. Conversion of sterigmatocystin to aflatoxin B1 by Aspergillus parasiticus. Biochem. Biophys. Res. Commun. 1973, 52, 992–997. [Google Scholar] [CrossRef]
- Yu, J.; Chang, P.K.; Cary, J.W.; Wright, M.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl. Environ. Microbiol. 1995, 61, 2365–2371. [Google Scholar]
- Bennett, J.W.; Papa, K.E. The aflatoxigenic Aspergillus spp. Adv. Plant Pathol. 1988, 6, 265–279. [Google Scholar]
- Bennett, J.W.; Silverstein, R.B.; Kruger, S.J. Isolation and characterization of two nonaflatoxigenic classes of morphological variants of Aspergillus parasiticus. J. Am. Oil Chem. Soc. 1981, 58, A952–A955. [Google Scholar] [CrossRef]
- Bhatnagar, D.; Ehrlich, K.C.; Cleveland, T.E. Oxidation-Reduction Reactions in Biosynthesis of Secondary Metabolites. In Mycotoxins in Ecological Systems; Bhatnagar, D., Lillehoj, E.B., Arora, D.K., Eds.; Marcel Dekker: New York, NY, USA, 1992; Volume 10, pp. 255–285. [Google Scholar]
- Chang, P.K. Lack of interaction between AFLR and AFLJ contributes to nonaflatoxigenicity of Aspergillus sojae. J. Biotechnol. 2004, 107, 245–253. [Google Scholar] [CrossRef]
- Chang, P.K.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Bennett, J.W.; Linz, J.E.; Woloshuk, C.P.; Payne, G.A. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl. Environ. Microbiol. 1993, 59, 3273–3279. [Google Scholar]
- Chang, P.K.; Cary, J.W.; Yu, J.; Bhatnagar, D.; Cleveland, T.E. The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B1 biosynthesis. Mol. Gen. Genet. 1995, 248, 270–277. [Google Scholar] [CrossRef]
- Chang, P.K.; Yu, J.; Bhatnagar, D.; Cleveland, T.E. The carboxy-terminal portion of the aflatoxin pathway regulatory protein AFLR of Aspergillus parasiticus activates GAL1: lacZ gene expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1999, 65, 2508–2512. [Google Scholar]
- Cleveland, T.E.; Lax, A.R.; Lee, L.S.; Bhatnagar, D. Appearance of enzyme activities catalyzing conversion of sterigmatocystin to aflatoxin B1 in late-growth-phase Aspergillus parasiticus cultures. Appl. Environ. Microbiol. 1987, 53, 1711–1713. [Google Scholar]
- Crawford, J.M.; Thomas, P.M.; Scheerer, J.R.; Vagstad, A.L.; Kelleher, N.L.; Townsend, C.A. Deconstruction of iterative multidomain polyketide synthase function. Science 2008, 320, 243–246. [Google Scholar]
- Ehrlich, K.C.; Cary, J.W.; Montalbano, B.G. Characterization of the promoter for the gene encoding the aflatoxin biosynthetic pathway regulatory protein AFLR. Biochim. Biophys. Acta 1999, 1444, 412–417. [Google Scholar] [CrossRef]
- Keller, N.P.; Dischinger, H.C., Jr.; Bhatnagar, D.; Cleveland, T.E.; Ullah, A.H. Purification of a 40-kilodalton methyltransferase active in the aflatoxin biosynthetic pathway. Appl. Environ. Microbiol. 1993, 59, 479–484. [Google Scholar]
- Ehrlich, K.C.; Yu, J. Aflatoxin-Like Gene Clusters and How They Evolved. In Mycotoxins in Food, Feed, and Bioweapons; Varma, A.K., Rai, M.K., Eds.; Springer Verlag: Heidelberg, Germany, 2009; pp. 65–76. [Google Scholar]
- Ehrlich, K.C. Predicted roles of uncharacterized clustered genes in aflatoxin biosynthesis. Toxins 2009, 1, 37–58. [Google Scholar] [CrossRef]
- Yu, J.; Chang, P.K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar]
- Chang, P.K.; Horn, B.W.; Dorner, J.W. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 2005, 42, 914–923. [Google Scholar] [CrossRef]
- Wilson, D.M. Analytical methods for aflatoxins in corn and peanuts. Arch. Environ. Contam. Toxicol. 1989, 18, 308–314. [Google Scholar]
- Trail, F.; Mahanti, N.; Rarick, M.; Mehigh, R.; Liang, S.H.; Zhou, R.; Linz, J.E. Physical and transcriptional map of an aflatoxin gene cluster in Aspergillus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Appl. Environ. Microbiol. 1995, 61, 2665–2673. [Google Scholar]
- Trail, F.; Mahanti, N.; Linz, J. Molecular biology of aflatoxin biosynthesis. Microbiol. 1995, 141, 755–765. [Google Scholar] [CrossRef]
- Townsend, C.A. Progress towards a biosynthetic rationale of the aflatoxin pathway. Pure Appl. Chem. 1997, 58, 227–238. [Google Scholar] [CrossRef]
- Yu, J.; Bhatnagar, D.; Cleveland, T.E. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 2004, 564, 126–130. [Google Scholar] [CrossRef]
- Bennett, J.W. Loss of norsolorinic acid and aflatoxin production by a mutant of Aspergillus parasiticus. J. Gen. Microbiol. 1981, 124, 429–432. [Google Scholar]
- Watanabe, C.M.; Wilson, D.; Linz, J.E.; Townsend, C.A. Demonstration of the catalytic roles and evidence for the physical association of type I fatty acid synthases and a polyketide synthase in the biosynthesis of aflatoxin B1. Chem. Biol. 1996, 3, 463–469. [Google Scholar] [CrossRef]
- Brown, D.W.; Yu, J.H.; Kelkar, H.S.; Fernandes, M.; Nesbitt, T.C.; Keller, N.P.; Adams, T.H.; Leonard, T.J. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 1996, 93, 1418–1422. [Google Scholar]
- Watanabe, C.M.; Townsend, C.A. Initial characterization of a type I fatty acid synthase and polyketide synthase multienzyme complex NorS in the biosynthesis of aflatoxin B1. Chem. Biol. 2002, 9, 981–988. [Google Scholar] [CrossRef]
- Crawford, J.M.; Dancy, B.C.; Hill, E.A.; Udwary, D.W.; Townsend, C.A. Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc. Natl. Acad. Sci. USA 2006, 103, 16728–16733. [Google Scholar]
- Crawford, J.M.; Vagstad, A.L.; Ehrlich, K.C.; Townsend, C.A. Starter unit specificity directs genome mining of polyketide synthase pathways in fungi. Bioorg. Chem. 2008, 36, 16–22. [Google Scholar] [CrossRef]
- Yabe, K.; Nakajima, H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2004, 64, 745–755. [Google Scholar] [CrossRef]
- Brown, D.W.; Adams, T.H.; Keller, N.P. Aspergillus has distinct fatty acid synthases for primary and secondary metabolism. Proc. Natl. Acad. Sci. USA 1996, 93, 14873–14877. [Google Scholar] [CrossRef]
- Mahanti, N.; Bhatnagar, D.; Cary, J.W.; Joubran, J.; Linz, J.E. Structure and function of fas-1A, a gene encoding a putative fatty acid synthetase directly involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 1996, 62, 191–195. [Google Scholar]
- Payne, G.A. Process of Contamination by Aflatoxin-Producing Fungi and Their Impacts on Crops. In Mycotoxins in Agriculture and Food Safety; Sinha, K.K., Bhatnagar, D., Eds.; Marcel Dekker: New York, NY, USA, 1998; pp. 279–306. [Google Scholar]
- Watanabe, C.M.; Townsend, C.A. Incorporation of molecular oxygen in aflatoxin B1 biosynthesis. J. Org. Chem. 1996, 61, 1990–1993. [Google Scholar] [CrossRef]
- Hitchman, T.S.; Schmidt, E.W.; Trail, F.; Rarick, M.D.; Linz, J.E.; Townsend, C.A. Hexanoate synthase, a specialized type I fatty acid synthase in aflatoxin B1 biosynthesis. Bioorg. Chem. 2001, 29, 293–307. [Google Scholar] [CrossRef]
- Crawford, J.M.; Vagstad, A.L.; Whitworth, K.P.; Ehrlich, K.C.; Townsend, C.A. Synthetic strategy of nonreducing Iterative polyketide synthases and the origin of the classical “Starter-Unit Effect”. Chembiochem 2008, 9, 1019–1023. [Google Scholar] [CrossRef]
- Bennett, J.W.; Bhatnagar, D.; Chang, P.K. The molecular genetics of aflatoxin biosynthesis. FEMS Symp. 1994, 51–58. [Google Scholar]
- Papa, G. Norsolorinic acid mutant of Aspergillus flavus. J. Gen. Microbiol. 1982, 128, 1345–1348. [Google Scholar]
- Lee, L.S.; Bennett, J.W.; Goldblatt, L.A.; Lundin, R.E. Norsolorinic acid from a mutant strain of Aspergillus parasiticus. J. Am. Oil Chem. Soc. 1971, 48, 93–94. [Google Scholar] [CrossRef]
- Detroy, R.W.; Freer, S.; Ciegler, A. Aflatoxin and anthraquinone biosynthesis by nitrosoguanidine-derived mutants of Aspergillus parasiticus. Can. J. Microbiol. 1973, 19, 1373–1378. [Google Scholar] [CrossRef]
- Chang, P.K.; Skory, C.D.; Linz, J.E. Cloning of a gene associated with aflatoxin B1 biosynthesis in Aspergillus parasiticus. Curr. Genet. 1992, 21, 231–233. [Google Scholar] [CrossRef]
- Zhou, R.; Linz, J.E. Enzymatic function of the nor-1 protein in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 1999, 65, 5639–5641. [Google Scholar]
- Trail, F.; Chang, P.-K.; Cary, J.; Linz, J.E. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl. Environ. Microbiol. 1994, 60, 4078–4085. [Google Scholar]
- Cary, J.W.; Wright, M.; Bhatnagar, D.; Lee, R.; Chu, F.S. Molecular characterization of an Aspergillus parasiticus dehydrogenase gene, norA, located on the aflatoxin biosynthesis gene cluste. Appl. Environ. Microbiol. 1996, 62, 360–366. [Google Scholar]
- Bennett, J.W.; Lee, L.S.; Shoss, S.M.; Boudreaux, G.H. Identification of averantin as an aflatoxin B1 precursor: Placement in the biosynthetic pathway. Appl. Environ. Microbiol. 1980, 39, 835–839. [Google Scholar]
- McCormick, S.P.; Bhatnagar, D.; Lee, L.S. Averufanin is an aflatoxin B1 precursor between averantin and averufin in the biosynthetic pathway. Appl. Environ. Microbiol. 1987, 53, 14–16. [Google Scholar]
- Yabe, K.; Ando, Y.; Hamasaki, T. A metabolic grid among versiconal hemiacetal acetate, versiconol acetate, versiconol and versiconal during aflatoxin biosynthesis. J. Gen. Microbiol. 1991, 137, 2469–2475. [Google Scholar] [CrossRef]
- Yabe, K.; Nakamura, Y.; Nakajima, H.; Ando, Y.; Hamasaki, T. Enzymatic conversion of norsolorinic acid to averufin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 1991, 57, 1340–1345. [Google Scholar]
- Yu, J.; Chang, P.K.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E. avnA, a gene encoding a cytochrome P-450 monooxygenase, is involved in the conversion of averantin to averufin in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 1997, 63, 1349–1356. [Google Scholar]
- Hsieh, D.P. Inhibition of aflatoxin biosynthesis of dichlorvos. J. Agric. Food Chem. 1973, 21, 468–470. [Google Scholar] [CrossRef]
- Singh, R.; Hsieh, D.P. Aflatoxin biosynthetic pathway: Elucidation by using blocked mutants of Aspergillus parasiticus. Arch. Biochem. Biophys. 1977, 178, 285–292. [Google Scholar] [CrossRef]
- Fitzell, D.L.; Hsieh, D.P.; Yao, R.C.; la Mar, G.N. Biosynthesis of averufin from acetate by Aspergillus parasiticus. J. Agric. Food Chem. 1975, 23, 442–444. [Google Scholar] [CrossRef]
- Lin, M.T.; Hsieh, D.P.; Yao, R.C.; Donkersloot, J.A. Conversion of averufin into aflatoxins by Aspergillus parasiticus. Biochemistry 1973, 12, 5167–5171. [Google Scholar] [CrossRef]
- Lin, M.T.; Hsieh, D.P. Averufin in the biosynthesis of aflatoxin B. J. Am Chem. Soc. 1973, 95, 1668–1669. [Google Scholar] [CrossRef]
- Keller, N.P.; Watanabe, C.M.; Kelkar, H.S.; Adams, T.H.; Townsend, C.A. Requirement of monooxygenase-mediated steps for sterigmatocystin biosynthesis by Aspergillus nidulans. Appl. Environ. Microbiol. 2000, 66, 359–362. [Google Scholar]
- Sakuno, E.; Yabe, K.; Nakajima, H. Involvement of two cytosolic enzymes and a novel intermediate, 5'-oxoaverantin, in the pathway from 5'-hydroxyaverantin to averufin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2003, 69, 6418–6426. [Google Scholar] [CrossRef]
- Chang, P.K.; Yu, J.; Ehrlich, K.C.; Boue, S.M.; Montalbano, B.G.; Bhatnagar, D.; Cleveland, T.E. adhA in Aspergillus parasiticus is involved in conversion of 5'-hydroxyaverantin to averufin. Appl. Environ. Microbiol. 2000, 66, 4715–4719. [Google Scholar]
- Ehrlich, K.C.; Chang, P.-K.; Scharfenstein, J.S.L.; Cary, J.W.; Crawford, J.M.; Townsend, C.A. Absence of the aflatoxin biosynthesis gene, norA, allows accumulation of deoxyaflatoxin B1 in Aspergillus flavus cultures. FEMS Microbiol. Lett. 2010, 305, 65–70. [Google Scholar] [CrossRef]
- Yu, J.; Woloshuk, C.P.; Bhatnagar, D.; Cleveland, T.E. Cloning and characterization of avfA and omtB genes involved in aflatoxin biosynthesis in three Aspergillus species. Gene 2000, 248, 157–167. [Google Scholar] [CrossRef]
- Yao, R.C.; Hsieh, D.P. Step of dichlorvos inhibition in the pathway of aflatoxin biosynthesis. Appl. Microbiol. 1974, 28, 52–57. [Google Scholar]
- Hsieh, D.P.; Wan, C.C.; Billington, J.A. A versiconal hemiacetal acetate converting enzyme in aflatoxin biosynthesis. Mycopathologia 1989, 107, 121–126. [Google Scholar] [CrossRef]
- Kusumoto, K.I.; Hsieh, D.P.H. Purification and characterization of the esterases involved in aflatoxin biosynthesis in Aspergillus parasiticus. Can. J. Microbiol. 1996, 42, 804–810. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Cole, R.J.; Grigsby, R.D.; Hein, H., Jr. Inhibition of aflatoxin production and tentative identification of an aflatoxin intermediate “versiconal acetate” from treatment with dichlorvos. Appl. Microbiol. 1974, 27, 394–399. [Google Scholar]
- Fitzell, D.L.; Singh, R.; Hsieh, D.P.; Motell, E.L. Nuclear magnetic resonance identification of versiconal hemiacetal acetate as an intermediate in aflatoxin biosynthesis. J. Agric. Food. Chem. 1977, 25, 1193–1197. [Google Scholar] [CrossRef]
- Yu, J.; Chang, P.K.; Bhatnagar, D.; Cleveland, T.E. Cloning and functional expression of an esterase gene in Aspergillus parasitcus. Mycopathologia 2002, 156, 227–234. [Google Scholar]
- Chang, P.K.; Yabe, K.; Yu, J. The Aspergillus parasiticus estA-encoded esterase converts versiconal hemiacetal acetate to versiconal and versiconol acetate to versiconol in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 3593–3599. [Google Scholar] [CrossRef]
- Yabe, K.; Chihaya, N.; Hamamatsu, S.; Sakuno, E.; Hamasaki, T.; Nakajima, H.; Bennett, J.W. Enzymatic conversion of averufin to hydroxyversicolorone and elucidation of a novel metabolic grid involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2003, 69, 66–73. [Google Scholar] [CrossRef]
- Lin, B.K.; Anderson, J.A. Purification and properties of versiconal cyclase from Aspergillus parasiticus. Arch. Biochem. Biophys. 1992, 293, 67–70. [Google Scholar] [CrossRef]
- McGuire, S.M.; Silva, J.C.; Casillas, E.G.; Townsend, C.A. Purification and characterization of versicolorin B synthase from Aspergillus parasiticus. Catalysis of the stereodifferentiating cyclization in aflatoxin biosynthesis essential to DNA interaction. Biochemistry 1996, 35, 11470–11486. [Google Scholar]
- Silva, J.C.; Minto, R.E.; Barry, C.E., III; Holland, K.A.; Townsend, C.A. Isolation and characterization of the versicolorin B synthase gene from Aspergillus parasiticus. Expansion of the aflatoxin B1 biosynthetic gene cluster. J. Biol. Chem. 1996, 271, 13600–13608. [Google Scholar]
- Silva, J.C.; Townsend, C.A. Heterologous expression, isolation, and characterization of versicolorin B synthase from Aspergillus parasiticu. A key enzyme in the aflatoxin B1 biosynthetic pathway. J. Biol. Chem. 1997, 272, 804–813. [Google Scholar]
- Zuckerman, A.J.; Rees, K.R.; Inman, D.; Petts, V. Site of action of aflatoxin on human liver cells in culture. Nature 1967, 214, 814–815. [Google Scholar]
- Yabe, K.; Matsuyama, Y.; Ando, Y.; Nakajima, H.; Hamasaki, T. Stereochemistry during aflatoxin biosynthesis: Conversion of norsolorinic acid to averufin. Appl. Environ. Microbiol. 1993, 59, 2486–2492. [Google Scholar]
- Kelkar, H.S.; Skloss, T.W.; Haw, J.F.; Keller, N.P.; Adams, T.H. Aspergillus nidulans stcL encodes a putative cytochrome P-450 monooxygenase required for bisfuran desaturation during aflatoxin/sterigmatocystin biosynthesis. J. Biol. Chem. 1997, 272, 1589–1594. [Google Scholar]
- Henry, K.M.; Townsend, C.A. Ordering the reductive and cytochrome P450 oxidative steps in demethylsterigmatocystin formation yields general insights into the biosynthesis of aflatoxin and related fungal metabolites. J. Am. Chem. Soc. 2005, 127, 3724–3733. [Google Scholar] [CrossRef]
- Skory, C.D.; Chang, P.K.; Cary, J.; Linz, J.E. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 1992, 58, 3527–3537. [Google Scholar]
- Keller, N.P.; Kantz, N.J.; Adams, T.H. Aspergillus nidulans verA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol. 1994, 60, 1444–1450. [Google Scholar]
- Keller, N.P.; Segnar, S.; Bhatnagar, D.; Adams, T.H. stcS, a putative P-450 monooxygenase, is required for the conversion of versicolorin A to sterigmatocystin in Aspergillus nidulans. Appl. Environ. Microbiol. 1995, 61, 3628–3632. [Google Scholar]
- Yabe, K.; Ando, Y.; Hashimoto, J.; Hamasaki, T. Two distinct O-methyltransferases in aflatoxin biosynthesis. Appl. Environ. Microbiol. 1989, 55, 2172–2177. [Google Scholar]
- Yabe, K.; Matsushima, K.; Koyama, T.; Hamasaki, T. Purification and characterization of O-methyltransferase I involved in conversion of demethylsterigmatocystin to sterigmatocystin and of dihydrodemethylsterigmatocystin to dihydrosterigmatocystin during aflatoxin biosynthesis. Appl. Environ. Microbiol. 1998, 64, 166–171. [Google Scholar]
- Yabe, K.; Nakamura, M.; Hamasaki, T. Enzymatic formation of G-group aflatoxins and biosynthetic relationship between G- and B-group aflatoxins. Appl. Environ. Microbiol. 1999, 65, 3867–3872. [Google Scholar]
- Motomura, M.; Chihaya, N.; Shinozawa, T.; Hamasaki, T.; Yabe, K. Cloning and characterization of the O-methyltransferase I gene (dmtA) from Aspergillus parasiticus associated with the conversions of demethylsterigmatocystin to sterigmatocystin and dihydrodemethylsterigmatocystin to dihydrosterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 1999, 65, 4987–4994. [Google Scholar]
- Kelkar, H.S.; Keller, N.P.; Adams, T.H. Aspergillus nidulans stcP encodes an O-methyltransferase that is required for sterigmatocystin biosynthesis. Appl. Environ. Microbiol. 1996, 62, 4296–4298. [Google Scholar]
- Yu, J.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Keller, N.P.; Chu, F.S. Cloning and characterization of a cDNA from Aspergillus parasiticus encoding an O-methyltransferase involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 1993, 59, 3564–3571. [Google Scholar]
- Yu, J.; Chang, P.K.; Payne, G.A.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E. Comparison of the omtA genes encoding O-methyltransferases involved in aflatoxin biosynthesis from Aspergillus parasiticus and A. flavus. Gene 1995, 163, 121–125. [Google Scholar] [CrossRef]
- Klich, M.A.; Yu, J.; Chang, P.K.; Mullaney, E.J.; Bhatnagar, D.; Cleveland, T.E. Hybridization of genes involved in aflatoxin biosynthesis to DNA of aflatoxigenic and non-aflatoxigenic aspergilli. Appl. Microbiol. Biotechnol. 1995, 44, 439–443. [Google Scholar] [CrossRef]
- Yabe, K.; Ando, Y.; Hamasaki, T. Biosynthetic relationship among aflatoxins B1, B2, G1, and G2. Appl. Environ. Microbiol. 1988, 54, 2101–2106. [Google Scholar]
- Prieto, R.; Yousibova, G.L.; Woloshuk, C.P. Identification of aflatoxin biosynthesis genes by genetic complementation in an Aspergillus flavus mutant lacking the aflatoxin gene cluster. Appl. Environ. Microbiol. 1996, 62, 3567–3571. [Google Scholar]
- Prieto, R.; Woloshuk, C.P. ord1, an oxidoreductase gene responsible for conversion of O-methylsterigmatocystin to aflatoxin in Aspergillus flavus. Appl. Environ. Microbiol. 1997, 63, 1661–1666. [Google Scholar]
- Yu, J.; Chang, P.K.; Ehrlich, K.C.; Cary, J.W.; Montalbano, B.; Dyer, J.M.; Bhatnagar, D.; Cleveland, T.E. Characterization of the critical amino acids of an Aspergillus parasiticus cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2. Appl. Environ. Microbiol. 1998, 64, 4834–4841. [Google Scholar]
- Ehrlich, K.C.; Chang, P.K.; Yu, J.; Cotty, P.J. Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl. Environ. Microbiol. 2004, 70, 6518–6524. [Google Scholar] [CrossRef]
- Price, M.S.; Yu, J.; Nierman, W.C.; Kim, H.S.; Pritchard, B.; Jacobus, C.A.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A. The aflatoxin pathway regulator AflR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. FEMS Microbiol. Lett. 2006, 255, 275–279. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Chang, P.-K.; Yu, J.; Cary, J.W.; Bhatnagar, D. Control of Aflatoxin Biosynthesis in Aspergilli. In Aflatoxins—Biochemistry and Molecular Biology; Guevara-Gonzalez, R.G., Ed.; Intech: Rijeka, Croatia, 2011; pp. 21–40. [Google Scholar]
- Yu, J.; Chang, P.K.; Bhatnagar, D.; Cleveland, T.E. Cloning of a sugar utilization gene cluster in Aspergillus parasiticus. Biochim. Biophys. Acta 2000, 1493, 211–214. [Google Scholar] [CrossRef]
- Cai, J.; Zeng, H.; Shima, Y.; Hatabayashi, H.; Nakagawa, H.; Ito, Y.; Adachi, Y.; Nakajima, H.; Yabe, K. Involvement of the nadA gene in formation of G-group aflatoxins in Aspergillus parasiticus. Fungal Genet. Biol. 2008, 45, 1081–1093. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Scharfenstein, J.S.L.; Montalbano, B.G.; Chang, P.-K. Are the genes nadA and norB involved in formation of aflatoxin G1. Int. J. Mol. Sci. 2008, 9, 1717–1729. [Google Scholar] [CrossRef]
- Rice, D.W.; Hsieh, D.P. Aflatoxin M1: In vitro preparation and comparative in vitro metabolism versus aflatoxin B1 in the rat and mouse. Res. Commun. Chem. Pathol. Pharmacol. 1982, 35, 467–490. [Google Scholar]
- Garrido, N.S.; Iha, M.H.; Santos Ortolani, M.R.; Duarte Fávaro, R.M. Occurrence of aflatoxin M1 and aflatoxin M2 in milk commercialized in Ribeirão Preto-SP, Brazil. J. Food Addit. Contam. 2003, 20, 70–73. [Google Scholar] [CrossRef]
- Hsieh, D.P.; Beltran, L.M.; Fukayama, M.Y.; Rice, D.W.; Wong, J.J. Production and isolation of aflatoxin M1 for toxicological studies. J. Assoc. Off. Anal. Chem. 1986, 69, 510–512. [Google Scholar]
- Hsieh, D.P.; Cullen, J.M.; Hsieh, L.S.; Shao, Y.; Ruebner, B.H. Cancer risks posed by aflatoxin M1. Princess Takamatsu Symp. 1985, 16, 57–65. [Google Scholar]
- Huang, J.H.; Hsieh, D.P. Comparative study of aflatoxins M1 and B1 production in solid-state and shaking liquid cultures. Proc. Natl. Sci. Counc. Repub. China 1988, 12, 34–42. [Google Scholar]
- Yabe, K.; Chihaya, N.; Hatabayashi, H.; Kito, M.; Hoshino, S.; Zeng, H.; Cai, J.; Nakajima, H. Production of M-/GM-group aflatoxins catalyzed by the OrdA enzyme in aflatoxin biosynthesis. Fungal Genet. Biol. 2012, 49, 744–754. [Google Scholar] [CrossRef]
- Woloshuk, C.P.; Prieto, R. Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol. Lett. 1998, 160, 169–176. [Google Scholar] [CrossRef]
- Meyers, D.M.; Obrian, G.; Du, W.L.; Bhatnagar, D.; Payne, G.A. Characterization of aflJ, a gene required for conversion of pathway intermediates to aflatoxin. Appl. Environ. Microbiol. 1998, 64, 3713–3717. [Google Scholar]
- Bok, J.-W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef]
- Perrin, R.M.; Fedorova, N.D.; Bok, J.W.; Cramer, R.A.; Wortman, J.R.; Kim, H.S.; Nierman, W.C.; Keller, N.P. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007, 3, e50. [Google Scholar] [CrossRef]
- Kato, N.; Brooks, W.; Calvo, A.M. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2003, 2, 1178–1186. [Google Scholar] [CrossRef]
- Calvo, A.M.; Bok, J.-W.; Brooks, W.; Keller, N.P. VeA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 2004, 70, 4733–4739. [Google Scholar] [CrossRef]
- Chang, P.K.; Ehrlich, K.C.; Yu, J.; Bhatnagar, D.; Cleveland, T.E. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl. Environ. Microbiol. 1995, 61, 2372–2377. [Google Scholar]
- Chang, P.K.; Yu, J.; Bhatnagar, D.; Cleveland, T.E. Repressor-AFLR interaction modulates aflatoxin biosynthesis in Aspergillus parasiticus. Mycopathologia 1999, 147, 105–112. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Montalbano, B.G.; Bhatnagar, D.; Cleveland, T.E. Alteration of different domains in AFLR affects aflatoxin pathway metabolism in Aspergillus parasiticus transformants. Fungal Genet. Biol. 1998, 23, 279–287. [Google Scholar] [CrossRef]
- Flaherty, J.E.; Payne, G.A. Overexpression of aflR leads to upregulation of pathway gene expression and increased aflatoxin production in Aspergillus flavus. Appl. Environ. Microbiol. 1997, 63, 3995–4000. [Google Scholar]
- Payne, G.A.; Nystrom, G.J.; Bhatnagar, D.; Cleveland, T.E.; Woloshuk, C.P. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl. Environ. Microbiol. 1993, 59, 156–162. [Google Scholar]
- Woloshuk, C.P.; Foutz, K.R.; Brewer, J.F.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 1994, 60, 2408–2414. [Google Scholar]
- Yu, J.H.; Wieser, J.; Adams, T.H. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 1996, 15, 5184–5190. [Google Scholar]
- Yu, J.H.; Butchko, R.A.; Fernandes, M.; Keller, N.P.; Leonard, T.J.; Adams, T.H. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 1996, 29, 549–555. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Montalbano, B.G.; Cary, J.W. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticu. Gene 1999, 230, 249–257. [Google Scholar] [CrossRef]
- Cary, J.W.; Ehrlich, K.C.; Wright, M.; Chang, P.K.; Bhatnagar, D. Generation of aflR disruption mutants of Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 2000, 53, 680–684. [Google Scholar] [CrossRef]
- Wilkinson, J.R.; Yu, J.; Bland, J.M.; Nierman, W.C.; Bhatnagar, D.; Cleveland, T.E. Amino acid supplementation reveals differential regulation of aflatoxin biosynthesis in Aspergillus flavus NRRL 3357 and Aspergillus parasiticus SRRC 143. Appl. Microbiol. Biotechnol. 2007, 74, 1308–1319. [Google Scholar] [CrossRef]
- Wilkinson, J.R.; Yu, J.; Abbas, H.K.; Scheffler, B.E.; Kim, H.S.; Nierman, W.C.; Bhatnagar, D.; Cleveland, T.E. Aflatoxin formation and gene expression in response to carbon source media shift in Aspergillus parasiticus. J. Food Addit. Contam. 2007, 24, 1051–1060. [Google Scholar] [CrossRef]
- Abnet, C.C. Carcinogenic food contaminants. Cancer Invest. 2007, 25, 189–196. [Google Scholar] [CrossRef]
- Du, W.; Obrian, G.R.; Payne, G.A. Function and regulation of aflJ in the accumulation of aflatoxin early pathway intermediate in Aspergillus flavus. J. Food Addit. Contam. 2007, 24, 1043–1050. [Google Scholar] [CrossRef]
- Bouhired, S.; Weber, M.; Kempf-Sontag, A.; Keller, N.P.; Hoffmeister, D. Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genet. Biol. 2007, 44, 1134–1145. [Google Scholar] [CrossRef]
- Sugui, J.A.; Pardo, J.; Chang, Y.C.; Zarember, K.A.; Nardone, G.; Galvez, E.M.; Mullbacher, A.; Gallin, J.I.; Simon, M.M.; Kwon-Chung, K.J. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 2007, 6, 1562–1569. [Google Scholar] [CrossRef]
- Bok, J.W.; Noordermeer, D.; Kale, S.P.; Keller, N.P. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 2006, 61, 1636–1645. [Google Scholar] [CrossRef]
- Kale, S.P.; Cary, J.W.; Hollis, N.; Wilkinson, J.R.; Bhatnagar, D.; Yu, J.; Cleveland, T.E.; Bennett, J.W. Analysis of aflatoxin regulatory factors in serial transfer-induced non-aflatoxigenic Aspergillus parasiticus. J. Food Addit. Contam. 2007, 24, 1061–1069. [Google Scholar] [CrossRef]
- Mooney, J.L.; Yager, L.N. Light is required for conidiation in Aspergillus nidulans. Genes Dev. 1990, 4, 1473–1482. [Google Scholar] [CrossRef]
- Duran, R.M.; Cary, J.W.; Calvo, A.M. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 2007, 73, 1158–1168. [Google Scholar]
- Stinnett, S.M.; Espeso, E.A.; Cobeno, L.; Araujo-Bazan, L.; Calvo, A.M. Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol. Microbiol. 2007, 63, 242–255. [Google Scholar] [CrossRef]
- Kale, S.P.; Cary, J.W.; Bhatnagar, D.; Bennett, J.W. Characterization of experimentally induced, nonaflatoxigenic variant strains of Aspergillus parasiticus. Appl. Environ. Microbiol. 1996, 62, 3399–3404. [Google Scholar]
- Kale, S.; Bennett, J.W. Strain Instability in Filamentous Fungi. In Handbook of Applied Mycology, Mycotoxins in Ecological Systems; Bhatnagar, D., Lillehoj, E.B., Arora, D.K., Eds.; Tylor and Francis: New York, NY, USA, 1991; Volume 5, pp. 311–332. [Google Scholar]
- Yabe, K.; Nakamura, H.; Ando, Y.; Terakado, N.; Nakajima, H.; Hamasaki, T. Isolation and characterization of Aspergillus parasiticus mutants with impaired aflatoxin production by a novel tip culture method. Appl. Environ. Microbiol. 1988, 54, 2096–2100. [Google Scholar]
- Demain, A.L. Induction of secondary metabolism. Int. Microbiol. 1998, 1, 259–264. [Google Scholar]
- Payne, G.A.; Brown, M.P. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 1998, 36, 329–362. [Google Scholar] [CrossRef]
- Guo, B.Z.; Holbrook, C.C.; Yu, J.; Lee, R.D.; Lynch, R.E. Application of Technology of Gene Expression in Response to Drought Stress and Elimination of Preharvest Aflatoxin Contamination. In Aflatoxin and Food Safety; Abbas, H.K., Ed.; CRC Press: Boca Raton, FL, USA, 2005; pp. 313–331. [Google Scholar]
- Feng, G.H.; Leonard, T.J. Culture conditions control expression of the genes for aflatoxin and sterigmatocystin biosynthesis in Aspergillus parasiticus and A. nidulans. Appl. Environ. Microbiol. 1998, 64, 2275–2277. [Google Scholar]
- Cuero, R.; Ouellet, T.; Yu, J.; Mogongwa, N. Metal ion enhancement of fungal growth, gene expression and aflatoxin synthesis in Aspergillus flavus: RT-PCR characterization. J. Appl. Microbiol. 2003, 94, 953–961. [Google Scholar]
- Adye, J.; Mateles, R.I. Incorporation of labelled compounds into aflatoxins. Biochim. Biophys. Acta 1964, 86, 418–420. [Google Scholar] [CrossRef]
- Bennett, J.W.; Rubin, P.L.; Lee, L.S.; Chen, P.N. Influence of trace elements and nitrogen sources on versicolorin production by a mutant strain of Aspergillus parasiticus. Mycopathologia 1979, 69, 161–166. [Google Scholar] [CrossRef]
- Luchese, R.H.; Harrigan, W.F. Biosynthesis of aflatoxin-the role of nutritional factors. J. Appl. Bacteriol. 1993, 74, 5–14. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Lewis, D.F. Regulation of aflatoxin biosynthesis: Effect of glucose on activities of various glycolytic enzymes. Appl. Environ. Microbiol. 1984, 48, 306–310. [Google Scholar]
- Woloshuk, C.P.; Cavaletto, J.R.; Cleveland, T.E. Inducers of aflatoxin biosynthesis from colonized maize kernels are generated by an amylase activity from Aspergillus flavus. Phytopathol 1997, 87, 164–169. [Google Scholar] [CrossRef]
- Fanelli, C.; Fabbri, A.A.; Brasini, S.; de Luca, C.; Passi, S. Effect of different inhibitors of sterol biosynthesis on both fungal growth and aflatoxin production. Nat. Toxins 1995, 3, 109–113. [Google Scholar] [CrossRef]
- Fanelli, C.; Fabbri, A.A. Relationship between lipids and aflatoxin biosynthesis. Mycopathologia 1989, 107, 115–120. [Google Scholar] [CrossRef]
- Fanelli, C.; Fabbri, A.A.; Finotti, E.; Passi, S. Stimulation of aflatoxin biosynthesis by lipophilic epoxides. J. Gen. Microbiol. 1983, 129, 1721–1723. [Google Scholar]
- Yu, J.; Mohawed, S.M.; Bhatnagar, D.; Cleveland, T.E. Substrate-induced lipase gene expression and aflatoxin production in Aspergillus parasiticus and Aspergillus flavus. J. Appl. Microbiol. 2003, 95, 1334–1342. [Google Scholar] [CrossRef]
- Davis, N.D.; Diener, U.L.; Agnihotri, V.P. Production of aflatoxins B1 and G1 in chemically defined medium. Mycopathol. Mycol. Appl. 1967, 31, 251–256. [Google Scholar] [CrossRef]
- Reddy, T.V.; Viswanathan, L.; Venkitasubramanian, T.A. High aflatoxin production on a chemically defined medium. Appl. Microbiol. 1971, 22, 393–396. [Google Scholar]
- Reddy, T.V.; Viswanathan, L.; Venkitasubramanian, T.A. Factors affecting aflatoxin production by Aspergillus parasiticus in a chemically defined medium. J. Gen. Microbiol. 1979, 114, 409–413. [Google Scholar] [CrossRef]
- Kachholz, T.; Demain, A.L. Nitrate repression of averufin and aflatoxin biosynthesis Aspergillus parasiticus. J. Nat. Prod. 1983, 46, 499–506. [Google Scholar] [CrossRef]
- Niehaus, W.G.J.; Jiang, W.P. Nitrate induces enzymes of the mannitol cycle and suppresses versicolorin synthesis in Aspergillus parasiticus. Mycopathologia 1989, 107, 131–137. [Google Scholar] [CrossRef]
- Chang, P.K.; Ehrlich, K.C.; Linz, J.E.; Bhatnagar, D.; Cleveland, T.E.; Bennett, J.W. Characterization of the Aspergillus parasiticus niaD and niiA gene cluster. Curr. Genet. 1996, 30, 68–75. [Google Scholar] [CrossRef]
- Chang, P.K.; Yu, J.; Bhatnagar, D.; Cleveland, T.E. Characterization of the Aspergillus parasiticus major nitrogen regulatory gene, areA. Biochim. Biophys. Acta 2000, 1491, 263–266. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Hein, H., Jr. Effect of diurnal temperature cycles on the production of aflatoxin. Appl. Microbiol. 1968, 16, 988–990. [Google Scholar]
- Diener, U.L.; Davis, N.D. Limiting temperature and relative humidity for aflatoxin production by Aspergillus flavus in stored peanuts. J. Am. Oil Chem. Soc. 1970, 47, 347–351. [Google Scholar] [CrossRef]
- Roy, A.K.; Chourasia, H.K. Effect of temperature on aflatoxin production in Mucuna pruriens seeds. Appl. Environ. Microbiol. 1989, 55, 531–532. [Google Scholar]
- OBrian, G.R.; Georgianna, D.R.; Wilkinson, J.R.; Yu, J.; Abbas, H.K.; Bhatnagar, D.; Cleveland, T.E.; Nierman, W.C.; Payne, G.A. The effect of elevated temperature on gene transcription and aflatoxin biosynthesis. Mycologia 2007, 99, 232–239. [Google Scholar] [CrossRef]
- Yu, J.; Fedorova, N.D.; Montalbano, B.G.; Bhatnagar, D.; Cleveland, T.E.; Bennett, J.W.; Nierman, W.C. Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol. Lett. 2011, 322, 145–149. [Google Scholar] [CrossRef]
- Sanders, T.H.; Blankenship, P.D.; Cole, R.J.; Hill, R.A. Effect of soil temperature and drought on peanut pod and stem temperatures relative to Aspergillus flavus invasion and aflatoxin contamination. Mycopathologia 1984, 86, 51–54. [Google Scholar] [CrossRef]
- Cotty, P.J.; Jaime-Garcia, R. Influences of climate on aflatoxin producing fungi and aflatoxin contamination. Int. J. Food Microbiol. 2007, 119, 109–115. [Google Scholar] [CrossRef]
- Keller, N.P.; Nesbitt, C.; Sarr, B.; Phillips, T.D.; Burow, G.B. pH regulation of sterigmatocystin and aflatoxin biosynthesis in Aspergillus spp. Phytopathol 1997, 87, 643–648. [Google Scholar] [CrossRef]
- Tilburn, J.; Sarkar, S.; Widdick, D.A.; Espeso, E.A.; Orejas, M.; Mungroo, J.; Penalva, M.A.; Arst, H.N., Jr. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline expressed genes by ambient pH. EMBO J. 1995, 14, 779–790. [Google Scholar]
- Espeso, E.A.; Tilburn, J.; Arst, H.N., Jr.; Penalva, M.A. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 1993, 12, 3947–3956. [Google Scholar]
- Espeso, E.A.; Arst, H.N., Jr. On the mechanism by which alkaline pH prevents expression of an acid-expressed gene. Mol. Cell Biol. 2000, 20, 3355–3363. [Google Scholar] [CrossRef]
- Hicks, J.K.; Yu, J.H.; Keller, N.P.; Adams, T.H. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 1997, 16, 4916–4923. [Google Scholar]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef]
- Reib, J. Development of Aspergillus parasiticus and formation of aflatoxin B1 under the influence of conidiogenesis affecting compounds. Arch. Microbiol. 1982, 133, 236–238. [Google Scholar] [CrossRef]
- Torres, J.; Guarro, J.; Suarez, G.; Suñe, N.; Calvo, M.A.; Ramírez, C. Morphological changes in strains of Aspergillus flavus Link ex Fries and Aspergillus parasiticus Speare related with aflatoxin production. Mycopathologia 1980, 72, 171–180. [Google Scholar] [CrossRef]
- Guzman-de-Pena, D.; Aguirre, J.; Ruiz-Herrera, J. Correlation between the regulation of sterigmatocystin biosynthesis and asexual and sexual sporulation in Emericella nidulans. Antonie Leeuwenhoek 1998, 73, 199–205. [Google Scholar] [CrossRef]
- Yu, J.H.; Keller, N. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 2005, 43, 437–458. [Google Scholar] [CrossRef]
- Shimizu, K.; Keller, N.P. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 2001, 157, 591–600. [Google Scholar]
- Jayashree, T.; Praveen Rao, J.; Subramanyam, C. Regulation of aflatoxin production by Ca(2+)/calmodulin-dependent protein phosphorylation and dephosphorylation. FEMS Microbiol. Lett. 2000, 183, 215–219. [Google Scholar] [CrossRef]
- Mahoney, N.; Molyneux, R.J. Phytochemical inhibition of aflatoxigenicity in Aspergillus flavus by constituents of walnut (Juglans regia). J. Agric. Food Chem. 2004, 52, 1882–1889. [Google Scholar] [CrossRef]
- Kim, J.H.; Campbell, B.C.; Molyneux, R.; Mahoney, N.; Chan, K.L.; Yu, J.; Wilkinson, J.R.; Cary, J.; Bhatnagar, D.; Cleveland, T.E. Gene targets for fungal and mycotoxin control. Mycotoxin Res. 2006, 22, 3–8. [Google Scholar] [CrossRef]
- Reverberi, M.; Zjalic, S.; Racelli, A.; Fabbri, A.A.; Fanelli, C. Oxidant/antioxidant balance in Aspergillus parasiticus affects aflatoxin biosynthesis. Mycotoxin Res. 2006, 22, 39–47. [Google Scholar] [CrossRef]
- Jayashree, T.; Subramanyam, C. Oxidative stress as a prerequisite for aflatoxin production by Aspergillus parasiticus. Free Radic. Biol. Med. 2000, 29, 981–985. [Google Scholar] [CrossRef]
- Reverberi, M.; Fabbri, A.A.; Zjalic, S.; Ricelli, A.; Punelli, F.; Fanelli, C. Antioxidant enzymes stimulation in Aspergillus parasiticus by Lentinula edodes inhibits aflatoxin production. Appl. Microbiol. Biotechnol. 2005, 69, 207–215. [Google Scholar] [CrossRef]
- Zeringue, H.J.; Bhatnagar, D. Neem and Control of Aflatoxin Contamination. In Neem and Environment; Singh, R.P., Chari, M.S., Raheja, A.K., Kraus, W., Eds.; Science Publishers, Inc.: Enfield, NH, USA, 1993; Volume 2, pp. 713–727. [Google Scholar]
- Zeringue, H.J., Jr.; Brown, R.L.; Neucere, J.N.; Cleveland, T.E. Relationships between C6-C12 alkanal and alkenal volatile contents and resistance of maize genotypes to Aspergillus flavus and aflatoxin production. J. Agric. Food Chem. 1996, 44, 403–407. [Google Scholar] [CrossRef]
- Zeringue, H.J., Jr. Effects of methyl jasmonate on phytoalexin production and aflatoxin control in the developing cotton boll. Biochem. Syst. Ecol. 2002, 30, 497–503. [Google Scholar] [CrossRef]
- Greene-McDowelle, D.M.; Ingber, B.; Wright, M.S.; Zeringue, H.J., Jr.; Bhatnagar, D.; Cleveland, T.E. The effects of selected cotton-leaf volatiles on growth, development and aflatoxin production of Aspergillus parasiticus. Toxicon 1999, 37, 883–893. [Google Scholar] [CrossRef]
- Wright, M.S.; Greene-McDowelle, D.M.; Zeringue, H.J.; Bhatnagar, D.; Cleveland, T.E. Effects of volatile aldehydes from Aspergillus-resistant varieties of corn on Aspergillus parasiticus growth and aflatoxin biosynthesis. Toxicon 2000, 38, 1215–1223. [Google Scholar] [CrossRef]
- Wilson, R.A.; Gardner, H.W.; Keller, N.P. Differentiation of aflatoxin-producing and non-producing strains of Aspergillus flavus group. Lett. Appl. Microbiol. 2001, 33, 291–295. [Google Scholar] [CrossRef]
- Xiulan, S.; Xiaolian, Z.; Jian, T.; Zhou, J.; Chu, F.S. Preparation of gold-labeled antibody probe and its use in immunochromatography assay for detection of aflatoxin B1. Int. J. Food Microbiol. 2005, 99, 185–194. [Google Scholar] [CrossRef]
- Chu, F.S.; Fan, T.S.; Zhang, G.S.; Xu, Y.C.; Faust, S.; McMahon, P.L. Improved enzyme-linked immunosorbent assay for aflatoxin B1 in agricultural commodities. J. Assoc. Off. Anal. Chem. 1987, 70, 854–857. [Google Scholar]
- Malloy, C.D.; Marr, J.S. Mycotoxins and public health: A review. J. Public Health Manag. Pract. 1997, 3, 61–69. [Google Scholar]
- Yu, J.; Cleveland, T.E. Aspergillus flavus Genomics for Discovering Genes Involved in Aflatoxin Biosynthesis. In Polyketides Biosynthesis, Biological Activity, and Genetic Engineering; Rimando, A.M., Baerson, S.R., Eds.; American Chemical Society: Washington, DC, USA, 2007; Volume 955, pp. 246–260. [Google Scholar]
- Yu, J.; Cleveland, T.E.; Nierman, W.C.; Bennett, J.W. Aspergillus flavus genomics: Gateway to human and animal health, food safety, and crop resistance to diseases. Rev. Iberoam. Micol. 2005, 22, 194–202. [Google Scholar] [CrossRef]
- Cleveland, T.E.; Yu, J.; Bhatnagar, D.; Chen, Z.-Y.; Brown, R.; Chang, P.K.; Cary, J.W. Progress in Elucidating the Molecular Basis of the Host Plant-Aspergillus flavus Interaction: A Basis for Devising Strategies to Reduce Aflatoxin Contamination in Crops. In Aflatoxin and Food Safety; Abbas, H.K., Ed.; CRC Press: Boca Raton, FL, USA, 2005; pp. 167–193. [Google Scholar]
- Cleveland, T.E.; Cary, J.W.; Brown, R.L.; Bhatnagar, D.; Yu, J.; Chang, P.K.; Chaln, C.A.; Rajasekaran, K. Use of biotechnology to eliminate aflatoxin in preharvest crops. Bull. Inst. Compr. Agric. Sci. Kinki Univ. 1997, 5, 75–90. [Google Scholar]
- Lillehoj, E.B.; Wall, J.H. Decontamination of Aflatoxin-Contaminated Maize Grain. In Proceedings of US Universities-CIMMYT Maize Aflatoxin Workshop, El Batan, Mexico, April 1987; pp. 260–279.
- Cotty, P.J.; Bayman, D.S.; Egel, D.S.; Elias, K.S. Agriculture, Aflatoxins and Aspergillus. In The Genus Aspergillus; Powell, K., Ed.; Plenum Press: New York, NY, USA, 1994; pp. 1–27. [Google Scholar]
- Tubajika, K.M.; Damann, K.E. Sources of resistance to aflatoxin production in maize. J. Agric. Food Chem. 2001, 49, 2652–2656. [Google Scholar] [CrossRef]
- Brown, R.L.; Chen, Z.Y.; Cleveland, T.E.; Russin, J.S. Advances in the development of host resistance in corn to aflatoxin contamination by Aspergillus flavus. Phytopathology 1999, 89, 113–117. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Brown, R.L.; Damann, K.E.; Cleveland, T.E. PR10 expression in maize and its effect on host resistance against Aspergillus flavus infection and aflatoxin production. Mol. Plant Pathol. 2010, 11, 69–81. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yu, J. Current Understanding on Aflatoxin Biosynthesis and Future Perspective in Reducing Aflatoxin Contamination. Toxins 2012, 4, 1024-1057. https://doi.org/10.3390/toxins4111024
Yu J. Current Understanding on Aflatoxin Biosynthesis and Future Perspective in Reducing Aflatoxin Contamination. Toxins. 2012; 4(11):1024-1057. https://doi.org/10.3390/toxins4111024
Chicago/Turabian StyleYu, Jiujiang. 2012. "Current Understanding on Aflatoxin Biosynthesis and Future Perspective in Reducing Aflatoxin Contamination" Toxins 4, no. 11: 1024-1057. https://doi.org/10.3390/toxins4111024



